Abstract
Enhancers, silencer and insulators are DNA elements that play central roles in regulation of the genome that are crucial for development and differentiation. In metazoans, these elements are often separated from target genes by distances that can reach 100 Kb. How regulation can be accomplished over long distances has long been intriguing. Current data indicate that although the mechanisms by which these diverse regulatory elements affect gene transcription may vary, an underlying feature is the establishment of close contacts or chromatin loops. With the generalization of this principle, new questions emerge, such as how the close contacts are formed and stabilized and, importantly, how they contribute to the regulation of transcriptional output at target genes. This review will concentrate on examples where a functional role and a mechanistic understanding has been explored for loops formed between genes and their regulatory elements or among the elements themselves.
Keywords: long range interactions, chromosome conformation capture, enhancers, silencers, insulators
INTRODUCTION
The paradigm for gene activation is the yeast recruitment model [1]. In this view, activators access the upstream activating sequence (UAS) of a gene and serve to recruit chromatin remodeling complexes that move or evict nucleosomes and histone modifying complexes that alter the N-terminal tails of histones by acetylation, methylation, phosphorylation or ubiquination. Together these co-activator complexes alter promoters so that the RNA polymerase II complex can bind and initiate transcription. However, yeast UASs are located quite close to transcription start sites, and they fail to function if they are experimentally moved away [2], while in metazoans enhancers, silencers and insulators are frequently very distant, as far as 100 Kb, from genes whose transcriptional output they influence.
The models put forward to explain the action of enhancers over such distances have generally been of two types [3]. One model envisions a signal emanating from a regulatory element and spreading along the chromatin until it encounters a proximal promoter (spreading) while the other envisions that the regulatory elements interact physically with gene promoters with extension of the chromatin in-between (looping). The first concrete evidence supporting long range chromatin looping showed that the β-globin locus control region (LCR) enhancer, which is located far upstream of the globin genes, exists in proximity to the actively transcribing genes while the inactive ones are distant [4, 5]. Although this model has been generalized to other enhancers as well as silencers and insulators, as is discussed below, there is no reason to suppose that looping occurs exclusive of spreading as the two mechanisms are not incompatible.
The most widely used experimental approach to document proximity between regions of chromatin is chromosome conformation capture (3C) which was developed to look at the organization of yeast chromosomes [6]. In 3C, chromatin is first cross-linked by formaldehyde in nuclei and then digested to completion with a restriction enzyme. A ligation step is then carried out under dilute conditions. Cleaved sites normally are re-ligated together with a low level of ligation to nearby non-adjacent sites that will fall off with increasing linear distance. However, two sequences that had been held in proximity by protein interactions will generate a novel ligation product that can be detected, after removal of cross-links and purification, by PCR with a forward anchor primer in a region of interest, such as an enhancer, and a reverse primer located at any position that it is desirable to interrogate.
Important caveats exist in interpreting 3C data in addition to very rigorous controls. Of note, 3C is performed on large numbers of pooled cells and thus yields an average of interactions across all cells that might not take place simultaneously in one cell. Furthermore, transient, dynamic interactions might be underrepresented. A variation of this approach, 4C, allows interrogation of all interactions that a chosen location engages in without bias [7–10]. However, the challenge of trying to understand the function of the observed 4C (and higher C) interactions is formidable. The issue of function is very important because some close contacts between chromosomal regions may be the result of the required compaction of the genome in the nucleus and may not be functionally relevant. Several recent reviews have critically considered issues related to chromatin looping [11–14]. The important question of whether long range gene contacts beyond those observed with regulatory regions—those that occur between chromosomes and in nuclear sub-domains—serve to meaningfully regulate gene output has also been reviewed [15].
ENHANCER AND LCR LOOPING
Enhancers are positive elements that activate gene expression from a position remote from the target gene. At least one mechanism by which enhancers function is by establishing contact with target genes (Figure 1). This was first shown for the β-globin LCR enhancer and gene using 3C as well as a different molecular approach called RNA-TRAP [4, 5]. The 3C interaction was observed in erythroid cells where the genes are expressed and not in brain cells where they are not expressed indicating that the looped conformation correlates with transcriptional activity and may be required for it [4]. Furthermore, the loops changed during development as the activity of the genes, which are sequentially expressed, changed [16].
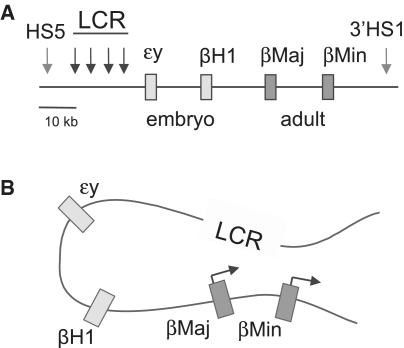
Figure 1:
Dynamic long range interaction between the β-globin LCR and gene. (A) The β-globin locus is depicted. Globin genes transcribed either in the embryo or adult are represented by colored rectangles. The DNase I hypersensitive sites of the LCR are represented by black arrows and the locus flanking hypersensitive sites, where the insulator protein CTCF is enriched, are indicated by red arrows. (B) The looped conformation of the β-globin locus is depicted with the LCR in proximity to the actively transcribed adult βmaj and βmin-globin genes while the earlier expressed genes are distant. The drawing represents findings described in references [4, 5].
It is generally assumed that proteins binding to enhancer and promoter sites will be involved in forming and stabilizing chromatin loops but this has been explored for only a limited number of long range interactions. Validating this concept, it was demonstrated that the LCR/β-globin loop did not form in the absence of the erythroid specific activators EKLF or GATA-1 [17, 18]. GATA-1 mediated recruitment of the chromatin remodeler Brg1 is also required for looping although the precise contribution of Brg1 to the loop remains to be further explored [19]. Restoring EKLF or GATA-1 leads to rescue of loop formation and β-globin expression supporting the idea that the loop is required for transcription activation. Not all factors binding to the regions that loop together in the β-globin locus have a role in looping. NF-E2 also binds to the LCR and β-globin gene but it is not apparently specifically involved in loop formation [20].
The widely expressed protein Ldb1 has emerged as a good candidate for bridging between the LCR and β-globin gene through dimerization. Ldb1 is not a DNA binding protein but is a component of a complex in erythroid cells including LMO2 and DNA binding components GATA-1 and SCL that occupies the β-globin LCR and gene in erythroid cells but not in brain cells [21]. An experiment combining ChIP and 3C provided evidence that Ldb1 was physically present at the juncture of the LCR/β-globin loop. Furthermore, reduction of Ldb1 using RNAi resulted in loss of β-globin transcription and of the LCR/β-globin loop. Restoration of Ldb1 rescued β-globin transcription in Ldb1 knock-down MEL cells, supporting the function of Ldb1 in long range enhancer interactions (IK and AD, unpublished data).
A recent report indicates that cohesin, physically and functionally connects enhancers and promoters of some genes in embryonic stem (ES) cells [22], possibly through encircling the sites held in proximity as it does sister chromatids during exchange. Importantly, genetic evidence in Drosophila supports the idea that enhancer–promoter communication is facilitated by Ldb1/Chip as well as NIPBL/Nipped B, the cohesin loading factor [23, 24].
Interestingly, Ldb1 is required for migration of the β-globin locus to nuclear transcription factories which is necessary to achieve high levels of transcription characteristic of this locus in erythroid cells [25]. The LCR is also required for this migration [26], linking LCR function to Ldb1 and suggesting that the salient feature for nuclear migration is looping (Figure 2). The emerging hypothesis is that looping may be important for transcription activation because it is required for nuclear migration [25]. An alternative model, that stabilization of the Ldb1 complex on chromatin allows migration but proximity is established within a transcription factory, cannot be ruled out at this point. Since Ldb1 is a widely expressed protein, it is attractive to consider the possibility that this protein may mediate long range interactions, via diverse DNA binding partners, perhaps with varying downstream outcomes.
Figure 2:
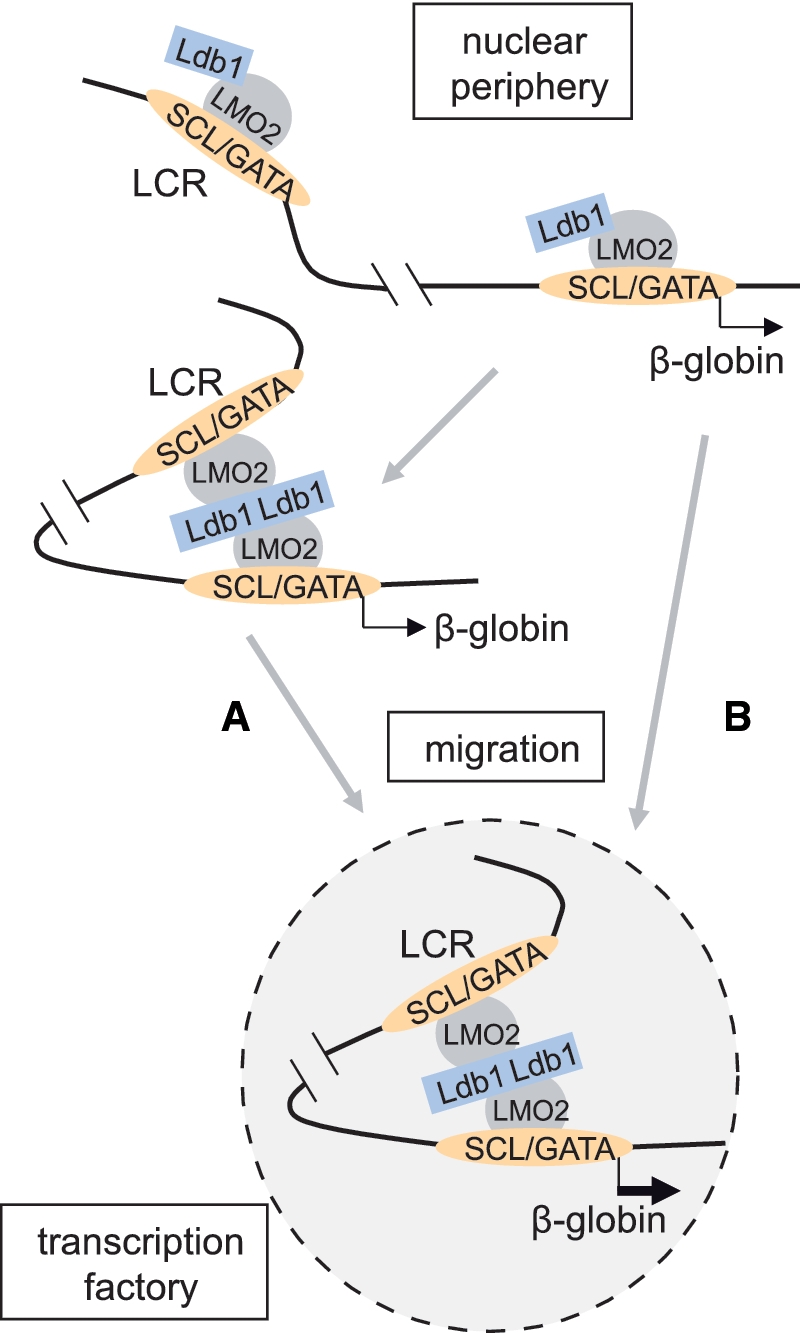
Models describing the relationship between LCR/β-globin looping and intra-nuclear migration of the β-globin locus. The enrichment of the Ldb1 complex (GATA-1/SCL/LMO2) at the LCR and β-globin gene is depicted. (A) Full occupancy may be sufficient to establish LCR/β-globin proximity through Ldb1 self-interaction, prior to nuclear migration and transcription factory localization for robust β-globin transcription. (B) Alternatively, once occupied by the Ldb1 complex, the locus might undergo re-localization to the nuclear interior and establish LCR/β-globin proximity as a result of positioning within a factory (B). Proximity would then be stabilized by protein–protein interactions, possibly dependent on Ldb1.
The looping paradigm has been generalized in other gene loci. For example, in the c-kit locus, GATA-2 stabilizes a loop between the gene and an enhancer located 114 Kb distant [27]. As this gene is down regulated during erythroid cell maturation, GATA-1 replaces GATA-2 with consequent loss of the enhancer loop in favor of a loop from the promoter to a site within the gene. Thus, loops can be reconfigured by different transcription factors with functional consequences to the gene involved. Two different kinds of long range interactions are observed in the TH2 cytokine gene locus. The first occurs between promoters of the Il4, Il5 and Il13 promoters early in T-cell differentiation. The LCR for these genes then loops into contact with the promoters later in T-cell specification to form a structure poised for rapid transcription activation. The transcription factors GATA-3 and STAT support these interactions [28].
LONG RANGE INTERACTIONS ENGAGED IN BY SILENCERS
Silencers are negative elements that act collectively to establish a repressed state that can be stably inherited. Silencers repress promoters at a distance by nucleating complexes, such as the polycomb (PcG) proteins that maintain the silent state of Hox genes through early development in Drosophila melanogaster [29]. PcG complexes are thought to bind to polycomb response elements (PREs) and subsequently spread across chromatin modifying histones by H3K27 tri-methylation across large regions. The outcome is inhibition of transcription of a target gene although the precise details of how this outcome is brought about are not clear. In the bithorax multi-gene locus of Drosophila the Mcp PRE controls the expression of the Abd-B gene over a distance of 60 Kb, similar to the range over which enhancers can act.
Recent investigations in Drosophila indicate that looping interactions are involved in silencing. PREs interact with each other to build up higher order structures in the bithorax locus [30]. All major polycomb-bound elements including PREs and core promoters participate in an extensive network of loops that can be detected by 3C. The higher order interactions corresponded to the repressed state of genes in the locus and to repressive histone modifications. Reduction of polycomb (PC) resulted in loss of a specific sub-set of the loops and de-repression of the homeotic genes while recovery of PC correlated with re-establishment of loops and repression of the genes, supporting the functional role of PC in repression through long range loop formation.
Polycomb silencing of the human GATA-4 gene provided an additional opportunity to explore chromatin organization [31]. The locus is broadly occupied by the polycomb protein EZH2 and by the polycomb repressive histone mark it catalyzes, H3K27me3, when the gene is silent. At the same time, multiple loops were demonstrated by 3C to correlate with the repressed state. The sites of long range interaction were particularly enriched for all PcG proteins that were assayed, providing evidence that they may collectively contribute to looping as do Ldb1 complex components. Reduction of EZH2 reduced loop formation and correlated with GATA-4 expression supporting a functional role for PcGs in the looping.
INSULATOR-MEDIATED LONG RANGE INTERACTIONS
Both the repressive effects of silencers and the activating effects of enhancers/LCRs, each with their respective histone marks, must be restricted to appropriate domains and not spill over to alter non-target genes. Insulators are elements that are proposed to fulfill this role [32, 33]. In ectopic assays, insulators appear to have two separable activities. Barrier insulators can block the spread of repressive chromatin from a silencer. Enhancer blocker insulators disrupt enhancer–promoter communication when placed between these two elements, and can be viewed as ‘antagonizing’ enhancers in the same sense that barrier activity ‘antagonizes’ silencer activity. Although it remains to be established, these activities of insulators may not be entirely distinct in vivo.
In contrast to flies where several insulator proteins exist [34], the CCCTC-binding factor CTCF is the only known insulator binding protein in vertebrates. CTCF binds to a specific site in the chicken β-globin HS4 insulator and CTCF binding is necessary for insulator function [35]. HS4 and an additional CTCF site at 3′HS1, downstream of the chicken β-globin genes are conserved in the mouse and human β-globin loci. The orthologous murine β-globin locus sites, HS5 and 3′HS1, bind CTCF [36] and loop to each other in progenitor cells (before globin gene expression) and erythroid cells (expressing globin genes) but not in brain cells, suggesting a requirement for insulation of the locus when it is transcriptionally active [4]. Mutation of the 3′HS1 site in human erythroid cells abrogated the loop indicating the direct involvement of CTCF [37]. Some data support self-interaction of CTCF molecules, which might be involved in loop formation but this issue deserves to be further explored [38]. Importantly, however, the functional significance of this loop had been called into question since deletion of HS5 from the mouse globin locus or mutation of the CTCF site in human 3′HS1 did not result in deregulation of the β-globin genes [37, 39, 40].
The significant question that arises is whether CTCF/insulator element interaction can, in fact, delimit enhancer function. In experiments using a transgene, an exogenous copy of HS5 placed between the LCR and downstream genes displayed enhancer blocking and reduced β-globin gene transcription by compromising looping between the LCR and β-globin gene [41, 42]. Furthermore, the endogenous and ectopic HS5 sites formed a new insulator loop that topologically isolated the LCR and nullified its activity on the globin genes. HS5 thus appears to have intrinsic and portable looping ability through which it can delimit enhancer activity. In further work, multiple CTCF-dependent loops that were primarily cell type specific were observed up and downstream of the globin locus [43] (Figure 3). The functional relevance of the loops was supported by knock down of CTCF in K562 cells resulting in loss of the extensive loops, repression of γ-globin transcription and invasion of the locus by repressive histone marks. The suggestion is that while an individual CTCF site, such as HS5 or 3′HS1, might be dispensable, general loss of cell type specific CTCF loops is severely detrimental to globin gene transcription.
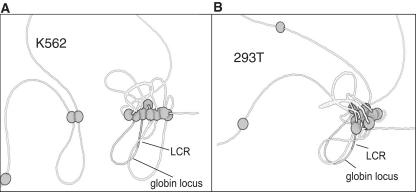
Figure 3:
Cell type specific long range CTCF site interactions. Sites occupied by CTCF up and downstream of the β-globin locus are represented by white spheres spaced proportionally along the chromatin fiber. Each sphere-to-sphere contact represents an interaction detected in 3C experiments [43]. (A) In K562 cells where the fetal γ-globin gene is transcriptionally active, 10 looping interactions were observed over a 2 Mb region of chromosome 11 around the β-globin locus. The interactions were primarily between adjacent CTCF sites with an average separation of 70 Kb. The largest interaction observed was over 185 Kb. (B) In 293 cells where the globin locus is transcriptionally silent, 10 chromatin loops were also detected but six were specific to 293 cells and not observed in K562 cells. There were many more clustered interactions, one including six sites all in contact with one another. The average distance between interacting sites was 190 Kb and the longest interval was 0.5 Mb.
CTCF also binds to specific sites in the Igf2/H19 imprinting control region (ICR) that loops to a second regulatory region, DMR1 on the maternal allele [44]. In this configuration, the Igf2 gene is within a loop, separated from enhancers it shares with H19 and is maintained in a silent state while H19 is transcribed. On the paternal allele the ICR is methylated, which prevents CTCF interaction, and loop formation, resulting in activation of Igf2 by the enhancers. Mutation of the CTCF sites within the ICR abrogated the loop on the maternal allele and resulted in Igf2 expression drawing a direct connection between CTCF binding, loop formation and regulatory function [45]. A 4C study revealed a chromosomal network of imprinted regions in contact with the Igf2/H19 ICR [7]. The network, observed in fetal liver cells, is largely absent in ES cells but is acquired during their maturation in vitro. Mutation of ICR CTCF sites decreased the frequency of the network interactions supporting a direct role for CTCF in these interactions.
Over 30K sites of CTCF enrichment exist genome-wide [46, 47]. Thus, a key unsolved puzzle is whether and how CTCF site interactions are specified among thousands of sites occupied by the protein. The cohesin complex may provide part of the answer. Cohesin co-occupies >80% of CTCF sites genome-wide and insulator function is affected by loss of cohesin even when CTCF occupancy is not affected [48, 49]. Furthermore, a regulatory role for cohesin interaction at CTCF sites is supported by studies at the Igh/Igκ, IFNG and IGF2-H19 loci [50–52]. However, in another example CTCF and cohesin were mutually lost if either component was reduced by RNAi [43]. Cohesin subunits play an important role in sister chromatin exchange by forming a ring-like structure around the sister DNA strands, suggesting how it might contribute to stabilizing insulator loops together with CTCF. In view of the number of CTCF binding sites genome-wide, it seems likely that CTCF will have other functional partners to collaborate with in determining loop specification [53].
In contrast to its role in insulator function, CTCF appears to participate in a looping interaction at HLA–DRB1 and HLA–DQA1 that is required for transcription of these genes. The locus has an enhancer element, XL9, between the two genes where CTCF interacts. The CTCF site loops to both promoters bringing them close together when they are transcribed and deletion of the element reduces transcription. CTCF cooperates with transcription factor RFX and co-activator CIITA to form the loop [54]. This result is consistent with the reported interaction between CTCF and RNA polymerase II at some genomic sites [55]. A mechanistic understanding of how this type of CTCF long range interaction differs from insulator function is quite important. CTCF, thus, emerges as a protein that organizes chromatin loops to mediate diverse downstream outcomes.
PERSPECTIVE
There remains a significant number of open questions concerning the molecular mechanisms underlying long range regulatory interactions of all types. First and foremost, it remains to define the cause and effect relationship between long range chromosomal looping and gene expression, either positive or negative. In the examples above, evidence supporting the role of specific proteins mediating loop formation has been emphasized. The repertoire needs to be expanded especially for silencer loops, for which there are only a few examples and for CTCF partners that might mediate specificity of loop interactions. In general, much would be gained from biochemical studies of the many mechanisms attributed to silencers and insulators through genetic experiments in Drosophila [56].
On this subject, understanding the functional versus structural role of CTCF will be very important. Genome-wide studies localize more than 30 000 CTCF sites that in many cases did not correspond to transitions in active versus repressive histone modifications [46, 47]. Of the sites that are unlikely to represent insulators, many may turn out to have an organizational role. Some evidence suggests the CTCF sites may generally maintain long range contact with each other in cell specific patterns that could have differing outcomes for gene transcription [43]. The problem will be telling one type of CTCF site interaction from another. Although time consuming, deletion of specific sites in a chromosomal milieu will ultimately contribute this information.
Equally important will be to integrate our understanding of activating enhancer loops with inhibitory insulator and silencer loops. In this regard, it may turn out that the long range looping interactions in the β-globin locus are among the simplest to understand. The CTCF sites flank the LCR and globin genes which can interact within this domain and no loops are in a position to block any other. However, in many other instances CTCF sites exist between enhancers and target genes but do not seem to interrupt activation. For example, a CTCF site exists between the c-kit gene and its –114 enhancer in mouse ES cells [57] even though this gene and enhancer engage in long range looping [27]. In another example, the α-globin enhancer can apparently bypass intergenic sites of CTCF interaction to activate a distant, unrelated gene in the circumstances of α-globin promoter mutation or deletion [58].
Finally, genome wide localization studies also indicate that >40% of CTCF sites are located in introns. Do CTCF sites in this type of location have insulator function and do the sites form long range contacts? Based on our current understanding of insulators, it is hard to imagine RNA polymerase transcribing through one. Perhaps such insulator sites within genes (or between enhancers and genes) are regulated so that they do not bind CTCF and/or form loops to other sites when the gene is active or the interaction occurs. PARylation appears to be one means of regulating CTCF function [59, 60] but more work remains to be done on insulator regulation.
Key Points.
Enhancers, silencers and insulators are DNA elements that regulate gene expression in both positive and negative ways over long distances. Their underlying mechanisms vary but a common theme is that these elements establish close proximity, or chromatin loops, with the genes they regulate.
It is assumed that the proteins interacting at the regulatory elements and genes that loop together are involved in formation or stabilization of the contacts but there are only a limited number of examples where experiments support the roles of specific factors in chromatin looping. Even in those cases, it is still difficult to determine the causal effects of loops from the correlative.
Insulators oppose the positive and negative effects of enhancers and silencers, respectively. However, the wide-spread occurrence of CTCF/insulator sites in the genome suggests an overall organizational role for insulators in genome conformation. There are hints that such nuclear organization may be cell type specific and influence transcription.
One of the means by which enhancer, silencer and insulator loops actually affect transcriptional output of target genes may be by facilitating migration of gene loci to particular nuclear domains.
FUNDING
Work in the author’s laboratory is supported by the Intramural Program of the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health.
Acknowledgements
I would like to thank past and present members of my laboratory for helpful discussion and Ryan Dale for Figure 3. I apologize to colleagues whose original work was not cited due to the brevity of the review.
Biography
Ann Dean is the Chief in the Section on Gene Regulation and Development in the Laboratory of Cellular and Developmental Biology, NIDDK, NIH, where she studies transcription regulation and chromatin structure.
References
- 1.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–77. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–74. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 3.Dean A. On a chromosome far, far away: LCRs and gene regulation. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Tolhuis B, Palstra RJ, Splinter E, et al. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 5.Carter D, Chakalova L, Osborne CS, et al. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–6. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 6.Dekker J, Rippe K, Dekker M, et al. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Z, Tavoosidana G, Sjolinder M, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 8.Simonis M, Klous P, Splinter E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 9.Dostie J, Richmond TA, Arnaout RA, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenfelder S, Sexton T, Chakalova L, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noordermeer D, de Laat W. Joining the loops: beta-globin gene regulation. IUBMB Life. 2008;60:824–33. doi: 10.1002/iub.129. [DOI] [PubMed] [Google Scholar]
- 13.Dean A. Close encounters of the 3C kind: long-range chromatin interactions and transcriptional regulation. Briefings in Functional Genomics and Proteomics. 2010;8:297–309. doi: 10.1093/bfgp/elp016. [DOI] [PubMed] [Google Scholar]
- 14.Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–55. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 16.Palstra RJ, Tolhuis B, Splinter E, et al. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–4. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 17.Drissen R, Palstra RJ, Gillemans N, et al. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–90. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vakoc CR, Letting DL, Gheldof N, et al. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–62. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 19.Kim SI, Bultman SJ, Kiefer CM, et al. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci USA. 2009;106:2259–64. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kooren J, Palstra RJ, Klous P, et al. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J Biol Chem. 2007;282:16544–52. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 21.Song S-H, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol Cell. 2007;28:810–22. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagey MH, Newman JJ, Bilodeau S, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morcillo P, Rosen C, Baylies MK, et al. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 1997;11:2729–40. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–93. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song SH, Kim A, Ragoczy T, et al. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood. 2010;116:2356–64. doi: 10.1182/blood-2010-03-272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ragoczy T, Bender MA, Telling A, et al. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–57. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing H, Vakoc CR, Ying L, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–42. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–27. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 29.Mateos-Langerak J, Cavalli G. Polycomb group proteins and long-range gene regulation. Adv Genet. 2008;61:45–66. doi: 10.1016/S0065-2660(07)00002-8. [DOI] [PubMed] [Google Scholar]
- 30.Lanzuolo C, Roure V, Dekker J, et al. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–74. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 31.Tiwari VK, McGarvey KM, Licchesi JD, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–27. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–7. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Brief Funct Genomic Proteomic. 2009;8:276–82. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–96. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 36.Bulger M, Schubeler D, Bender MA, et al. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. MolCell Biol. 2003;23:5234–44. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Splinter E, Heath H, Kooren J, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pant V, Kurukuti S, Pugacheva E, et al. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrell CM, Grinberg A, Huang SP, et al. A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse β-globin locus. Proc Natl Acad Sci USA. 2000;97:14554–9. doi: 10.1073/pnas.97.26.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bender MA, Byron R, Ragoczy T, et al. Flanking HS-62.5 and 3' HS1, and regions upstream of the LCR, are not required for beta-globin transcription. Blood. 2006;108:1395–401. doi: 10.1182/blood-2006-04-014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanimoto K, Sugiura A, Omori A, et al. Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol Cell Biol. 2003;23:8946–52. doi: 10.1128/MCB.23.24.8946-8952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou C, Zhao H, Tanimoto K, et al. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci USA. 2008;105:20398–403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci USA. 2010;107:3651–6. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 45.Kurukuti S, Tiwari VK, Tavoosidana G, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Wendt KS, Yoshida K, Itoh T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 49.Parelho V, Hadjur S, Spivakov M, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Degner SC, Wong TP, Jankevicius G, et al. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–8. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadjur S, Williams LM, Ryan NK, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–3. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nativio R, Wendt KS, Ito Y, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci. 2009;122:1275–84. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 54.Majumder P, Gomez JA, Chadwick BP, et al. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–98. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chernukhin I, Shamsuddin S, Kang SY, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–48. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 58.Lower KM, Hughes JR, De Gobbi M, et al. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci USA. 2009;106:21771–6. doi: 10.1073/pnas.0909331106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34:271–84. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furlan-Magaril M, Rebollar E, Guerrero G, et al. An insulator embedded in the chicken {alpha}-globin locus regulates chromatin domain configuration and differential gene expression. Nucleic Acids Res. 2010;39:89–103. doi: 10.1093/nar/gkq740. [DOI] [PMC free article] [PubMed] [Google Scholar]