Abstract
MicroRNAs (miRNAs) guide Argonaute (AGO)-containing microribonucleoprotein (miRNP) complexes to target mRNAs. It has been assumed that miRNAs behave similarly to each other with regard to mRNA target recognition. The usual assumptions, which are based on prior studies, are that miRNAs target preferentially sequences in the 3′UTR of mRNAs, guided by the 5′ “seed” portion of the miRNAs. Here we isolated AGO- and miRNA-containing miRNPs from human H4 tumor cells by co-immunoprecipitation (co-IP) with anti-AGO antibody. Cells were transfected with miR-107, miR-124, miR-128, miR-320, or a negative control miRNA. Co-IPed RNAs were subjected to downstream high-density Affymetrix Human Gene 1.0 ST microarray analyses using an assay we validated previously—a “RIP-Chip” experimental design. RIP-Chip data provided a list of mRNAs recruited into the AGO-miRNP in correlation to each miRNA. These experimentally identified miRNA targets were analyzed for complementary six nucleotide “seed” sequences within the transfected miRNAs. We found that miR-124 targets tended to have sequences in the 3′UTR that would be recognized by the 5′ seed of miR-124, as described in previous studies. By contrast, miR-107 targets tended to have ‘seed’ sequences in the mRNA open reading frame, but not the 3′ UTR. Further, mRNA targets of miR-128 and miR-320 are less enriched for 6-mer seed sequences in comparison to miR-107 and miR-124. In sum, our data support the importance of the 5′ seed in determining binding characteristics for some miRNAs; however, the “binding rules” are complex, and individual miRNAs can have distinct sequence determinants that lead to mRNA targeting.
Introduction
MicroRNAs (miRNAs) regulate gene expression of their target mRNAs through many different mechanisms. These mechanisms including inhibition of translation at initiation and/or post-initiation, deadenylation, nucleolysis and others.1–5 Argonaute (AGO) proteins, which bind directly to mature miRNAs, are a central component of most mammalian microribonucleoparticles (miRNPs).1,6–8 As miRNAs regulate gene expression through differing mechanisms, and most of those mechanisms seem to still involve AGO-miRNPs, there must be differing subtypes of miRNPs that subserve somewhat different functions. To date, the full variety of miRNP subytpes and miRNA-related mechanisms have not been completely elucidated.
In addition to the complexity of miRNA mechanisms, researchers are challenged with predicting exactly which miRNAs will bind to which individual mRNA targets. This is a formidable task because metazoan miRNAs tend to bind to mRNA targets through partial sequence complementarity. Much has been learned about the principles that govern those interactions, relating to the complementary sequence determinants between miRNAs and target mRNAs. Computational algorithms, based on prior experimental studies, can help to predict mRNA target binding.9–13 However, the biochemistry underlying miRNA:mRNA interactions still needs to be fully elucidated.
The two issues described above—different miRNA functional mechanisms on the one hand, and the sequence determinants of miRNA:mRNA targeting principles on the other—may be linked. The sequence determinants for mRNA targeting according to one mechanism may not be identical to that for another mechanism across all species and cell types. In order to better understand and test these complex ideas, more experiments are required that involve direct determination of miRNA targets.
A promising method for directly characterizing miRNPs is co-immunoprecipitation (co-IP) that pulls down AGO proteins along with associated molecules.7,14 Using AGO co-IP assays, researchers have isolated multiple proteins, miRNAs, and mRNA targets from miRNPs.15–21 A subset of AGO co-IP experiments involve “RIP-Chip”22,23 techniques that integrate miRNP co-IP with downstream high-density microarrays to study target mRNAs systematically.
Here we describe results using a RIP-Chip assay that uses Anti-AGO to co-IP mRNA targets. The monoclonal antibody (“Anti-AGO”, which was also termed “2A8”) was raised against human AGO2, and recognizes a C-terminal epitope on AGO2.24 Multiple human and mouse AGO paralogs are also recognized by Anti-AGO, both in co-IPs and on western blots. Here, the co-IP was coupled with downstream high-throughput microarray analyses of mRNAs that are associated with miRNPs. Applying this method to H4 glioneuronal cell line25 enabled us to evaluate some of the sequence determinants of miRNA:mRNA interactions.23 The RIP-Chip experimental assay, coupled with computational analyses, were used to provide new information about miRNA targeting.
Results
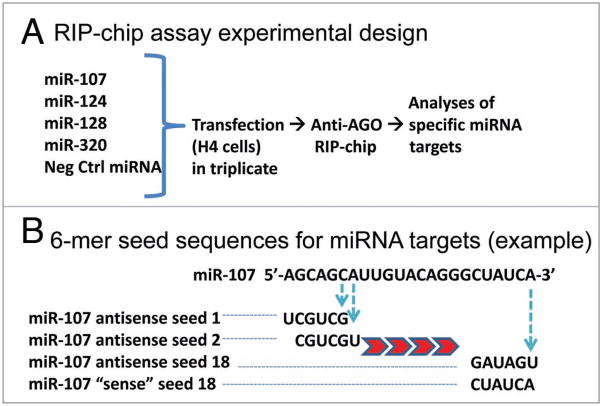
The study design is indicated schematically in Figure 1. The putative miRNA targets (PmiTs) were experimentally identified by applying a threshold of least two-fold enriched in the co-IPed miRNPs following the transfection of individual miRNAs. The results of three biological replicates were used in each case. Less-enriched targets, or targets that exhibited decreased levels in the lysates, were not included in this study. A list of PmiTs identified with RIP-Chip is shown in Table 1. Note that miR-320 had the most PmiTs identified (57 total), relative to the other miRNAs studied. Several PmiTs in this list have been validated previously.23
Figure 1.
(A) Experimental design. Following miRNA transfections (performed with three biological replicates), Anti-AGO RIP-Chip was performed. Analyses of Affymetrix Human Gene 1.0 ST microarray data were used to identify mRNA targets enriched following miRNA transfections. (B) Further analyses were performed on putative miRNA targets (PmiTs). The 5′ UTR, open reading frames, and 3′ UTR of PmiTs were evaluated for ‘seed’ sequences corresponding to the complementary 6mers from different portions of transfected miRNAs. For example, “seed 1” for miR-107 corresponds to the 6 nucleotides at the 5′ end of miR-107, whereas “seed 18” corresponds to the 6 nucleotides at the 3′ end of miR-107. Most experiments were performed using the “anti-sense” seeds as shown, but for control experiments, “sense” sequences were evaluated.
Table 1.
Targets identified using Anti-AGO RIP-Chip
| miR-107 | miR-124 | miR-128 | miR-120 | ||
|---|---|---|---|---|---|
| GRN | PTBP1 | LSM10 | LSM2 | NRN1 | CTDSP1 |
| INSIG1 | APEX2 | TXNIP | ANXA2P2 | AP2A1 | OSR2 |
| RPLPO | SNAI2 | DBI | ARPC1A | TSPAN13 | LY6E |
| C22orf13 | CTDSP1 | RAB24 | SEPW1 | CPSF6 | ANXA2P1 |
| ZDHHC4 | LAGE3 | CDH11 | MT2A | NR1H2 | SALL1 |
| SAT1 | ATP6VOE1 | PARS2 | ANXA2 | CTF8 | PRR13 |
| E I R3IP1 | TMEM69 | LDLR | TRAPPC1 | BHLHB2 | UBA52 |
| PPIB | SLC16A13 | ANXA7 | E2F4 | SRF | YKT6 |
| RPL1O | ERAL1 | RPS15 | PGK1 | TEX261 | |
| RHOG | S100A10 | FTL | RPL41 | ||
| STX1O | MT1G | STUB1 | TCEA2 | ||
| NME4 | FAM127C | ITFG3 | HRB | ||
| PPAP2B | UROS | MEA1 | DULLARD | ||
| ZFP36L2 | C6orf125 | BCL7B | TBCA | ||
| PCOLCE | DYNLRB1 | CAPNS1 | MT2A | ||
| UFC1 | MT2A | PPIL1 | SYNGR2 | ||
| CCNJL | FLJ20489 | SCN1B | CSF1 | ||
| PPIA | FAM89A | RNASEK | PRR13 | ||
| CCL2 | CLDN12 | PPP1R11 |
Putative miRNA targets (PmiTs) for miR-107, miR-124, miR-128 and miR-320. These were selected on the basis of being enriched more than two-fold in the AGO-miRNPs following anti-AGO RIP-Chip.
PmiTs identified using the RIP-Chip assay served as the bases to analyze some of the sequence determinants that are correlated with miRNA targeting in these cultured cells. As shown in Figure 1B, the mRNA sequences of the PmiTs were evaluated for the presence of 6mer sequences that were antisense complementary to portions of the transfected miRNAs. Sequences across the full length of the miRNAs were evaluated; for example, 18 different 6mers were studied in the case of miR-107. Complementary sites in the sequences of the 5′UTRs, open reading frames, and 3′UTRs were searched separately. Thus we could test hypotheses about how miRNAs target mRNAs in terms of the 6mer sequences in different portions of the miRNAs and mRNAs.
For miR-124, there was a clear and strong tendency for sequences complementary to the 5′ seed (nts #2–7) of miR-124 to be present in the 3′UTR of PmiTs. For example, the 6mer seed complementary to miR-124 nts #2–7 was present, on average, once per 410 nts in the 3′ UTR of miR-124 PmiTs, in contrast to once per 1,604 nts in the 3′ UTR of miR-107 PmiTs. See Table 2 for specific information about miR-124 seed sequences in PmiTs. Statistical analyses were performed using the Fisher’s exact test to evaluate the likelihood that these sequence elements would have been present in the PmiTs by chance (Fig. 2). These data are presented using a chart with the −log(p value) on the Y axis to demonstrate the tendency for the sequences complementary to corresponding 6mers on the miR-NAs to be present in 5′UTR, open reading frame, and 3′UTR of PmiTs. Note that the p values for 5′ seeds starting at nts #1, #2 and #3 were significant at the level of p < 10−6, p < 10−12 and p < 10−4, respectively. By contrast, there was no indication of tendency for miR-124 complementary 6mer sequences to be present in the 5′ UTR or open reading frame of mRNAs that were miR-124 PmiTs.
Table 2.
Number of 5′ seed complementary sequences per 1,000 nts observed in 5′UTR, open reading frame (ORF), or 3′UTR of putative miRNA targets for miR-107, miR-128 and miR-320.
| miR-107 5′ seed seqs/1,000 nts | |||
|---|---|---|---|
| PmiTs | 5′UTR | ORF | 3′UTR |
| miR-107 | 1.50 | 4.63 | 0.51 |
| miR-124 | 0.86 | 1.19 | 0.71 |
| miR-128 | 1.64 | 0.59 | 0.10 |
| miR-320 | 0.39 | 0.97 | 1.02 |
| miR-124 5′ seed seqs/1,000 nts | |||
| PmiT’s | 5′UTR | ORF | 3′UTR |
| miR-107 | 0.00 | 0.51 | 0.64 |
| miR-124 | 0.58 | 0.65 | 2.32 |
| miR-128 | 0.00 | 0.39 | 0.39 |
| miR-320 | 0.10 | 0.40 | 1.02 |
Figure 2.
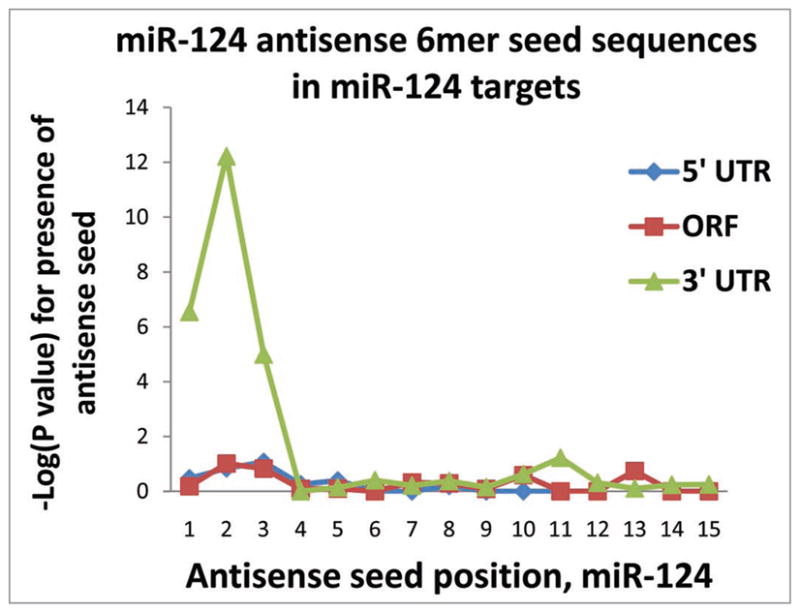
As has been shown previously, miR-124 targets tend to have “seed” sequences corresponding to nucleotides #2–7 in miR-124, specifically in the 3′ UTR. This chart shows the −log(p value) of Fisher’s exact test analyses to determine the likelihood that the number of sequences found in the putative miRNA targets would have occurred by chance. Separate analyses were performed for the 5′ UTR, open reading frame (“Coding”), and 3′ UTR. Note that the 5′ seeds (6mer sequences corresponding to #1–6, #2–7 and #3–8) of miR-124 are enriched in miR-124 putative miRNA targets, but only in the 3′ UTR.
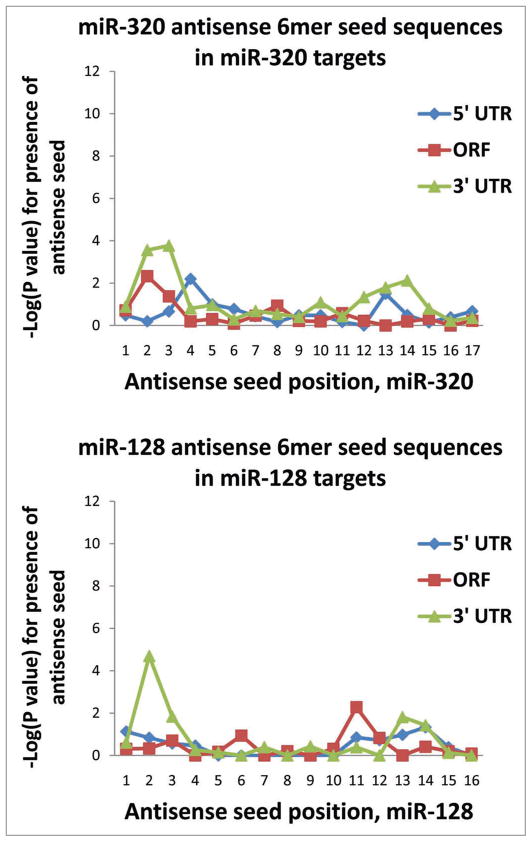
Figure 3 shows that the mRNAs identified as miR-320 and miR-128 PmiTs also showed significant enrichment for 5′ seed sequences for their respective miRNAs within the 3′UTRs, again as assessed using the Fisher’s exact test. However, these miRNAs’ PmiTs did not have the same degree of enrichment as seen in miR-124 PmiTs. Whereas the enrichment was significant to p < 10−12 for miR-124, the significance was at p < 10−4 for miR-128 and p < 10−3 for miR-320. Note that for both miR-128 and miR-320 there were 3′ miRNA seed regions, corresponding in antisense orientation to nts #11–14, with lower p values (~p < 0.01), indicating a possible contributory binding via 3′ sequences on the miRNAs.
Figure 3.
As was the case for miR-124, putative miR-128 and miR-320 putative targets tend to have sequences corresponding to nucleotides #2–7 in the 3′UTR. However, the tendency for these two miRNAs is much less strong (p < 10−5 versus p < 10−11). Moreover, unlike miR-124, both miR-128 and miR-320 have a tendency—albeit far weaker than the 5′ seed—for the 3′ sequences of the miRNAs to have possible effects (enrichment of complementary 6mers in putative targets). Since the current study was limited to studying 6mer seeds, these data do not test whether sequence determinants with fewer hybridizing nucleotides (5mers and smaller) may be more important in the case of these other miRNAs.
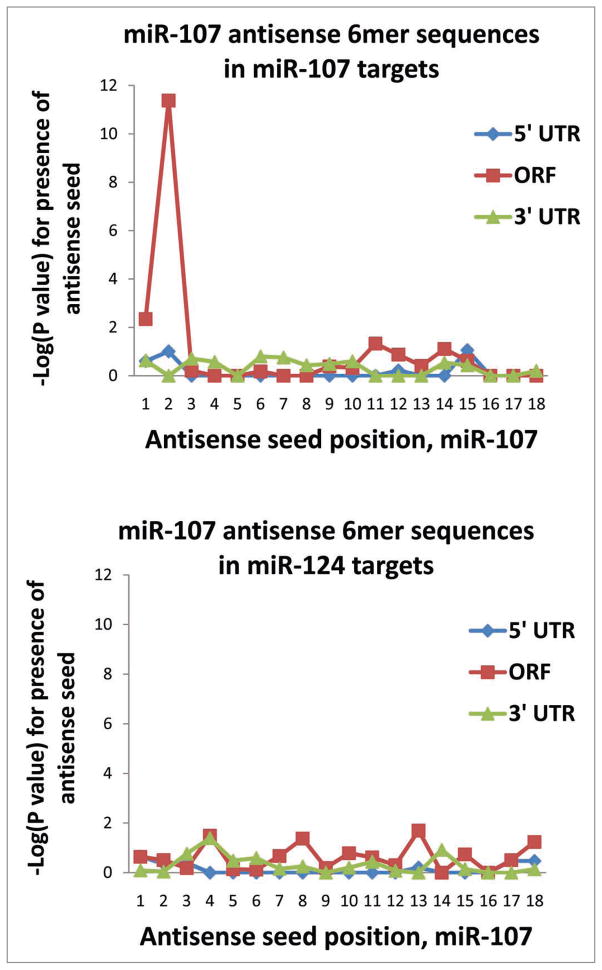
The results for miR-107 were quite different in comparison to those of miR-124, miR-128 or miR-320. For miR-107 there was a marked tendency for the miR-107 6mer complementary seed sequences to be present in the PmiTs’ open reading frames (Table 2). Also represented in Figure 4, it was the 5′ seed (nts #2–7) that was by far the strongest associational determinant of this binding (p < 10−11). Remarkably, there was no enrichment among miR-107 PmiTs for 6-mer 3′UTR sequences corresponding to miR-107.
Figure 4.
(A) Unlike for miR-124 and the other miRNAs tested in the current study, putative miR-107 targets have a marked tendency to have “seed” sequences corresponding to miR-107 nucleotides #2–7 in the open reading frame (“Coding” sequence) as opposed to the 3′ UTR. Equally remarkable is the apparent lack of a tendency for miR-107 seed sequences to be enriched in the 3′ UTR of miR-107 targets identified experimentally using anti-AGO RIP-Chip. (B) To evaluate whether miR-107 seed sequences are present in other mRNAs (i.e., nonspecifically), we tested other groups of mRNAs and such was not the case. For example, we sampled those mRNAs that were recruited into the AGO-miRNP according to anti-AGO RIP-Chip after miR-124 transfection. There is no “nonspecific” tendency for miR-107 nucleotides #2–7 (in antisense) to be present in these or other mRNAs that were analyzed.
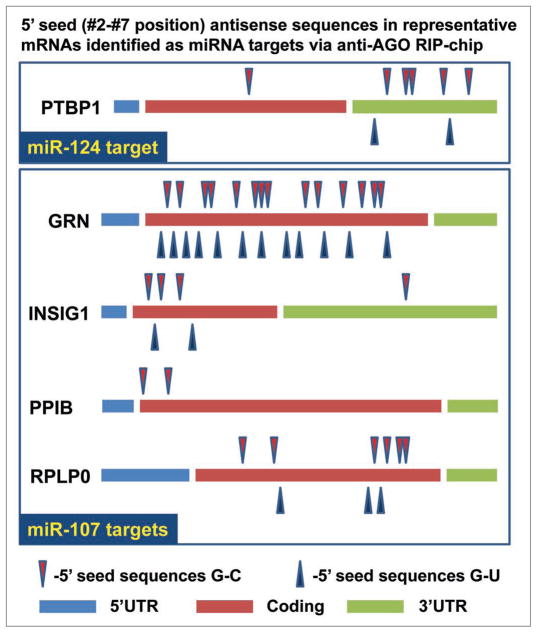
Figure 5 provides a schematic/cartoon depiction of representative PmiTs for miR-107 in contrast to miR-124. PTBP1 is chosen as representative for miR-124 target because it was identified in the current RIP-Chip assay and has also been shown previously to be a validated miR-124 target.26 As miR-107 PmiTs for this figure, we chose GRN, PPIB, INSIG1 and RPLP0 as being representative. GRN is the strongest target identified using RIP-Chip in H4 cells and is the topic of a separate paper (Wang W-X, et al. In Press). For Figure 5 we included ‘seed’ sequences separately that would pair through G-U bindings (blue triangles), in addition to the “canonical”, but completely separate, G-C binding (red triangles).
Figure 5.
A cartoon to demonstrate that mRNAs strongly recruited into the AGO-microribonucleoparticle following transfection with miR-107 tended to have “seed” sequences corresponding to miR-107 nucleotides #2–7 in the open reading frame. miR-124 ‘seed’ sequences on the PTBP1 are shown at top because this gene has been shown previously to be a miR-124 target and it was a strong miR-124 target on the RIP-Chip assay. Note that the seed sequences corresponding to miR-124 (in antisense) are present mostly within the 3′ UTR of miR-124 targets. Even those miR-124 seed sequences that would rely on G-U binding are mostly in the 3′UTR. By contrast, the miR-107 putative targets tend to contain 5′ seed sequences of miR-107 in the open reading frame. Again, this tendency is also seen for 6mers that would theoretically cause binding with the 5′ seed of miR-107 through G-U binding.
Immunoblots of GRN, INSIG1 and PPIB were performed after miRNA transfections to assess whether the surprising miR-107 RIP-Chip results were truly indicating physiological miR-107 targets. Transfections were performed with both miR-107 and with the negative control miRNA (Fig. 6). All of the assessed miR-107 PmiTs (but not the control, beta-Actin) were indeed knocked down following miR-107 transfection.
Figure 6.

The putative miR-107 targets identified experimentally by RIP-Chip were subjected to biochemical validation. These proteins are GRN, INSIG1 and PPIB, which are targeted by miR-107 in the open reading frame as shown in Figure 5. Immunoblot analyses of these proteins’ expression following transfections of miR-107 (versus control miRNA) show that the expression of these genes are effectively suppressed by miR-107. Beta-Actin immunoblots following identical transfections provides a negative control.
To provide some idea of signal-to-noise ratio for the RIP-Chip assay, we assessed the presence of “sense-oriented” 6mer sequences (see Fig. 1B) in the PmiTs. These sequences would not be predicted to be recognized by their respective miRNAs, so they serve in this context as control sequences. Data from these analyses are shown in Figure 7 which again depict the −log(p value) from multiple Fisher’s exact tests. Note that the p values indicate a much less significant enrichment in these cases than for the 6mer sequences in the antisense orientation, which suggest that the results for the “antisense”-oriented 6mer seeds are specific.
Figure 7.
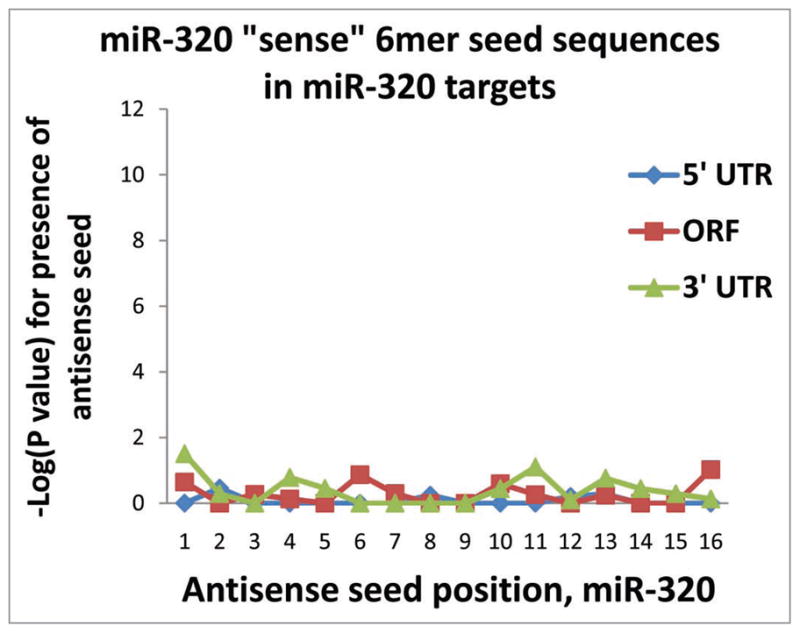
Control experiments show that there is relatively low background “noise” in the anti-AGO RIP-Chip results. These analyses were performed testing for the presence of 6mer sequences corresponding to portions of miRNAs in the “sense” orientations that are predicted NOT to bind to the mRNA targets. Shown here are the results for miR-320, which are representative. As in Figures 2–4 and 7, the Y axis shows the results of −log(p value) for Fisher’s exact test to assess the likelihood that the sequences occurred by chance.
Luciferase reporter assays were employed to demonstrate that a specific sequence in the open reading frame INSIG1 could be specifically targeted by miR-107. We cloned a segment of INSIG1 open reading sequence (nts 370–518) that contains two TGCTGC iterations (INSIGMRE), and its mutated counterpart (INSIGMREmut, see Fig. 8) in pRL-TK reporter plasmid. Co-transfection of reporter plasmids and miRNAs demonstrated that INSIGMRE reporter, but not its mutant, was effectively knock-downed by miR-107, indicating the specificity of miR-107 targeting on the seed sequence in INSIG1 mRNA. Similar reports were obtained for reporter assays referent to a GRN mRNA open reading frame sequence (Wang W-X, et al. In Press).
Figure 8.

Dual luciferase reporter assays were used to validate miR-107 target sites on INSIG1 mRNA. A sequence in the INSIG1 mRNA open reading frame (nts 370–518) that contains two TGCTGC iterations (top) was subcloned into pRL-TK reporter plasmid (INSIGMRE). The pRL-TK plasmid containing the mutated sequence of INSIGMRE (INSIGMREmut, which is only subtly different as shown) was constructed in parallel. Plasmid transfection, miRNA transfection, and dual luciferase activity assay were followed our published protocols (See Methods section). The figure showed that INSIGMRE specifically responds to miR-107 transfection but not to miR-320, or to a negative control miRNA. “*”-Student’s one-tailed t-test p < 0.001.
Discussion
RIP-Chip experiments show that the mRNAs targeted by individual miRNAs may be recruited according to distinct sequence determinants. In line with prior studies, our data analyses confirm that miRNA 5′ seeds12,27 are critical determinants with target sites usually in the 3′UTRs of PmiTs for miR-124, miR-128 and miR-320. As has also been shown previously,28 there are 3′ seed sequences in miRNAs that also appear to contribute to target binding, and sites outside the 3′UTR where miRNA seeds are found in PmiTs. Most notably, in our current study, miR-107 appears far more likely than other tested miRNAs to target the open reading frame of mRNAs. In contrast, miR-107 complementary 6mer seeds are remarkably less enriched in the 3′UTR of miR-107 PmiTs. Immunoblot analyses and reporter assays provide further validation of the miR-107 targets. Together these data demonstrate the importance of evaluating individual miRNAs experimentally instead of relying solely on a universal prediction algorithm.
Prior studies have shown that the open reading frame of mRNAs can be targeted by miRNAs. For example, the genes Nanog, Oct4, Sox2 and Dicer are regulated through miRNA targeting in their mRNAs’ open reading frames.28,29 In high-throughput, direct experimental evaluation of miRNA target with “HITS-CLIP” in mouse brain, some 25% of miRNA target sequences resided in open reading frames of targeted mRNA.30 Further, it has been known for years that small interfering RNAs target open reading frame of mRNAs using the same or very similar molecular machinery.31 A recent review summarizes evidence that a miRNA may recognize target mRNAs’ open reading frames.28 However, it has not been shown previously that a given miRNA—such as miR-107—tend to target mRNAs selectively in the open reading frame.
If there are idiosyncratic determinants of specific miRNA and mRNA targets, there must be corresponding biological explanations. The differences in targeting characteristics between miRNAs may be due to individual miRNAs existing in differing cell compartments. Some miRNAs are spatially trafficked specifically to particular parts of the cell (nucleus versus cytoplasm, and dendrites versus axons, for example) and these compartmental differences might be accompanied by differing target determinants.32–36 Alternatively, particular miRNAs may interact selectively with different proteins—miRNAs comprise only a small portion of miRNPs and AGO is only one of the many miRNP-associated proteins.7,8,21,37 MiR-107 has been shown to share an unusual sequence motif with the miRNA let-7. Through this motif, miR-107 may interact with mRNA targets through a novel mechanism associated with the uridylyl transferase protein “TUTase 4” (TUT4).38 In future studies, some distinct characteristics of miR-107 may be linked to this interaction.
There are limitations to the current study. H4 “neuroglioma” cells derive from an aneuploid cancer cell line comprising an imperfect model of any particular “normal” cell.25 We have shown previously that the miRNP is functional in H4 cells23 but there may be currently uncharacterized idiosyncrasies. Aside from the inherent limitations of the cultured tumor cells, RIP-Chip is an assay that does not provide all the information about miRNA:mRNA interactions. AGO-miRNPs have been shown to exert gene expression regulation via multiple mechanisms.1,3 For the mRNA target to be detected by a microarray downstream of the anti-Ago co-IP, the target mRNA must be stably bound to the miRNP. This may not necessarily be the case if the miRNA:mRNA interaction is transient or if the main mechanism for the translational inhibition involves mRNA degradation. In other words, our experimental design probably only describes a subset of miRNA:mRNA interactions.
In summary, analyses of RIP-Chip experiments in H4 cells suggest that individual miRNAs bind to target mRNA using different sequence determinants. Future studies may help explore how and why such short RNA molecules can exhibit distinct binding tendencies. In addition, these data also indicate the need for more “cross-talk” between high-throughput experimental studies and computational algorithms used to predict miRNA targets.
Materials and Methods
Co-IP of miRNPs using anti-AGO antibodies
The Anti-AGO RIP-Chip methodology has been validated and described in detail.23 Briefly, Protein G agarose beads (Invitrogen, Carlsbad, CA) were incubated with monoclonal anti-AGO.24 Beads were washed 3 times in PBS and twice in lysis buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM MgCl2, 0.5% NP-40 and 5 mM DTT). 48 hours after the final transfection(s), the cells were rinsed twice in PBS, then lysed on ice for 10 minutes in fresh lysis buffer with protease inhibitors (1 tablet/10 ml lysis buffer, Complete Protease Inhibitor Cocktail Tablets, EDTA-free, Roche Applied Science, Mannheim, Germany) and RNasin (250 U/ml; Promega, Madison, WI). Lysates were precleared with pre-blocked Protein G beads at 4°C for 60 min and then co-IP was performed with 2A8,24-Protein G beads at 4°C for 90 min. After co-IP, the beads were washed twice with lysis buffer; three times with lysis buffer containing 900 mM NaCl and 1% NP-40; twice more with lysis buffer, followed by a final wash with lysis buffer containing 0.05% NP-40. Beads were then incubated with 250 μl of DNA digestion solution (40 mM Tris-HCl, pH 8.0, 10 mM MgSO4, 1 mM CaCl2, 200 U/ml RNasin, and 40 U/ml DNase I (Promega, Madison, WI)) at 37°C for 20 min with shaking. RNA was extracted using Trizol LS (Invitrogen, Carlsbad, CA) as described.39
Microarray analysis and RT-qPCR
Microarray analysis of RNAs isolated from co-IP or from total lysates were performed using Affymetrix Human Gene 1.0 ST™ chip at University of Kentucky Microarray Core Facility. Three biological replicates were carried out in each treatment.
Downstream data analyses
Criteria for selecting PmiTs according to our anti-AGO RIP-Chip data (based on the averaged results of the three biological replicates on the array data for each transfection condition) were as follows:
The mRNA was enriched more than two-fold higher in the AGO-miRNPs after the cells were transfected with a particular miRNA, relative to negative control miRNA; and
The same mRNA was not upregulated more than two-fold in the lysate after transfection with the miRNA.
Following the identification of PmiTs using these criteria, the 5′UTR, open reading frame, and 3′UTR sequences were analyzed for 6-mer sequences correlating to portions of the miR-NAs (in antisense and sense orientation). The total number of complementary sequences in each location was assessed using the Fisher’s exact test to determine whether this number would be expected to occur by chance.
Immunoblot analysis of miRNA targets
RIP-Chip experiments identified miR-107 PmiTs with miR-107 binding elements in the open reading frames. A subset of these targets were subjected to further biochemical validation. Full-length cDNA plasmid that express human granulin (GRN, NM_002087.2), and insulin induced gene 1 (INSIG1, NM_005542.3) were purchased from OriGene (OriGene Technologies, Inc., Rockville, MD). H4 cells were first transfected with either GRN, or INSIG1, or empty vector plasmids for 24 hr using LF-2000 (Invitrogen, Carlsbad, CA); and were then transfected with 25 nM of Pre-miR-107 or “Pre-miR negative control #1” (Ambion, Austin, TX) for additional 48 hr. For GRN and PPIB analysis, total soluble protein was isolated and used for immunoblot analysis; For INSIG1, membrane proteins were isolated and used in western blot analysis. Goat anti-human GRN antibody (R&D systems, Minneapolis, MN); rabbit anti-human INSIG1, mouse anti-human PPIB (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-β-Actin antibody (Rockland, Gilbertsville, PA) were used at 1:500, 1:200, 1:2,000 and 1:4,000, respectively.
Reporter assays of INSIG1 mRNA putative miR-107 target sites
To test whether a sequence (nts 370–518) in INSIG1 mRNA open reading frame that contains two antisense miR-107 seed sequences are miR-107 target sites, we constructed pRL-TK reporters bearing either the original sequence (designed as INSIGMRE), or its mutated sequence (designed as INSIGMREmut), where the seed region was altered. Reporter assay procedure was described in detail previously.39 Briefly, pRL-TK reporter plasmids (INSIGMRE and INSIGMREmut) along with a control reporter plasmid pGL3 were first transfected in H4 cells. Transfected cells were washed in antibiotic-free media after 6 hr of transfection, and the cells were allowed to recover overnight. Twenty-four hours after the first transfection, cells were further transfected with 25 nM of Pre-miR-107, Pre-miR-320, or a negative control for Pre-miRNAs (Ambion). Luciferase reporter activity was measured 48 hr after the Pre-miRNA transfection. Each transfection was performed in triplicate and data shown represent two individual experiments.
Acknowledgments
We thank Ms. Willa Huang for technical and collegial assistance in the project. This study was supported by grants R01 NS061933 and K08 NS050110 from the National Institutes of Health, Bethesda, MD and NIRG-89917 from the Alzheimer Association.
References
- 1.Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–21. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Nelson PT, Tang G. WILEY ENCYCLOPEDIA OF CHEMICAL BIOLOGY. John Wiley & Sons, Inc; 2008. RNA Interference, Mechanisms and Proteins Involved. [Google Scholar]
- 4.Orom UA, Lund AH. Experimental identification of microRNA targets. Gene. 2009 doi: 10.1016/j.gene.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Hausser J, Landthaler M, Jaskiewicz L, Gaidatzis D, Zavolan M. Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C-miRNA complexes and the degradation of miRNA targets. Genome Res. 2009;19:2009–20. doi: 10.1101/gr.091181.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–9. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 7.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steitz JA, Vasudevan S. miRNPs: versatile regulators of gene expression in vertebrate cells. Biochem Soc Trans. 2009;37:931–5. doi: 10.1042/BST0370931. [DOI] [PubMed] [Google Scholar]
- 9.Saito T, Saetrom P. MicroRNAs-targeting and target prediction. N Biotechnol. doi: 10.1016/j.nbt.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–94. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA (New York, NY) 2008;14:2440–51. doi: 10.1261/rna.1139508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA biology. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 17.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA (New York, NY) 2007;13:1198–204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3:2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, et al. A biochemical approach to identifying microRNA targets. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19291–6. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA (New York, NY) 2008 doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–60. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–7. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 23.Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA. 2010;16:394–404. doi: 10.1261/rna.1905910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O’Doherty U, et al. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–92. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnstein P, Taylor DO, Nelson-Rees WA, Huebner RJ, Lennette EH. Propagation of human tumors in antithymocyte serum-treated mice. J Natl Cancer Inst. 1974;52:71–84. doi: 10.1093/jnci/52.1.71. [DOI] [PubMed] [Google Scholar]
- 26.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–78. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–8. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 29.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 30.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 32.Smalheiser NR. Regulation of mammalian microRNA processing and function by cellular signaling and subcellular localization. Biochim Biophys Acta. 2008;1779:678–81. doi: 10.1016/j.bbagrm.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smalheiser NR, Lugli G. microRNA Regulation of Synaptic Plasticity. Neuromolecular Med. 2009 doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, Cuellar TL, et al. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–34. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science (New York, NY) 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 37.Gu SG, Pak J, Barberan-Soler S, Ali M, Fire A, Zahler AM. Distinct ribonucleoprotein reservoirs for microRNA and siRNA populations in C. elegans. RNA. 2007;13:1492–504. doi: 10.1261/rna.581907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–23. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]