Abstract
Human PTEFb is a protein kinase composed by CDK9 and Cyclin T that controls the elongation phase of RNA Pol II. This complex also affects the activation and differentiation program of lymphoid cells. In this study we found that several head and neck tumor cell lines overexpress PTEFb. We also established that Cyclin T1 is able to induce transformation in vitro, as we determined by foci and colony formation assays. Nu/nu mice s.c. injected with stable transfected Cyclin T1 cells (NIH 3T3 Cyclin T1) developed tumors faster than animals injected with control cells (NIH 3T3 β-gal). In vitro, NIH 3T3 Cyclin T1 cells show increased proliferation and CDK4-Rb phosphorylation. Even more, silencing E2F1 expression (shRNA E2F1) in NIH 3T3 cells resulted in a dramatic inhibition of Cyclin T1-induced foci. All these data demonstrate for the first time the Cyclin T1 oncogenic function and suggest a role for this protein in controlling cell cycle probably via Rb/E2F1 pathway.
Key words: cyclin T1, CDK9, PTEFb
Introduction
Cell cycle is regulated by cyclin-dependent kinases (CDK) which form heterodimer complexes with cyclins, cofactors required for the CDK activity. There are at least 9 different CDK (CDK1-CDK9)1 and many more cyclins (Cyclin A through T).2 When CDK are activated by the binding of its partner cyclin, phosphorylation cascades are turned on to orchestrate the coordinated entry in the next phase of the cell cycle. Different cyclin-CDK combinations determine the downstream proteins targeted. CDKs are constitutively expressed in cells whereas cyclins are synthesized at specific stages of the cell cycle, in response to various molecular signals.3
Cell cycle deregulation is a common feature of human cancer. Cancer cells frequently display unscheduled proliferation, genomic instability (increased DNA mutations and chromosomal aberrations) and chromosomal instability (changes in chromosome number).3 Constitutive and deregulated CDK activation may contribute not only to unscheduled proliferation but also to genomic and chromosomal instability in cancer cells.
More than 90% of human neoplasias have abnormalities in some component of the cell cycle. These abnormalities are due to hyperactivation of CDK as a result of amplification/overexpression of positive cofactors, cyclins/CDKs or downregulation of negative factors.4 These changes promote deregulated S-phase progression in a way that ignores growth factor signals, with loss of G1 checkpoints.5 Previous reports clearly demonstrated that aberrant induction of Cyclin D1 activity, an essential regulator of cell cycle progression, is involved in human tumorigenesis5,6 and its deregulation has been documented in a variety of cancer types.7 Even more, Cyclin D1 activity was shown to be critical for tumor formation induced by other oncogenes (e.g., Ras), as mice deficient in Cyclin D1 are resistant to tumorigenesis.8
Human PTEFb is a protein kinase composed by CDK9 and Cyclin T (T1, T2a or T2b) that controls the elongation phase of RNA Pol II. This complex is a cellular cofactor required for the transcriptional activation of the viral transactivator Tat in the HIV-1 genome.9 Cdk9 gene is widely expressed in human and murine tissues, with higher levels found in terminally differentiated cells.10 Cyclin T1 and T2 mRNAs were detected in adult human tissues with higher levels in the muscle, spleen, thymus, testis, ovary and peripheral blood lymphocytes. In HeLa nuclear extracts, roughly 80% of CDK9 is complexed with Cyclin T1 and 10% with Cyclin T2a and Cyclin T2b.11,12
PTEFb was also found to be involved in the phosphorylation and regulation of the carboxy-terminal domain (CTD) of the largest RNA Pol II subunit.13 It was also reported that the CDK9/Cyclin T1 complex affected the activation and differentiation program of lymphoid cells.14 However, the molecular mechanism through which the CDK9/Cyclin T1 complex is altered in malignant transformation needs to be elucidated.
In the present study, we demonstrated that Cyclin T1 expression levels are increased in head and neck carcinoma cell lines. A striking finding of our investigation was that Cyclin T1 is directly involved in malignant transformation and is able to induce tumor growth in vivo. Furthermore, we present herein evidence that Cyclin T1 effect on cellular proliferation is probably mediated by Rb/E2F1 pathway.
Results
Cyclin T1 and CDK9 are overexpressed in head and neck human tumor cell lines.
We first analyzed Cyclin T1 and CDK9 protein expression in different head and neck tumor cell lines (HaCaT, IHOK, 15b, HN4, HN6, HN12, HN13, HN30 and HN31) by western blot. As control, we used a normal cell line (NHEK). Cyclin T1 and CDK9 protein levels were found to be increased in most of the tumor cell lines analyzed (Fig. 1).
Figure 1.
PTEFb is increased in head and neck human tumor cell lines. Head and neck tumor cell lines (HaCaT, IHOK, 15b, HN4, HN6, HN12, HN13, HN30 and HN31) or normal keratinocytes (NHEK) were analyzed by western blot using anti-Cyclin T1, anti-CDK9 or anti-HSP90 antibodies.
Cyclin T1 induces transformation in vitro.
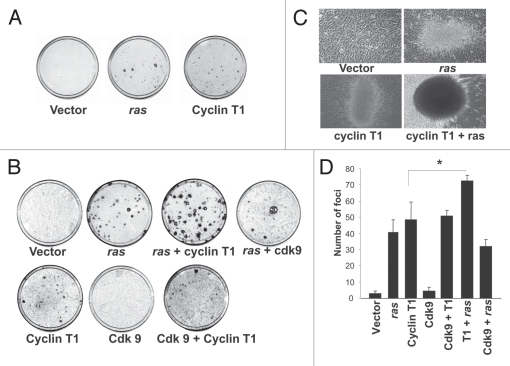
In order to investigate whether PTEFb induces transformation in vitro, we performed the focus formation assay transfecting NIH 3T3 cells with expression vectors for Cyclin T1 or CDK9. In addition cells were transfected with Ras as positive control. As shown in Figure 2A and B, Cyclin T1 but not CDK9 induces foci formation. The number of foci generated by Cyclin T1 were similar to the foci generated by Ras (Fig. 2B–D).
Figure 2.
Cyclin T1 induces foci formation in NIH 3T3 cells. (A) pcDNA3 β-gal (vector), pcDNA3 Ras or pcDNA3 Cyclin T1 were transfected in NIH 3T3 cells for the focus formation assay as described in Materials and Methods. (B) pcDNA3 β-gal (vector), pcDNA3 Ras, pcDNA3 Cyclin T1 or pcDNA3 Cdk9 and the combinations detailed were transfected in NIH 3T3 cells for the focus formation assay as described in Materials and Methods. (C) Morphology foci were observed at the microscope and photographed. (D) Average ± Standard deviation of number of foci from three independent experiments were plotted.
We also co-transfected NIH 3T3 cells with the Cyclin T1 or CDK9 in combination with Ras and we determined the number of foci. As shown in Figure 2B and D the number of foci is significantly increased when a very low amount of Ras is cotransfected with Cyclin T1. Furthermore, Cyclin T1 and Ras combination foci are morphological different and bigger than the foci produced by Cyclin T1 or Ras alone (Fig. 2C). These results suggest that Cyclin T1 is sufficient to induce transformation; however a synergistic effect is achieved in combination with Ras.
Cyclin T1 induces colony and tumor formation.
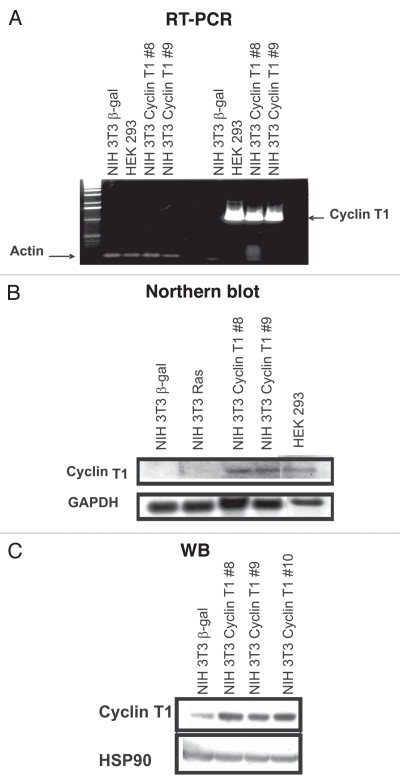
To further investigate the transformation process induced by Cyclin T1, we generated NIH 3T3 stable transfected cell lines with Cyclin T1 expression vector (NIH 3T3 Cyclin T1) or β-galactosidase (NIH 3T3 β-gal) as control. Cyclin T1 overexpression was confirmed by RT-PCR and northern blot analysis using human specific primers or a PCR generated probe, respectively (Fig. 3A and B). We also used RNA from HEK 293 cell line as human positive control. As shown in Figure 3A and B, we obtained two different clones (8 and 9) overexpressing human Cyclin T1. Even more, we also detected the protein overexpression by western blot using a Cyclin T1 antibody that recognizes both, human and mouse protein (Fig. 3C).
Figure 3.
NIH 3T3 Cyclin T1 clones overexpresses Cyclin T1. RNA from stable transfected cell lines (NIH 3T3 Cyclin T1, NIH 3T3 Ras or NIH 3T3 β-gal) was isolated and (A) RT-PCR; or (B) Northern blot analysis was performed. Human specific primers and PCR generated probe were designed, respectively. Human HEK 293 cell line also was used as positive control. (C) NIH 3T3 Cyclin T1, NIH 3T3 Ras and NIH 3T3 β-gal were analyzed by western blot using anti-Cyclin T1 and anti-HSP90 antibodies.
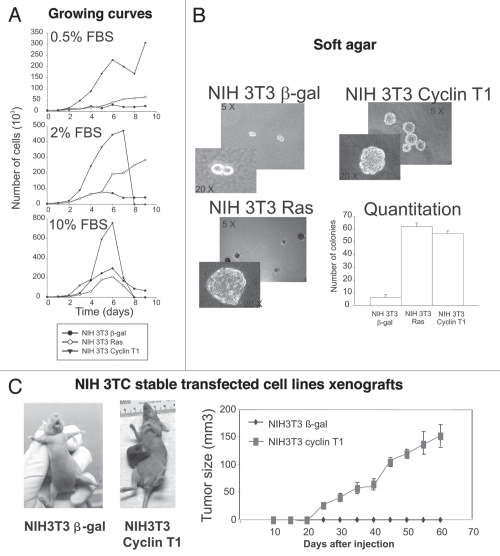
Upon establishment of the Cyclin T1 expressing cells, we studied the growth properties of these transfectants. NIH 3T3 Cyclin T1 cells were found to grow faster in low serum (0.5% and 2%) compared to controls (NIH 3T3 Ras and NIH 3T3 β-gal) (Fig. 4A). Furthermore, the doubling time was shorter for NIH 3T3 Cyclin T1 (31 h) in comparison with NIH 3T3 Ras (42 h) or NIH 3T3 β-gal (65 h). Altogether, these results suggest that Cyclin T1 overexpression highly increases proliferation in vitro.
Figure 4.
NIH 3T3 Cyclin T1 clones show increased proliferation, colony formation and tumor growth. (A) NIH 3T3 Cyclin T1, NIH 3T3 Ras or NIH 3T3 β-gal clones were grown in the presence of different percentage of serum (0.5; 2 or 10%) and number of cells was determined every day during 10 days. (B) Colony formation assays were performed for the NIH 3T3 stable transfected clones as described in Materials and Methods. Average ± Standard deviation of number of colonies from three independent experiments were plotted. (C) NIH 3T3 Cyclin T1 or NIH 3T3 β-gal clones were s.c. injected in nu/nu mice as described in Materials and Methods. Average ± standard deviation of the tumor volumes were determined.
Next we tested Cyclin T1 clones ability to grow in soft agar. Cyclin T1 and Ras overexpression strongly increased the cells capacity to form colonies in soft agar (Fig. 4B). These results suggest that Cyclin T1 may function as an oncogene in vitro.
We then analyzed the in vivo tumorigenicity of NIH 3T3 Cyclin T1, NIH 3T3 β-gal and NIH 3T3 Ras cells inoculated in immunodeficient nude mice. As shown in Figure 4C, subcutaneous injection of Cyclin T1 transfected cells into nude mice resulted in tumor growth within 3 weeks (five out of five animals). None of the mice injected with the NIH 3T3 β-gal induced tumors along the experimental time (60 days) assayed (five out of five animals). Animals injected with NIH 3T3 Ras developed tumors 8 days after injection (data not shown). These results strikingly demonstrate that Cyclin T1 also functions as an oncogene in vivo.
Cyclin T1 induces cell proliferation and CDK4-Rb phosphorylation.
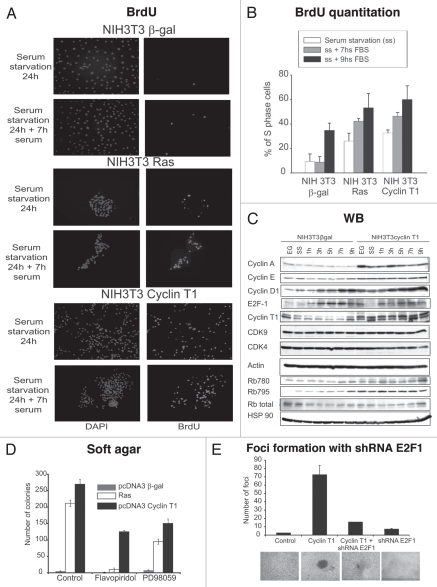
To further investigate the increased proliferation of NIH 3T3 Cyclin T1 cells, we performed BrdU assay. The cells were starved for 24 h and then grew in the absence (0 h) or presence of serum (7 or 9 h). As shown in Figure 5A and B, Cyclin T1 and Ras significantly enhanced the percentage of S phase cells in all the conditions assayed, clearly confirming that the proliferation is induced.
Figure 5.
NIH 3T3 Cyclin T1 clones show increased proliferation and Rb phosphorylation. (A) NIH 3T3 Cyclin T1, NIH 3T3 Ras or NIH 3T3 β-gal clones were starved and then grown in the absence or presence of serum (7 h). BrdU analysis was performed as described in Materials and Methods. (B) BrdU quantitation is shown as % of S-phase cells. (C) NIH 3T3 Cyclin T1 or NIH 3T3 β-gal clones were grown in complete medium (10% FBS) (EG); serum starved (ss) or starved and then grown in the presence of serum for different times (1, 3, 5, 7 or 9 h). Western blot were performed using the detailed antibodies. (D) Colony formation assays were performed for the NIH 3T3 stable transfected clones as described in Materials and Methods and the cells were incubated in media with Flavopiridol (3 nM), PD98059 (500 nM) or DMSO. Average ± Standard deviation of number of colonies from two independent experiments were plotted. (E) pcDNA3 β-gal (vector), pcDNA3 Cyclin T1 and/or shRNA E2F1 plasmids were transfected in NIH 3T3 cells for the focus formation assay as described in Materials and Methods.
To understand the mechanism that triggers Cyclin T1-induced proliferation, cells were grown under starvation for 24 h and afterwards serum was added during different times (1, 3, 5, 7 or 9 h). Proteins involved in cell cycle regulation were analyzed by immunobloting. As shown in Figure 5C, Cyclin A, D1 and E2F1 proteins levels were induced in the NIH 3T3 Cyclin T1 clone even after an hour of serum addition. Furthermore, we detected an increase in Rb hyperphosphorylation at the CDK4 specific sites (780 and 795) suggesting that the proliferation induction in the NIH 3T3 Cyclin T1 clones is probably due to an increase in Rb phosphorylation by CDK4. Even more, Flavopiridol, a potent cdks inhibitor, significantly decreased the number of colonies induced by Cyclin T1 (Fig. 5D). Similar results were obtained when the colony formation assay was performed in the presence of PD98059, a potent ERK pathway inhibitor. These results suggest that Cyclin T1 oncogenic function is probably mediated by Ras pathway and cdks activity.
Due to Cyclin T1 overexpressing cells show increased proliferation and Rb phosphorylation, we next analyzed Rb/E2F1 pathway role in the Cyclin T1 transformation mechanism. We performed focus formation assay transfecting NIH 3T3 cells with Cyclin T1 or pcDNA3 β-gal control plasmids with or without the shRNA E2F1 plasmid to silence E2F1 expression and in consequence to turn off Rb/E2F1 pathway. As shown in Figure 5E, E2F1 silencing abolished the Cyclin T1 foci strongly suggesting that Cyclin T1 induces transformation via Rb/E2F1 pathway.
Discussion
It is well documented that cell cycle deregulation leads to uncontrolled proliferation, one of the hallmarks of cancer cells. However, more critical players in this process remained unidentified. In this paper, we demonstrate for the first time that Cyclin T1 functions as an oncogene in vitro and in vivo.
During the cell cycle, an important role is played by cyclins, a family of proteins named for their cyclical expression and degradation.15 Cyclins act as regulatory subunits of complexes together with the family of related protein kinases (CDKs) that function as catalytic subunits. Interaction between the cyclins and the CDKs occur at specific stages of the cell cycle, and the progression through the cell cycle requires their activities.15 Cyclin T1 is not a typical cell cycle regulator since its levels does not oscillate at any phase during the cell cycle (Fig. 5). This suggests cyclin T1 involvement in specialized functions, such as peripheral blood monocytes and lymphocytes differentiation.14,16,17
Among the cyclins of the T-family, cyclin T1 appears to play the most important role in regulating Cdk9 activity. Cyclin T1 is ubiquitously expressed, but in contrast to Cdk9 its expression level is not constantly high, as the protein is synthesized in response to specific stimuli.14,16 Interestingly, cyclin T1 appears to be conserved from Drosophila to human, especially in the cyclin box.18 Amino acid substitutions in specific regions of the protein can be responsible for the loss of its activity. A cysteine residue at position 261 seems to be of particular importance for cyclin T1 activity.17,19 The most investigated function of the complex Cdk9/cyclin T1 is the phosphorylation of the carboxyl-terminal domain (CTD) of the RNA Polymerase II, that activates transcription elongation.
A required step during transcription initiation is the phosphorylation of serine 5 in the CTD of Pol II by the kinase subunit of TFIIH, CDK7,20,21 after which Pol II pauses to ensure proper pre-mRNA capping.22,23 The transition from pausing to elongation is facilitated by the PTEFb elongation complex, which also mediates efficient elongation.24 PTEFb consists of two subunits, Cyclin T1 and the kinase CDK9, which phosphorylates serine 2 of the CTD, required for the productive elongation and the recruitment of complexes involved in mRNA processing (splicing and polyadenylation).12,24–26 PTEFb appears to function as a crucial CTD kinase for RNA Pol II transcribing immediate early genes (IEG).27,28 Recently, Fujita el al.29 found that Thyrotropin-releasing hormone (TRH)-induced recruitment of PTEFb abolished the pausing of Pol II and enhanced phosphorylation of CTD serine 2, resulting in transcription elongation. In addition the authors determined that the ERK signaling pathway is decisive for TRH-induced recruitment of PTEFb to IEG and subsequent transcript elongation and processing.29 In concordance with these findings, in this work we show that the transformation capacity of cyclin T1 is induced by Ras (Fig. 2). Notably, we also observed that the colony formation was dramatically decreased in the presence of PD98059, an ERK pathway inhibitor (Fig. 5D). Even more, stable cell lines that overexpress cyclin T1 show increased CDK4/Rb phosphorylation (Fig. 5). Altogether these evidences indicate that ERK signaling pathway favors assembly of Cyclin T1/CDK9 heterodimers, which form the active PTEFb enhancing transformation through cell cycle deregulation and provide novel functional insights for PTEFb pathway.
Furthermore, CDK9 and Cyclin T1 are important regulators of several cellular processes.14 A deregulation in the CDK9-related pathway may also be implicated in the establishment and/or maintenance of a transformed cell phenotype.14,16,17,30–33 For instance, one of the functions of CDK9 and its Cyclin partners consists of protecting cells from apoptotic injuries in normal tissues.34,35 In this context, CDK9 and type T cyclins may promote the expression of anti-apoptotic factors such as myeloid cell leukemia 1 (MCL-1).12,36,37 A deregulation in the antiapoptotic CDK9-signaling system may lead to malignant cell transformation, as reported in a number of other systems, such as insulin-like growth factor-I receptor, epidermal growth factor receptor, autocrine/paracrine secreted Frizzled-related protein 2, AKT-related pathway and surviving.38–41
A deregulated CDK9-related pathway was observed in several human tumors: lymphomas,14,30,42 neuroblastoma,17 primary neuroectodermal tumor (PNET),17 rhabdomyosarcoma31 and prostate cancer.32 For instance, an imbalance in CDK9 and Cyclin T1 mRNA was observed in several hematopoietic malignancies: follicular lymphoma, diffuse large B-cell lymphoma with germinal center phenotype, Burkitt's lymphoma and cell lines of classical Hodgkin's lymphoma.14 In addition, high levels of CDK9/Cyclin T1 expression were detected in lymphomas derived from B and T cell precursors, follicular lymphomas and activated T cells (i.e., anaplastic large cell lymhpoma), whereas Hodgkin and Reed-Sternberg cells of classical Hodgkin's lymphoma exhibited strong nuclear staining for both proteins.14
Our findings that Cyclin T1 induces transformation probably by CDK4-Rb mediated cell proliferation may represent a potential therapeutic target. Hence, it is critical to dissect the mechanism for Cyclin T1 transformation and to identify the downstream targets and their crosstalk with other signaling networks in order to activate its oncogenic role. In this regard, very recently, Mueller et al.43 demonstrated that aberrant transcriptional elongation mediated by the Cyclin T1 containing PTEFb complex is a novel mechanism for oncogenic transformation and more recent studies have identified Cyclin T and PTEFb as part of high molecular weight super complexes that may mediate aberrant transcriptional elongation in mixed lineage leukemia.44 This evidence in addition to our results, clearly demonstrate that targeting Cyclin T and other Cyclin T containing complexes like PTEFb should be considered in new alternative options in cancer therapy.
Materials and Methods
Cell culture.
NHEK, HaCaT, IHOK, 15b, HN4, HN6, HN12, HN13, HN30 and HN31 human keratinocytes cells were grown in D-MEM containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, California) in a 5% CO2 humidified atmosphere at 37oC. NIH 3T3 cells were maintained in D-MEM containing 10% calf serum (Invitrogen).
Immunoblot analysis.
Cell lysates were obtained as described previously.45 The following primary antibodies were used: Cyclin T1 (clone T18), CDK9 (clone H169), Hsp 90 (clone H114), Cyclin A (C19), Cyclin D1 (C20), Cyclin E (E4), E2F1 (C20) and Rb (C15) from Santa Cruz Biotechnology, Inc., (Santa Cruz, CA); actin (Chemicon International, Temecula, CA) and Phospho-Rb antibodies (Ser-780, Ser-795) were from New England Biolabs, Inc., (Beverly, MA). Reactions were detected by horseradish peroxidase conjugated secondary antibodies and enhanced chemiluminescence (Pierce, Rockford, IL) following manufacturer's directions.
Stable transfected cell lines generation.
G418 concentration for selection (100 µg/ml) or maintains (50 µg/ml) were determined performing a concentration curve in NIH 3T3 cells between 10 to 500 µg/ml. NIH 3T3 stable cell lines were generated transfecting with 10 µg of pcDNA3 Cyclin T1 wt (NIH 3T3 Cyclin T1), pcDNA3 Ras (NIH 3T3 Ras) or pcDNA3 β-galactosidase vectors (NIH 3T3 β-gal) by calcium phosphate method in a 100 mm plate. Twenty four hours post-transfection, G418 was added at the concentration for selection. After 10 days, single clones were amplified and Cyclin T1 expression was determined by western blot, northern blot and RT-PCR. Clones with the highest Cyclin T1 expression were used. pcDNA3 Cyclin T1 wt and pcDNA3 CDK9 plasmids were a gift from Dr. Matija Peterlin (University of California, San Francisco, USA). pcDNA3 Ras and pcDNA3 β-galactosidase plasmids were a gift from Dr. Silvio Gutkind (NIDCR, NIH, Bethesda, USA).
Focus formation assays.
NIH 3T3 cells were grown up to 20% confluence in 10-cm plates and transfected with the plasmids detailed by calcium-phosphate precipitation technique as previously described.46 The day after transfection, cells were washed and incubated in DMEM supplemented with 5% calf serum. The cultures were maintained in the same medium, with medium changes every 3 days, until the appearance of foci from transformed cells was evident (usually 15 to 25 days after transfection). Plasmid used in the transfection were pcDNA3 Cyclin T1 wt, pcDNA3 CDK9, pcDNA3 Ras and pcDNA3 β-galactosidase described above and shRNA E2F1 A, shRNA E2F1 B and pGIPS obtained from OpenBiosystems.
Northern blot analysis.
Total RNA was extracted using TRIZOL (Invitrogen) following manufacturer's recommendations. Northern blot analysis was performed as described.45 Full length Cyclin T1 probe (2.2 Kb) was generated by PCR from pcDNA3 Cyclin T1 plasmid. GAPDH probe was obtained from Ambion Inc, Austin, Texas.
Colony formation assay.
A bottom layer of media with 10% of calf serum and 1% of agar (4 ml) was poured in a 6 cm plate. After solidifying another layer containing a lower amount of agar (0.5%) and serial dilutions of cells (1 × 103 to 1 × 105) was poured over the bottom layer. The plates were placed in the incubator, feed once per week with 200–400 µl of media with 10% calf serum and after two weeks colonies were counted.
Xenograft model.
All animal experiments were done in accordance with institutional guidelines for animal welfare. NIH 3T3 β-gal, NIH 3T3 Cyclin T1 or NIH 3T3 Ras (5 × 106) cells were injected s.c. into 5 to 6-week-old male immunodeficient nu/nu mice. Tumors were measured with a caliper twice a week, and their volumes were calculated using the formula π/6 × a × b2, where a is the longest dimension of the tumor and b is the width. Tumors injected with NIH 3T3 β-gal or NIH 3T3 Cyclin T1 were allowed to grow 60 days and then mice were sacrificed.
BrdU assay.
NIH 3T3 stable transfected cell lines were seeded on glass coverslips and serum starved for 24 h. Then, media was replaced with media with 10% of calf serum and after 7 hs or 9 hs, incubation with BrdU staining was performed using BrdU Immunofluorescence Assay Kit (Roche Applied Science), following the manufacturer protocol. Coverslips were mounted in Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Inc., Burlingame, CA), and viewed using a Leica TCS-SP2 confocal system (Heidelberg, Germany).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12526
References
- 1.Morgan DO. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 2.MacLachlan TK, Sang N, Giordano A. Cyclins, cyclin-dependent kinases and cdk inhibitors: implications in cell cycle control and cancer. Crit Rev Eukaryot Gene Expr. 1995;5:127–156. doi: 10.1615/critreveukargeneexpr.v5.i2.20. [DOI] [PubMed] [Google Scholar]
- 3.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 6.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 7.Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, et al. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- 8.Robles AI, Rodriguez-Puebla ML, Glick AB, Trempus C, Hansen L, Sicinski P, et al. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, et al. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bagella L, MacLachlan TK, Buono RJ, Pisano MM, Giordano A, De Luca A. Cloning of murine CDK9/PITALRE and its tissue-specific expression in development. J Cell Physiol. 1998;177:206–213. doi: 10.1002/(SICI)1097-4652(199811)177:2<206::AID-JCP2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 12.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gegonne A, Weissman JD, Lu H, Zhou M, Dasgupta A, Ribble R, et al. TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc Natl Acad Sci USA. 2008;105:5367–5372. doi: 10.1073/pnas.0801637105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellan C, De Falco G, Lazzi S, Micheli P, Vicidomini S, Schurfeld K, et al. CDK9/CYCLIN T1 expression during normal lymphoid differentiation and malignant transformation. J Pathol. 2004;203:946–952. doi: 10.1002/path.1588. [DOI] [PubMed] [Google Scholar]
- 15.Senderowicz AM. Small-molecule cyclin-dependent kinase modulators. Oncogene. 2003;22:6609–6620. doi: 10.1038/sj.onc.1206954. [DOI] [PubMed] [Google Scholar]
- 16.De Luca A, De Falco M, Baldi A, Paggi MG. Cyclin T: three forms for different roles in physiological and pathological functions. J Cell Physiol. 2003;194:101–107. doi: 10.1002/jcp.10196. [DOI] [PubMed] [Google Scholar]
- 17.De Falco G, Giordano A. CDK9: from basal transcription to cancer and AIDS. Cancer Biol Ther. 2002;1:342–347. [PubMed] [Google Scholar]
- 18.Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujinaga K, Taube R, Wimmer J, Cujec TP, Peterlin BM. Interactions between human cyclin T, Tat and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz BE, Larochelle S, Suter B, Lis JT. Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol Cell Biol. 2003;23:6876–6886. doi: 10.1128/MCB.23.19.6876-6886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 26.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 27.Byun JS, Wong MM, Cui W, Idelman G, Li Q, De Siervi A, et al. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci USA. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryser S, Fujita T, Tortola S, Piuz I, Schlegel W. The rate of c-fos transcription in vivo is continuously regulated at the level of elongation by dynamic stimulus-coupled recruitment of positive transcription elongation factor b. J Biol Chem. 2007;282:5075–5084. doi: 10.1074/jbc.M607847200. [DOI] [PubMed] [Google Scholar]
- 29.Fujita T, Ryser S, Piuz I, Schlegel W. Upregulation of P-TEFb by the MEK1-extracellular signal-regulated kinase signaling pathway contributes to stimulated transcription elongation of immediate early genes in neuroendocrine cells. Mol Cell Biol. 2008;28:1630–1643. doi: 10.1128/MCB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu TJ, Peng J, Lee G, Price DH, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem. 1999;274:34527–34530. doi: 10.1074/jbc.274.49.34527. [DOI] [PubMed] [Google Scholar]
- 31.Simone C, Giordano A. Abrogation of signal-dependent activation of the cdk9/cyclin T2a complex in human RD rhabdomyosarcoma cells. Cell Death Differ. 2007;14:192–195. doi: 10.1038/sj.cdd.4402008. [DOI] [PubMed] [Google Scholar]
- 32.Lee DK, Duan HO, Chang C. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J Biol Chem. 2001;276:9978–9984. doi: 10.1074/jbc.M002285200. [DOI] [PubMed] [Google Scholar]
- 33.Gan DD, Macaluso M, Cinti C, Khalili K, Giordano A. How does a normal human cell become a cancer cell? J Exp Clin Cancer Res. 2003;22:509–516. [PubMed] [Google Scholar]
- 34.Falco GD, Neri LM, Falco MD, Bellan C, Yu Z, Luca AD, et al. Cdk9, a member of the cdc2-like family of kinases, binds to gp130, the receptor of the IL-6 family of cytokines. Oncogene. 2002;21:7464–7470. doi: 10.1038/sj.onc.1205967. [DOI] [PubMed] [Google Scholar]
- 35.Foskett SM, Ghose R, Tang DN, Lewis DE, Rice AP. Antiapoptotic function of Cdk9 (TAK/P-TEFb) in U937 promonocytic cells. J Virol. 2001;75:1220–1228. doi: 10.1128/JVI.75.3.1220-1228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Napolitano G, Majello B, Licciardo P, Giordano A, Lania L. Transcriptional activity of positive transcription elongation factor b kinase in vivo requires the C-terminal domain of RNA polymerase II. Gene. 2000;254:139–145. doi: 10.1016/s0378-1119(00)00278-x. [DOI] [PubMed] [Google Scholar]
- 37.Chen R, Keating MJ, Gandhi V, Plunkett W. Transcription inhibition by flavopiridol: mechanism of chronic lymphocytic leukemia cell death. Blood. 2005;106:2513–2519. doi: 10.1182/blood-2005-04-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romano G, Prisco M, Zanocco-Marani T, Peruzzi F, Valentinis B, Baserga R. Dissociation between resistance to apoptosis and the transformed phenotype in IGF-I receptor signaling. J Cell Biochem. 1999;72:294–310. doi: 10.1002/(sici)1097-4644(19990201)72:2<294::aid-jcb14>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Peruzzi F, Prisco M, Dews M, Salomoni P, Grassilli E, Romano G, et al. Multiple signaling pathways of the insulin-like growth factor 1 receptor in protection from apoptosis. Mol Cell Biol. 1999;19:7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen MW, Pedersen N, Damstrup L, Villingshoj M, Sonder SU, Rieneck K, et al. Analysis of the epidermal growth factor receptor specific transcriptome: effect of receptor expression level and an activating mutation. J Cell Biochem. 2005;96:412–427. doi: 10.1002/jcb.20554. [DOI] [PubMed] [Google Scholar]
- 41.Lee JL, Lin CT, Chueh LL, Chang CJ. Autocrine/paracrine secreted Frizzled-related protein 2 induces cellular resistance to apoptosis: a possible mechanism of mammary tumorigenesis. J Biol Chem. 2004;279:14602–14609. doi: 10.1074/jbc.M309008200. [DOI] [PubMed] [Google Scholar]
- 42.Bettayeb K, Tirado OM, Marionneau-Lambot S, Ferandin Y, Lozach O, Morris JC, et al. Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases. Cancer Res. 2007;67:8325–8334. doi: 10.1158/0008-5472.CAN-07-1826. [DOI] [PubMed] [Google Scholar]
- 43.Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/PTEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Siervi A, Marinissen M, Diggs J, Wang XF, Pages G, Senderowicz A. Transcriptional activation of p21(waf1/cip1) by alkylphospholipids: role of the mitogenactivated protein kinase pathway in the transactivation of the human p21(waf1/cip1) promoter by Sp1. Cancer Res. 2004;64:743–750. doi: 10.1158/0008-5472.can-03-2505. [DOI] [PubMed] [Google Scholar]
- 46.Monje P, Marinissen MJ, Gutkind JS. Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol Cell Biol. 2003;23:7030–7043. doi: 10.1128/MCB.23.19.7030-7043.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]