Abstract
We describe here a method to generate combinatorial libraries of oligonucleotides mutated at the codon-level, with control of the mutagenesis rate so as to create predictable binomial distributions of mutants. The method allows enrichment of the libraries with single, double or larger multiplicity of amino acid replacements by appropriate choice of the mutagenesis rate, depending on the concentration of synthetic precursors. The method makes use of two sets of deoxynucleoside-phosphoramidites bearing orthogonal protecting groups [4,4′-dimethoxytrityl (DMT) and 9-fluorenylmethoxycarbonyl (Fmoc)] in the 5′ hydroxyl. These phosphoramidites are divergently combined during automated synthesis in such a way that wild-type codons are assembled with commercial DMT-deoxynucleoside-methyl-phosphoramidites while mutant codons are assembled with Fmoc-deoxynucleoside-methyl-phosphoramidites in an NNG/C fashion in a single synthesis column. This method is easily automated and suitable for low mutagenesis rates and large windows, such as those required for directed evolution and alanine scanning. Through the assembly of three oligonucleotide libraries at different mutagenesis rates, followed by cloning at the polylinker region of plasmid pUC18 and sequencing of 129 clones, we concluded that the method performs essentially as intended.
INTRODUCTION
Rational design of protein structure and function is highly dependent on knowledge of the three-dimensional structure of the protein of interest and its corresponding mechanism of action. Moreover, even in the cases where such information is available, accurate predictions of perturbations caused by limited amino acid substitutions are highly uncertain with current methodology. One powerful approach towards the solution of these problems is based on semi-rational design and random combinatorial chemistry, a strategy that takes advantage of the skill of the experimenter to identify important regions (windows) for the function of the protein and accessibility to generate controlled amino acid variation over these regions (1). The resulting mutant libraries can be selected or screened for the desired property. Inherent limitations on the size of those libraries and on the feasibility of selecting or screening them suggest the need for developing more efficient mutagenesis protocols.
There are two extremes in typical random mutagenesis protocols for protein analysis and improvement. One extreme is where random base changes are introduced all along the desired gene at low multiplicity (e.g. <1% per base), and there are several ways to achieve this [e.g. spiked or degenerate oligonucleotides (2), chemical (3) and enzymatic (4,5) methods]. At the other extreme, a few triplets of bases (codons) are saturated with mutations utilizing synthetic DNA (6). The first method provides a simple way to sample the closest area of sequence space for the whole protein and the second permits a larger departure, but on a very limited set of amino acid positions. It is very important to realize that non-saturating regimes at the level of individual bases can only sample between 15 and 40% of the whole set of amino acids, depending on the starting codon, due to the structure of the genetic code (7) (a low level of mutagenesis will only sample single base mutations within any particular codon).
Ideally, one would like to extend the low-level mutagenesis protocols to sample all possible replacements. This can be done by synthesizing the mutagenic oligonucleotides (oligos) with trinucleotides as synthetic units, where the degeneracy can be introduced with a mixture of codons (8) (as opposed to a mixture of bases). We recently published an automatable procedure to achieve such a goal consisting of the doping of wild-type sequences with mixtures of 9-fluorenylmethoxycarbonyl (Fmoc)-trinucleotide-phosphoramidites at concentrations adjusted for the appropriate level of mutagenesis (hereafter referred to as mutagenesis rate) (9). This method circumvents the limitations imposed by the genetic code and allows the exploration of substitutions to all 20 natural amino acids or any subset of them (e.g. hydrophobics, polars, etc.). The method is versatile, but it requires a stock of Fmoc-trinucleotide-phosphoramidites, which will be hard to maintain unless they become commercially available. An alternative approach involves the resin-splitting method (10) whereby the oligo synthesis column is disassembled at the beginning of the codon to be substituted, and the controlled pore glass (CPG) support containing the growing oligo is split into two portions (in proportions dictated by the desired mutagenesis rate) and repacked into a mutant and a wild-type column, where degenerate and wild-type codons, respectively, are synthesized in parallel. Repetition of this cycle after re-mixing and re-splitting of the column results in the introduction of controlled amounts of degenerate codons comprising the whole set of amino acids. Unfortunately, the method is laborious, tedious, difficult to automate, deleterious to the overall yield and requires large amounts of CPG support and hence large amounts of deoxynucleoside-phosphoramidites (11). It is well suited only for short windows and high mutagenesis rates.
Here we describe a mutagenic method that we have termed orthogonal combinatorial mutagenesis (OCM), which makes use of two sets of deoxynucleoside-phosphoramidites protected in their 5′ hydroxyl with two orthogonal protecting groups, 4,4′-dimethoxytrityl (DMT) and Fmoc, for the synthesis of oligonucleotide libraries with controlled mutagenesis rates at the codon level. The wild-type sequence component is assembled with commercial DMT-deoxynucleoside-methyl-phosphoramidites (DMT-phosphoramidites) while the degenerate codon component is assembled with Fmoc-deoxynucleoside-methyl-phosphoramidites (Fmoc-phosphoramidites), in an NNG/C fashion, using a single synthesis column. The method affords a binomial distribution of variants equivalent to that obtained by the resin-splitting method. Subsets of amino acids, or a single one (e.g. alanine) can be favored if appropriate combinations of Fmoc-phosphoramidites are used for assembly of the mutagenic codons, as proposed by Youvan and Arkin (6). In contrast to the Fmoc-trinucleotide-phosphoramidites (9), all four Fmoc-phosphoramidites are easily synthesized (12–15) at a low cost.
MATERIALS AND METHODS
1H and 31P NMR analyses were obtained at 300 and 121 MHz on a Varian VXR spectrometer. The samples were tested on CDCl3 containing tetramethylsilane as internal reference or 85% H3PO4 as external reference, respectively. The oligonucleotide libraries were assembled on two DNA synthesizers (model 391) from Applied Biosystems using a 0.2 µmol standard protocol as recommended by the manufacturer. HPLC analyses were done on a System Gold chromatograph from Beckman, using a 218TP C18 analytical column (4.6 × 250 mm) from Vydac with detection at 260 nm. N-acyl-deoxynucleosides, thymidine and DMT-phosphoramidites were purchased from Glen Research and used without additional treatments. 9-fluorenylmethoxycarbonyl chloride (Fmoc-Cl), 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU), N,N-diisopropylethylamine (DIPEA) and all ancillary reagents for oligonucleotide synthesis were purchased from Aldrich. The chemical reactions in solution-phase were followed by thin-layer chromatography (TLC) on aluminum-backed silica gel 60 F254 sheets (Merck) using CHCl3/MeOH (9:1 v/v) as the elution system. 5′-O-Fmoc-deoxynucleosides were purified by flash column chromatography using silica gel 60 H (Merck, 5–40 µm) as the stationary phase, while their corresponding phosphoramidites were purified on silica gel 60 (Merck, 40–63 µm). The chloro(diisopropylamino)methoxyphosphine used to phosphitylate the 5′-O-Fmoc-deoxynucleosides was prepared according to Atkinson and Smith (16). Reagents for electrophoresis including acrylamide, agarose and buffers were all purchased from Sigma or Bio-Rad. Restriction endonucleases, T4 DNA ligase, Klenow polymerase, pUC18 plasmid and deoxynucleoside-triphosphates (dNTPs) were bought from Boehringer Mannheim and used according to standard protocols.
Synthesis of Fmoc-phosphoramidites
All four Fmoc-phosphoramidites were synthesized by the procedure reported by Lehmann et al. (14), with only minor changes. Fmoc-Cl was added to the N-acyl-deoxynucleosides or thymidine, previously dissolved in pyridine, at room temperature instead of at 0°C and the reaction was quenched after 5 min instead of 30 min. In all cases, tiny amounts of the 3′- and 3′,5′-byproducts were generated and easily removed by flash column chromatography. 5′-O-Fmoc-deoxynucleosides were recovered in high yields (60–70%) using methanol gradients in dichloromethane for the elution process. Phosphitylation of the 5′-Fmoc-deoxynucleosides was accomplished with chloro(diisopropylamino)methoxyphosphine and DIPEA in tetrahydrofuran (THF) as solvent, to give the corresponding Fmoc-phosphoramidites. These were then purified by column chromatography with 5% of pyridine in dichloromethane and obtained with at least 90% purity (as assessed by HPLC and 31P NMR analysis), with yields ranging from 50 to 70%. A key step that we found to be deleterious for the overall yield of Fmoc-phosphoramidites was the alkaline washing with sodium bicarbonate commonly used in the work-up procedure, since a big proportion of the Fmoc group was removed. Instead, we performed the washing steps only with brine and water in the presence of pyridine to avoid hydrolysis of the compounds. The complete 1H and 31P NMR spectroscopic characterization of the four compounds is as follows.
5′-O-(9-Fluorenylmethoxycarbonyl)-6N-benzoyldeoxyadenosine-3′-O-methyl-N,N-diisopropylphosphoramidite (FmocdA-Me-phosphoramidite). 1H NMR (CDCl3) of the diastereomeric mixture: δ 9.00 (NH of dA, s), 8.79 (H8 of dA, 1H, s), 8.26 and 8.26 (H2 de dA, 1H, 2s), 7.99–7.26 (aromatics of Fmoc + aromatics of benzoyl, 13H, m), 6.52 (H1′, 1H, dT, J = 2.4 and 9 Hz), 4.81–4.74 (H3′, 1H, m), 4.55–4.39 (H4′ and H5′ of dG + CH2 of Fmoc, 4H, m), 4.24 (CH of Fmoc, 1H, t, J = 7.2 Hz), 3.67–3.59 (CH of isopropyl, 2H, m), 3.45 and 3.40 (MeOP, 3H, 2d, J = 3.3 and 3 Hz), 2.95–2.87 (H2′β, 1H, m), 2.78–2.65 (H2′α, 1H, m), 1.24–1.20 (CH3 of isopropyl, 12H, m). 31P NMR (CDCl3): δ 150.27 and 150.1 (2s).
5′-O-(9-Fluorenylmethoxycarbonyl)-4N-benzoyldeoxycytidine-3′-O-methyl-N,N-diisopropylphosphoramidite (FmocdC-Me-phosphoramidite). 1H NMR (CDCl3) of the diastereomeric mixture: δ 8.7 (NH of dC, 1H, bs), 8.11–7.29 (H5 and H6 of dC + aromatics of Fmoc + aromatics of benzoyl, 15H, m), 6.29 (H1′, 1H, dd, J = 6 and 18 Hz), 4.51–4.33 (H3′, H4′ and 2H5′ + CH2 of Fmoc, 6H, m), 4.26 (CH of Fmoc, 1H, t, J = 7.5 Hz), 3.69–3.55 (CH of isopropyl, 1H, m), 3.40 and 3.54 (MeOP, 3H, 2d, J = 7.5 and 6.9 Hz), 2.85–2.72 (H2′β, 1H, m), 2.20–2.12 (H2′α, 1H, m), 1.26–1.16 (CH3 of isopropyl, 12H, m). 31P NMR (CDCl3): δ 150.57 and 149.97 (2s).
5′-O-(9-Fluorenylmethoxycarbonyl)-2N-isobutyryldeoxyguanosine-3′-O-methyl-N,N-diisopropylphosphoramidite (FmocdG-Me-phosphoramidite). 1H NMR (CDCl3) of the diastereomeric mixture: δ 11.8 (H1, 1H, bs), 8.9 (NH of dG, 1H, bs), 7.76 (H8, 1H, bs), 7.58–7.21 (aromatics of Fmoc, 8H, m), 6.24 (H1′, 1H, t, J = 7.5 Hz), 4.69–4.64 (H3′ + 1H5′, 2H, m), 4.5–4.4 (H4′ + 1H5′ + CH2 of Fmoc, 4H, m), 4.25 (CH of Fmoc, 1H, t, J = 9 Hz), 3.68–3.6 (CH of isopropyl, 2H, m), 3.44 and 3.39 (MeOP, 3H, 2s) 2.89–2.8 (CH of isobutyryl, 1H, m), 2.62–2.47 (H2′, 2H, m), 1.27–1.16 (CH3 of isopropyl + CH3 of isobutyryl, 18H, m). 31P NMR (CDCl3): δ 150.06 and 149.8 (2s).
5′-O-(9-Fluorenylmethoxycarbonyl)thymidine-3′-O-methyl-N,N-diisopropylphosphoramidite (FmocdT-Me-phosphoramidite). 1H NMR (CDCl3) of the diastereomeric mixture: δ 8.6 (NH of dT, 1H, bs), 7.77–7.25 (H6 of dT + aromatics of Fmoc, 9H, m), 6.33 (H1′, 1H, dT, J = 2.7 and 6.9 Hz), 4.59–4.38 (H3′, H4′ and 2H5′ + CH2 of Fmoc, 6H, m), 4.27–4.22 (CH of Fmoc, 1H, m), 3.65–3.55 (CH of isopropyl, 2H, m), 3.40 and 3.36 (MeOP, 3H, 2d, J = 6.6 and 6.9 Hz), 2.54–2.41 (H2′β, 1H, m), 2.12–2.02 (H2′α, 1H, m), 1.8 (H7 of dT, 3H, s), 1.2–1.16 (CH3 of isopropyl, 12H, m). 31P NMR (CDCl3): δ 150.4 and 150.02 (2s).
Synthesis of oligonucleotide libraries
Three oligonucleotide libraries with the sequence 5′-TAG GAG GAT CCC CGG GTA CCG AGC TCG AAT TCA CTC GGA C-3′ (codons subjected to mutagenesis are underlined) were synthesized in order to evaluate two different doping protocols, which we termed ‘pre-addition’ and ‘on-line mixing’ protocols and three different mutagenesis rates (α). These syntheses, depicted in Figure 1, required the use of two oligonucleotide synthesizers, which were labeled as synthesizer A and B, respectively. For all oligonucleotide libraries, synthesizer B was prepared with an equimolar acetonitrile solution of the four Fmoc-phosphoramidites (25 mM each) in vial 1, and an equimolar acetonitrile solution of DMT-dG and DMT-dC-phosphoramidite (50 mM each) in vial 2. A 100 mM DBU solution in acetonitrile was fitted instead of the detritylating reagent, while the other ancillary reagents were conventional. Synthesizer A was prepared with individual 100 mM acetonitrile solutions of each DMT-phosphoramidite and conventional ancillary reagents, except vial X, which was prepared with an equimolar 20 mM acetonitrile solution of the four Fmoc-phosphoramidites (5 mM each) for library I and a 50 mM solution (12.5 mM each) for library II. These two libraries were assembled with the pre-addition protocol. Library III was assembled with the on-line mixing protocol using an equimolar 50 mM solution of the four Fmoc-phosphoramidites (12.5 mM each) in vial X. With the described setup, the wild-type codons and the first base of each mutant codon were assembled on synthesizer A, but completion of the mutant codons was performed on synthesizer B through Fmoc synthesis. Thus, assembly of the oligonucleotide libraries requires switching of the synthesis column between both synthesizers to complete each mutant codon. Since the third position of the mutant codons was completed with DMT-phosphoramidites instead of Fmoc-phosphoramidites, all sequences were programmed to remain tritylated. Synthesis of libraries I and II started with assembly of the sequence 5′-AGC X TCG AAT TCA CTC GGA C-3′on synthesizer A, using the conventional DMT-dCbz-CPG support as starting material and jumping the capping step during addition of the X. Next, the synthesis column was transferred to synthesizer B and the sequence N and G/C was added to the growing fragment to complete the first set of 32 mutant codons. This fragment also contains the EcoRI restriction site. The column was re-transferred to synthesizer A and the sequence CCGX was added to the previous fragment, jumping again the capping step during addition of the X. The column was re-transferred to synthesizer B and submitted to coupling with N and G/C as before. The process was repeated twice again with the appropriate wild-type codons to complete the mutagenesis window and finally the sequence 5′-TAG GAG GAT CCC-3′ containing the BamHI restriction site was added on synthesizer A to the growing oligonucleotide fragment to complete the two oligonucleotide libraries.
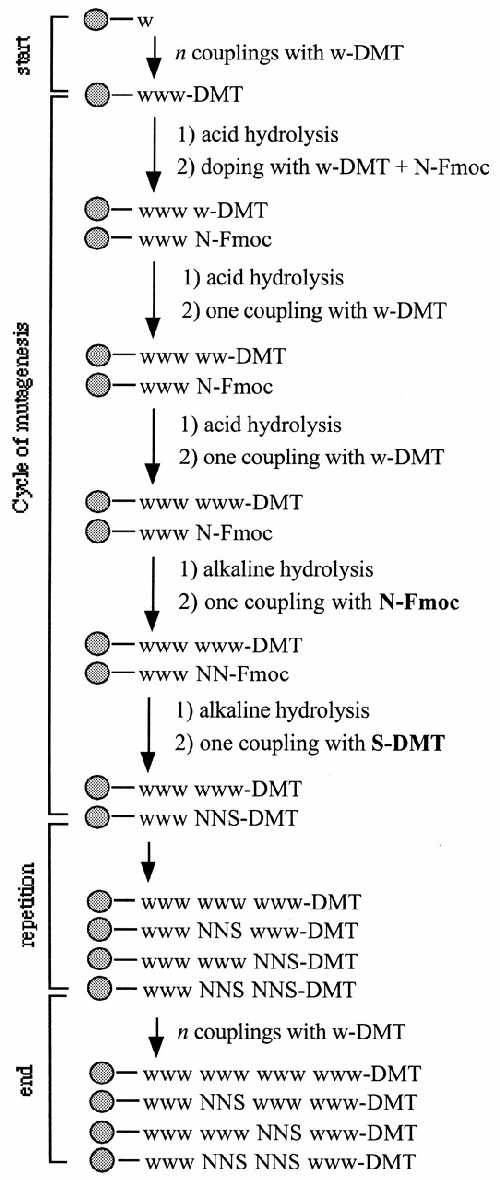
Figure 1.
The OCM method using the on-line mixing mode and exemplified with mutagenesis of two adjacent codons. Start: assembly of a wild-type sequence 3′ to the mutagenesis window, using only conventional DMT-phosphoramidites (w-DMT). Cycle of mutagenesis: doping of the growing oligo with a combination of an appropriate DMT-phosphoramidite and a diluted mixture of the four Fmoc-phosphoramidites (N-Fmoc); two sequential couplings with pure DMT-phosphoramidites to complete the wild-type codon; assembling the second base of the mutant codons with a concentrated mixture of the four Fmoc-phosphoramidites (N-Fmoc); assembling the third base of the mutant codons with a concentrated mixture of DMT-dGib and DMT-dCbz-phosphoramidites. Repetition: the cycle of mutagenesis is repeated as many times as codons to be mutated. End: assembly of a wild-type sequence 5′ region to the mutagenesis window, using conventional DMT-phosphoramidites. A detailed description of this protocol is given in the text. w, the CPG-support with the starting wild-type base, contained in only one synthesis column.
Assembly of library III started on synthesizer A with synthesis of the fragment 5′-AG(CX) TCG AAT TCA CTC GGA C-3′, where bases in parenthesis were simultaneously added to the growing oligonucleotide from different vials. Next the column was transferred to synthesizer B and the bases N and G/C were coupled as before. The column was returned to synthesizer A and the sequence CC(GX) was added to the fragment. Again the column was transferred to synthesizer B, and the mutant codons were completed with N and G/C to reach the level of the wild-type codon. The two following codons were mutagenized in the same way and this oligonucleotide library was finished in the same way as libraries I and II. The mutagenesis rate for each mutated codon was estimated by the difference in absorbance of the DMT cation released from the previous deoxynucleotide of the codon to be substituted and the cation released from the first deoxynucleotide of the substituted codon. The CPG containing the fully protected oligonucleotide was treated with thiophenol for 1 h to remove the internucleotidic methyl groups and subsequently treated with concentrated ammonium hydroxide to remove all remaining protecting groups. The oligonucleotide libraries were purified on 15% polyacrylamide gels containing 8 M urea and recovered in deionized water after n-butanol desalting.
In addition to the three oligonucleotide libraries, an oligo with the sequence 5′-GTC CGA GTG AAT TCG-3′ was also assembled on synthesizer A using DMT-phosphoramidites in order to use it as a primer for the construction of mutant cassettes.
Recombinant DNA methods
According to established recombinant DNA methods, we independently processed and cloned the three oligonucleotide libraries to generate the corresponding mutant libraries. Fifty picomoles of each gel-purified oligonucleotide library and primer were annealed by incubating for a few minutes at 95°C and cooling to 37°C. Full duplex molecules or cassettes were obtained by the action of Klenow fragment and dNTPs. The mutant cassettes were digested with restriction enzymes EcoRI and BamHI and ligated to plasmid pUC18 previously digested with the same enzymes. The ligation mixture was electroporated into Escherichia coli JM101 and the transformed cells were plated in selective media containing ampicillin and incubated overnight at 37°C. Forty-two, 38 and 49 clones for libraries I, II and III respectively were randomly picked and submitted to DNA sequencing by PCR, employing the thermosequenase radiolabeled terminator cycle sequencing kit from Amersham and following the recommended protocol.
Synthesis of the model dinucleotides dApC, dGpC, dTpC and dCpC in solution-phase.
The title compounds were synthesized in a 1 mmol scale in solution-phase by the phosphite-triester approach, mixing 1.2 equivalents of the appropriate 5′-DMT-deoxynucleoside-β-cyanoethyl-phosphoramidite, three equivalents of tetrazol and 1 mmol of 3′-O-DMTdCbz [prepared as reported by Gaytan et al. (9)] in anhydrous acetonitrile and nitrogen atmosphere. After 30 min of reaction, an excess of t-butylhydroperoxide was added to oxidize the phosphite-triester into the corresponding phosphate-triester (1 h). The reactions were worked-up in the usual manner and the fully protected products were purified by column chromatography in silica gel 60 H utilizing gradients of ethyl-acetate in hexane. Deblocking of the dinucleotides was accomplished by an overnight treatment with a mixture of concentrated ammonium hydroxide and methanolic ammonia in a 7:1 proportion (v/v) at 55°C. The dinucleotides were then desalted and detritylated inside an omnifit column packed with 10 g of Jordi support, which displays high affinity for tritylated compounds and retains the removed trityl cation. The elution protocol was similar to that suggested for polypak cartridges from Glen Research. HPLC analysis of the dinucleotides showed products that were at least 96% pure (see Supplementary Material) and of identical retention times with those of dimers independently prepared by conventional solid-phase synthesis with DMT-deoxynucleoside-β-cyanoethyl-phosphoramidites. These pure compounds were used to prepare HPLC calibration curves of each component and to measure their molar extinction coefficients at 260 nm and their particular maximum absorbance wavelength at pH 7.2, utilizing concentrations ranging from 10 to 100 µM.
Reactivity of Fmoc-phosphoramidites
An equimolar solution of the four Fmoc-phosphoramidites (25 mM each) in anhydrous acetonitrile was prepared and fitted in the X vial position of one DNA synthesizer. Dinucleotides with the sequence XC were synthesized by triplicate using the conventional DMTdCbz-CPG support as starting material and the coupling protocol recommended by the manufacturer for β-cyanoethylphosphoramidites. The Fmoc group was removed with 100 mM DBU in acetonitrile for 1 min and the dimers were sequentially submitted to demethylation with thiophenol for 1 h and complete deprotection using a 12 h treatment with concentrated NH4OH at 55°C. The three mixtures of dimers were analyzed by reverse-phase (RP)-HPLC in a manner similar to that reported by Ward and Juehne (17).
Reactivity of Fmoc-phosphoramidites versus DMT-phosphoramidites
Two solutions of mixed phosphoramidites (FmocdA/FmocdC/DMTdG/DMTdT and DMTdA/DMTdC/FmocdG/FmocdT) in acetonitrile, 25 mM for each component, were prepared and used in a similar way to the previous experiment. Dinucleotides with the sequence XC were synthesized with both solutions, in triplicate, and HPLC quantified.
RESULTS AND DISCUSSION
The rationale for the OCM method proposed here implies two sets of deoxynucleoside-phosphoramidites, one protected with the conventional DMT group and the other with a group (R) orthogonal to DMT to be used in solid-phase oligonucleotide synthesis. Wild-type codons were assembled with DMT-phosphoramidites, while the mutant codons were assembled by judicious use of the R-phosphoramidites, in an alternating fashion, but in the same synthesis column at a controlled, non-saturating rate (Fig. 1, where Fmoc is the R group). The synthesis cycles were controlled by selectively removing the DMT or the R protecting group, and codon-level mutagenesis was achieved, generating a binomial distribution of mutants. This was easily predicted by the equation P = [n!/x!(n – x)!]αx(1 – α)n–x, where P represents the proportion of each set of mutants in the library, n the number of wild-type codons to explore in the window, x the type of mutant in the library (e.g. for single mutants x = 1, for double mutants x = 2, etc.) and α the mutagenesis rate (18).
We considered potential protecting groups that were orthogonal to the DMT group, some of which had been previously described in solid-phase oligonucleotide syntheses aimed at avoiding the depurinating acidic conditions required for the removal of the DMT group. These groups include t-butyldiphenylsilyl (19), t-butyldimethylsilyl (19), Fmoc (12–15), levulinyl (20) and 2-dansylethoxycarbonyl (21). In the case of the silyl derivatives, complete removal of these groups requires long reaction times (15 min) of the growing oligonucleotide with tetrabutylammonium fluoride (TBAF) per cycle of synthesis, conditions that are unacceptable for automated DNA synthesis. Furthermore, TBAF could also attack the CPG support due to its silicate nature, diminishing the overall yield of the mutant oligonucleotide libraries. With regard to the levulinyl group, it reacts with a poor regioselectivity toward the 5′ hydroxyl of N-acyl-deoxynucleosides or thymidine and a large amount of the 3′ substituted byproduct is obtained, complicating the chromatographic purification of the product of interest and reducing its overall yield. 2-dansylethoxycarbonyl was a potential candidate as an orthogonal group to DMT, since it is removed with DBU and gives a yellow solution, useful for visually following the yield of the mutagenic couplings. However this latter reagent is not yet commercially available. Out of these potential orthogonal protecting groups we selected Fmoc for further work due to its commercial availability, proven performance in biopolymer synthesis, stability, high regioselectivity to the 5′ hydroxyl of nucleosides and rapid removal in mild alkaline conditions (12–15).
The first point in this work was to synthesize the four Fmoc-phosphoramidites as depicted in Scheme 1, following the general protocol reported by Lehmann et al. (14) with only minor changes, as described in the Materials and Methods. The most important contribution we made to this protocol was the elimination of the sodium bicarbonate washing step during work-up of the reactions, a process that we found to be deleterious to the overall yield of all Fmoc-containing compounds due to the basic pH of the saturated solution that partially removes the Fmoc group. Regarding protection of the phosphate group, we decided to use the methyl group instead of the more conventional β-cyanoethyl group, which is labile to the alkaline conditions used to remove the Fmoc group. Each Fmoc-phosphoramidite was purified by column chromatography and isolated with at least 90% of purity as assessed by RP-HPLC (see Supplementary Material). Additionally, they displayed very good solubility in anhydrous acetonitrile, the solvent commonly used for oligonucleotide synthesis. Its chemical characterization was performed by 1H and 31P NMR analysis.
We tested the OCM method with two different protocols. In the pre-addition mode, a diluted mixture of Fmoc-phosphoramidites (e.g. equimolar A, G, C and T) was reacted with the growing oligo in the synthesis column in order to contaminate the wild-type sequence; excess reagents were washed off and then the DMT-phosphoramidite corresponding to the first base of the wild-type sequence, was added at the usual concentration (details given in Materials and Methods). In the on-line mixing mode, the first base of both the mutant and the wild-type sequence were introduced together by simultaneously coupling the Fmoc-phosphoramidite mixture and DMT-phosphoramidite. The end result of these two methods was the same, as depicted in Figure 1; the additional synthetic cycle needed for the pre-addition mode in comparison with the on-line mode, is justified by the lower quantity of Fmoc-phosphoramidites required to achieve equivalent mutagenesis rates.
The growing oligo then had a controlled quantity of Fmoc degenerate base. The wild-type codon was completed by two cycles of conventional DMT-phosphoramidite chemistry. Then, to complete the mutant component, Fmoc was removed with alkali (DBU) and the second degenerate base was introduced as a mixture of Fmoc-phosphoramidites. Fmoc was removed again and the third degenerate base was added as a mixture of DMT-phosphoramidites. The growing oligo was then a mixture of wild-type and a controlled amount of degenerate codons, both ending with a DMT group. The synthesis cycle was repeated as many times as codons were in the window targeted for mutagenesis.
The OCM method was tested by synthesizing oligonucleotides where four adjacent codons were subjected to various mutagenesis rates. The actual distribution of wild-type and mutated sequences was determined from clones derived from the oligos. Three oligonucleotide libraries were synthesized with the sequence 5′-TAG GAG GAT CCC CGG GTA CCG AGC TCG AAT TCA CTC GGA C-3′, where the underlined triplets denote mutagenized codons. Additionally, an oligo with the sequence 5′-GTCCGAGTGAATTCG-3′, complementary to the 3′ region of the oligonucleotide libraries, was synthesized with DMT-phosphoramidites to use it as primer in the generation of mutant cassettes. The fully protected oligonucleotide libraries and primer were submitted to internucleotidic demethylation with thiophenol, deprotection with concentrated ammonium hydroxide and, finally, they were purified by electrophoresis under denaturing conditions and recovered in sterile deionized water. After electrophoretic analysis, the purified primer appeared as a single band, while the purified oligonucleotide libraries appeared as slightly broadened bands, as expected for the sequence degeneracy.
Libraries I and II were assembled with the pre-addition protocol, using equimolar mixtures of the four Fmoc-phosphoramidites at total concentrations of 20 and 50 mM, respectively, for the doping step.
Any other phosphoramidite or mixture of phosphoramidites were used at 100 mM concentration (details given in Materials and Methods). Library III was also assembled with the 50 mM mixture of Fmoc-phosphoramidites, but in this case with the on-line mixing protocol. Assembly of the libraries required the use of two DNA synthesizers, one to couple the wild-type bases and the first base of the mutant codons and the other to complete the mutant codons. The experimental average mutagenesis rate per codon was 49.5, 78.9 and 10.61% for libraries I, II and III respectively.
Our protocol employed switching of the synthesis column between both synthesizers to complete each mutant codon. Such switching was necessary with the equipment at our disposal, but will not be necessary with synthesizers capable of performing DNA and RNA synthesis, such as models 380B, 392 and 394 from Applied Biosystems, which are equipped with eight vials (this strategy only requires seven) and the ammonium hydroxide bottle that can be used for the DBU solution to remove the Fmoc group. An appropriate synthesis program on those synthesizers would permit the automation of the process to generate complete hands-off codon-level mutagenic oligonucleotide libraries.
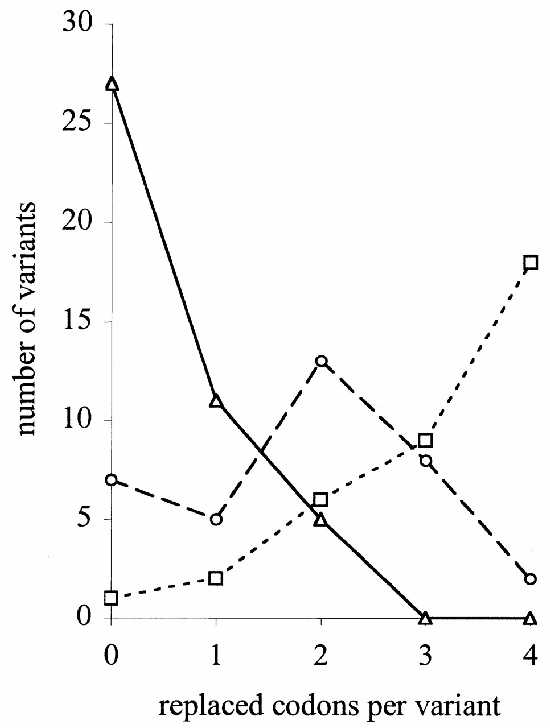
After electroporation into E.coli cells each combinatorial library of mutants was analyzed by direct colony sequencing 42, 38 and 49 clones for libraries I, II and III, respectively; Table 1 summarizes the results. The distribution of multiplicity of replacement is illustrated in Figure 2. An approximately binomial distribution can be observed, concordant with the level of mutagenesis, indicating that the method of mutagenesis we are describing is indeed capable of delivering the intended outcome. With the pre-addition protocol, high mutagenesis rates were easily achieved, but this mode may be less adequate for lower rates and good reproducibility. The on-line mixing protocol would be the method of choice, especially for lower rates; it is more wasteful with the Fmoc-phosphoramidites, but it is also easier to implement in a fully automated fashion (see Materials and Methods).
Table 1. Distribution of mutants obtained in the three oligonucleotide libraries.
| Clone genotype |
Library I, α = 49.5% |
Library II, α = 78.9% |
Library III,
α = 10.6% |
| Wild-type | 7 | 1 | 27 |
| Single mutants | 5 | 2 | 11 |
| Double mutants | 13 | 6 | 5 |
| Triple mutants | 8 | 9 | 0 |
| Quadruple mutants | 2 | 18 | 0 |
| Deletions | 5 | 1 | 4 |
| Insertions | 2 | 1 | 2 |
| Total | 42 | 38 | 49 |
Figure 2.
Distribution of replacements obtained in three oligonucleotide libraries mutated at different mutagenesis rates (α). Library I: α = 49.5% (circles); library II: α = 78.9% (squares); library III: α = 10.61% (triangles).
Inspection of the DNA sequence also reveals that, as prescribed by the experimental implementation, codons with 1, 2 and 3 bp replacements were present, thus allowing sampling of the whole spectrum of amino acid replacements, unlike all the methods that operate at the base pair level (2,4,7). This is achieved while mutagenesis rate is easily controlled. The resin splitting method, which can also sample all amino acids is, as pointed out before, technically tedious, and is suitable for relatively high mutagenesis rates, as was clearly exemplified in the original paper describing it [>10% per codon, using a large amount of resin (10)].
In order to move forward in solving some of the optimization requirements, we decided to determine the relative reactivities of DMT and Fmoc-phosphoramidites and compare them.
The reactivity assay was performed in a similar manner to that reported by Ward and Juehne (20), preparing pools of the dimers dApC, dCpC, dGpC and dTpC by coupling an equimolar mixture of the four Fmoc-phosphoramidites or two Fmoc-phosphoramidites + two DMT-phosphoramidites to dCbz-CPG support. In contrast to the method of Ward and Juehne (17), molar ratios of the dimers were determined by HPLC calibration curves of each dimer [independently prepared in solution-phase with DMT-deoxynucleoside-β-cyanoethyl-phosphoramidites and 3′-O-(4,4′-dimethoxytrityl)6N-benzoyl-deoxycytidine], and not by standards where concentrations were calculated taking into consideration questionable molar extinction coefficients. Ward and Juehne used the values 10.6, 7.3, 8.8 and 8.1 mM–1cm–1 as molar extinction coefficients for dimers dApC, dCpC, dGpC and dTpC respectively (22), while experimentally we obtained the values 18.3, 13.1, 16.8 and 14.3 mM–1cm–1 (pH 7.2 and 32°C), which are closer to those theoretically calculated by Cantor et al. (23). In the case of the four Fmoc-phosphoramidites, we calculated a relative reactivity of 31.4, 22.4, 20.8 and 25.4% for dA, dC, dG and dT, respectively, while in the crossed experiments against the four DMT-phosphoramidites, we observed a relative reactivity of 12.7, 12.2, 13.3, 12.5, 14.2, 9.9, 12.2 and 13.0% for DMT-dA, DMT-dC, DMT-dG, DMT-dT, Fmoc-dA, Fmoc-dC, Fmoc-dG and Fmoc-dT, respectively. Since all Fmoc-phosphoramidites showed different reactivities, an even distribution of variants would require the proper adjustment of concentrations as is done for libraries assembled with β-cyanoethyl-phosphoramidites, where a larger amount of A and C are used to compensate their lower reactivity with respect to the other bases (24).
When we analyzed the distribution of the four bases in the first two positions of the 197 mutant codons obtained in the three oligonucleotide libraries, we found 128 dAs, 76 dGs, 80 dCs and 110 dTs, which represented a relative abundance of 32.5, 19.3, 20.3 and 27.9% respectively. These percentages deviate from an equal distribution to a degree that is similar to other methods (2). The results are also consistent with the relative reactivities measured by the HPLC experiments. In the third position of each mutant codon, performed with an equimolar mixture of DMT-dC and DMT-dG-methyl-phosphoramidites, we found a high bias toward the dCs [a result similar to that of Dunn et al. (1)], obtaining 60.4% of this latter base, 33.0% of dG and, unexpectedly, 4.6% of dA and 2.0% of dT. Additionally, out of the 129 analyzed clones, 10 contained nucleotide deletions and five contained nucleotide insertions. An unexpectedly high rate of insertions and deletions is not uncommon when using mutagenic oligonucleotides, as other research groups have observed (2,25,26), but is certainly a drawback. Recent results from our laboratory (J.Osuna, personal communication) indicate that fresher Fmoc preparations contribute to lowering the rate of insertions. From these results, it is evident that the method we are proposing is not yet optimized, and requires improvements to address current limitations, nevertheless, in its current status, OCM constitutes an improvement over previous approaches.
CONCLUSIONS
The results shown here demonstrate that the OCM method we have developed is a viable alternative for making oligonucleotide libraries mutagenized at a codon level in non-saturating conditions, generating a binomial distribution roughly concordant with the mutagenesis rate. This method offers a powerful potential application since the mutagenic units, that is, Fmoc-phosphoramidites, are easily synthesized and can be combined in conventional oligonucleotide synthesizers with commercial DMT-phosphoramidites. Further, with the use of specific Fmoc-phosphoramidites it should be possible to generate combinatorial oligonucleotide libraries of specific subsets of amino acids (6) or specific amino acid (such as alanine) scanning (27). In the future, we will analyze the use of β-cyanoethyl-phosphoramidites instead of the methyl-phosphoramidites for this mutagenic method, and also additional protecting groups that would add to the power and versatility of the approach.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
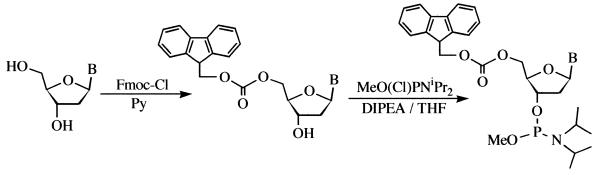
Scheme 1. Synthesis of Fmoc-phosphoramidites. B = 6N-benzoyladenine, 4N-benzoylcitosine, 2N-isobutyrylguanine or thymine. Fmoc-Cl = 9-fluorenylmethoxycarbonyl chloride. Py = pyridine. DIPEA= N,N-diisopropylethylamine.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Eugenio López, Beatriz Quiroz and Javier Pérez for their technical assistance in the laboratory, NMR analysis and FAB-MS analysis, respectively. We are also indebted to Gloria Saab-Rincón, Antonia Olivares and Mario Trejo for their critical review of the manuscript. We gratefully acknowledge the financial support from CONACyT (Mexico), grant no. G030-N9607, to X.S.
References
- 1.Dunn I.S., Cowan,R. and Jennings,P.A. (1988) Improved peptide function from random mutagenesis over short ‘windows’. Protein Eng., 2, 283–291. [DOI] [PubMed] [Google Scholar]
- 2.Hermes J.D., Parekh,S.H, Blacklow,S.C., Köster,H. and Knowles,J.R. (1989) A reliable method for random mutagenesis: the generation of mutant libraries using spiked oligodeoxyribonucleotide primers. Gene, 84, 143–151. [DOI] [PubMed] [Google Scholar]
- 3.Botstein D. and Shortle,D. (1985) Strategies and applications of in vitro mutagenesis. Science, 229, 1193–1201. [DOI] [PubMed] [Google Scholar]
- 4.Lehtovaara P.M., Koivula,A.K., Bamford,J. and Knowles,J.K.C. (1988) A new method for random mutagenesis of complete genes: enzymatic generation of mutant libraries in vitro. Protein Eng., 2, 63–68. [DOI] [PubMed] [Google Scholar]
- 5.Cadwell R.C. and Joyce,G.F. (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl., 2, 28–33. [DOI] [PubMed] [Google Scholar]
- 6.Arkin A.P. and Youvan,D.C. (1992) Optimizing nucleotide mixtures to encode specific subsets of amino acids for semi-random mutagenesis. Biotechnology, 10, 297–300. [DOI] [PubMed] [Google Scholar]
- 7.Sirotkin K. (1986) Advantages to mutagenesis techniques generating populations containing the complete spectrum of single codon changes. J. Theor. Biol., 123, 261–279. [DOI] [PubMed] [Google Scholar]
- 8.Sondek J. and Shortle,D. (1992) A general strategy for random insertion and substitution mutagenesis: substoichiometric coupling of trinucleotide phosphoramidites. Proc. Natl Acad. Sci. USA, 89, 3581–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaytán P., Yañez,J., Sánchez,F., Mackie,H. and Soberón,X. (1998) Combination of DMT-mononucleotide and Fmoc-trinucleotide phosphoramidites in oligonucleotide synthesis affords an automatable codon-level mutagenesis method. Chem. Biol., 5, 519–527. [DOI] [PubMed] [Google Scholar]
- 10.Glaser S.M., Yelton,D.E. and Huse,W.D. (1992) Antibody engineering by codon-based mutagenesis in a filamentous phage vector system. J. Immunol., 149, 3903–3913. [PubMed] [Google Scholar]
- 11.Cormack B.P. and Struhl,K. (1993) Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science, 262, 244–248. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y. and Sonveaux,E. (1987) The 9-fluorenylmethyloxycarbonyl group as a 5′-OH protection in oligonucleotide synthesis. Nucl. Nucl., 6, 491–493. [DOI] [PubMed] [Google Scholar]
- 13.Balgobin N., and Chattopadhyaya,J. (1987) Solid phase synthesis of DNA under a non-depurinating condition with a base labile 5′-protecting group (Fmoc) using phosphiteamidite approach. Nucl. Nucl., 6, 461–463. [Google Scholar]
- 14.Lehmann C., Xu,Y.Z., Christodoulou,C., Tan,Z.K. and Gait,M.J. (1989) Solid-phase synthesis of oligoribonucleotides using 9-fluorenyl-methoxycarbonyl (Fmoc) for 5′-hydroxyl protection. Nucleic Acids Res., 17, 2379–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shchepinov M.S., Chalk,R. and Southern,E.M. (2000) Trityl tags for encoding in combinatorial synthesis. Tetrahedron, 56, 2713–2724. [DOI] [PubMed] [Google Scholar]
- 16.Atkinson T. and Smith,M. (1984) Solid-phase synthesis of oligodeoxyribonucleotides by the phosphite-triester method. In Gait,M.J. (ed.), Oligonucleotide Synthesis-A Practical Approach. IRL Press, Oxford, UK, pp. 39–45.
- 17.Ward B. and Juehne,T. (1998) Combinatorial library diversity: probability assessment of library populations. Nucleic Acids Res., 26, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del-Río G., Osuna,J. and Soberón,X. (1994) Combinatorial libraries of proteins: analysis of efficiency of mutagenesis techniques. Biotechniques, 6, 1132–1139. [PubMed] [Google Scholar]
- 19.Wu T. and Ogilvie,K.K. (1990) A study on the alkylsilyl groups in oligoribonucleotide synthesis. J. Org. Chem., 55, 4717–4724. [Google Scholar]
- 20.Iwai S. and Ohtsuka,E. (1988) 5′-Levulinyl and 2′-tetrahydrofuranyl protection for the synthesis of oligoribonucleotides by the phosphoramidite approach. Nucleic Acids Res., 16, 9443–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergmann F. and Pfleiderer,W. (1994) The 2-Dansylethoxycarbonyl(=2-{[5-(dimethylamino)naphtalen-1-yl]sulfonyl}ethoxycarbonyl; Dnseoc) group for protection of the 5′-hydroxy function in oligodeoxyribonucleotide synthesis. Helv. Chim. Acta, 77, 203–215. [Google Scholar]
- 22.Borer P.N. (1975) Optical properties of nucleic acids, absorption, and circular dichroism spectra. In Fasman,G.D. (ed.), Handbook of Biochemistry and Molecular Biology, Nucleic Acids, 3rd edn. CRC Press, Boca Raton, Vol. 152, pp. 589. [Google Scholar]
- 23.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 24.Conrad R.C., Giver,L., Tian,Y. and Ellington,A.D. (1996) In vitro selection of nucleic acid aptamers that bind proteins. Methods Enzymol., 267, 336–367. [DOI] [PubMed] [Google Scholar]
- 25.Osinga K., Van der Bliek,A., Van der Horst,G., Groot Koerkamp,M. and Tabak,H. (1983) In vitro site-directed mutagenesis with synthetic DNA oligonucleotides yields unexpected deletions and insertions at high frequency. Nucleic Acids Res., 11, 8595–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ner S.S., Goodin,D.B. and Smith,M. (1988) A simple and efficient procedure for generating random point mutations and for codon replacements using mixed oligodeoxynucleotides. DNA, 7, 127–134. [DOI] [PubMed] [Google Scholar]
- 27.Chatellier J., Mazza,A., Brousseau,R. and Vernet,T. (1995) Codon-based combinatorial alanine scanning site-directed mutagenesis: design, implementation, and polymerase chain reaction screening. Anal. Biochem., 229, 282–290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.