Abstract
The insect chemoreceptor superfamily in Drosophila melanogaster is predicted to consist of 62 odorant receptor (Or) and 68 gustatory receptor (Gr) proteins, encoded by families of 60 Or and 60 Gr genes through alternative splicing. We include two previously undescribed Or genes and two previously undescribed Gr genes; two previously predicted Or genes are shown to be alternative splice forms. Three polymorphic pseudogenes and one highly defective pseudogene are recognized. Phylogenetic analysis reveals deep branches connecting multiple highly divergent clades within the Gr family, and the Or family appears to be a single highly expanded lineage within the superfamily. The genes are spread throughout the Drosophila genome, with some relatively recently diverged genes still clustered in the genome. The Gr5a gene on the X chromosome, which encodes a receptor for the sugar trehalose, has transposed from one such tandem cluster of six genes at cytological location 64, as has Gr61a, and all eight of these receptors might bind sugars. Analysis of intron evolution suggests that the common ancestor consisted of a long N-terminal exon encoding transmembrane domains 1-5 followed by three exons encoding transmembrane domains 6-7. As many as 57 additional introns have been acquired idiosyncratically during the evolution of the superfamily, whereas the ancestral introns and some of the older idiosyncratic introns have been lost at least 48 times independently. Altogether, these patterns of molecular evolution suggest that this is an ancient superfamily of chemoreceptors, probably dating back at least to the origin of the arthropods.
Keywords: odorant receptor, gustatory receptor, olfaction, taste, gustation
Chemoreception in insects has long been a major focus of insect chemical ecology; however, despite many efforts, it was only with the sequencing of the genome of Drosophila melanogaster that candidate receptor proteins mediating olfaction and gustation were identified. These discoveries depended on the use of bioinformatic methods to identify genes encoding novel candidate G protein-coupled seven-transmembrane receptor proteins (1-5).
Members of the first family of these genes, the odorant receptor (Or) genes, were found to be expressed in subsets of olfactory neurons in the antenna and maxillary palp, the olfactory organs of this fly (1, 3, 6, 7). Completion of the genome sequence allowed extension of the Or family to 60 receptors with a unified naming system based on their chromosomal location (8). Immunolocalization showed expression in dendrites, as expected of odorant receptors (9, 10). Functional evidence for a role in odor reception was provided by Wetzel et al. (11) and Stortkuhl and Kettler (12), who used heterologous expression in Xenopus oocytes and overexpression in the Drosophila antenna, respectively, to show that Or43a mediates responses to a subset of odorants. Recently Dobritsa et al. (10) have shown through mutant and transgenic rescue analysis that Or22a is required in vivo for response to ethyl butyrate and certain other odorants. Moreover, several other Or genes were shown to confer response to particular odorants or were mapped to particular functional classes of neurons, either by receptor substitution experiments in a mutant neuron or by analysis of strains in which Or promoters were used to drive reporter genes (10). Finally, we note that this family is conserved in other insects. Anopheles gambiae contains as many as 79 Or genes (13, 14), with few simple orthologs of Drosophila Or genes and largely species-specific expansion of gene subfamily lineages. In a study of four of these genes, all were detected exclusively in the antenna, and one is female-specific and down-regulated after a bloodmeal, as expected of a receptor for host odors (13).
Clyne et al. (2) subsequently used bioinformatics to identify another set of 42 seven-transmembrane genes, the gustatory receptor (Gr) genes. A role in gustatory reception was suggested by their expression profile. RT-PCR analysis revealed expression primarily in the proboscis but also in other organs containing gustatory neurons; moreover, expression was absent in mutants lacking gustatory neurons. After completion of the genome sequence, Scott et al. (15) and Dunipace et al. (16) extended the Gr family to at least 54 and 56 members, respectively, and used in situ hybridization and reporter gene constructs to reveal the detailed expression patterns of a subset of the Gr genes. Members of this family are expressed in subsets of neurons in proboscis, pharynx, and leg as well as in larval chemosensory organs. Some members are expressed in the antenna, suggesting a role for some members of the Gr family in olfaction. Evidence that a Gr gene functions in taste perception was provided by genetic analysis of Gr5a, which showed that it is required for response to the sugar trehalose (17, 18), and heterologous expression experiments show that Gr5a is a taste receptor tuned to trehalose (19).
Dunipace et al. (16) noted that the Gr proteins are distantly related to Or83b, whereas Scott et al. (15) suggested that the Or and Gr families belong together in a superfamily of insect chemoreceptors based on conservation of a few amino acid residues in transmembrane domain 7 (TM7). Here we extend the Or family to 62 receptors and the Gr family to 68 receptors, and we analyze their molecular evolution in an insect chemoreceptor superfamily.
Materials and Methods
The public DNA database of the Drosophila genome sequences at National Center for Biotechnology Information (20) was searched with all available Or and Gr proteins by using tblastn (21) to find additional genes encoding proteins in these families, which were in turn used in searches to find more genes in an iterative process. These searches included the updated genomic sequences available as Release 3.1 of the genome (22). Multiple psi-blastp searches were initiated with divergent Ors and Grs to find any additional already annotated proteins that might belong to these families, and up to 10 iterations were used. The genes were reconstructed manually in the paup editor (23) by using the expected exon/intron structures as guides and the spl program (Softberry, www.softberry.com/berry.phtml) to locate predicted introns. In addition, protein alignments were used to indicate instances of unusual gene structure, as were comparisons with orthologs in the draft Drosophila pseudoobscura genome sequence. Proteins were aligned by using clustalx (24), with considerable testing of alternative settings (see ref. 25). To facilitate alignment, the unusually long extracellular loop 2 between TM4 and -5 was removed from Or83a, 83b, and 85e, and Gr33a, -43a, and -66a, as were the long N and C termini of Gr5a, -21a, -32a, -61a, -63a, and Gr64a, -e, and -f. Relaxing the pairwise and multiple alignment gap and extension penalties by 10% to 9 and 0.09, respectively, yielded the best alignment of the seven TMs. Amino acid distances calculated between each pair of proteins were corrected for multiple amino acid changes in the past by using the maximum likelihood model in tree-puzzle Ver. 5 (26), with the blosum62 amino acid exchange matrix and uniform rates based on the actual sequences. A phylogenetic tree was constructed by using neighbor joining followed by a heuristic search for better trees by using tree-bisection-reconnection branch-swapping in paup* Ver. 4.0b10 (23). Bootstrap analysis was performed by using 1,000 neighbor-joining replications with uncorrected distances. RT-PCR was performed as in Clyne et al. (1).
Results
The Or Family. Fifty-four members of the Or family had been annotated by Celera Genomics and the Drosophila Annotation Jamboree (27) when this work was begun in mid-2000, whereas six more were recognized subsequently (6, 8). In the original Celera Genomics scaffold sequences, there were two identical copies of Or19a in inverted orientation ≈50 kb apart; however, resequencing of the genome by the Berkeley Drosophila Genome Project showed that these copies differ at seven nucleotide positions, yielding three changes in amino acid sequence; the proximal gene retains the Or19a name, and the distal gene we have named Or19b. The duplication extends ≈850 bp beyond the predicted N termini and ≈700 bp beyond the predicted C termini. This gene pair represents an unusually recent segmental duplication of the kind that might have been responsible for some of the expansion of the family; however, the separated and inverted nature of the duplication is not typical of the tandem pairs and triplets of related genes seen for the rest of the family. One additional divergent Or gene, Or67d, was discovered in tblastn searches, located near the three known Or genes in chromosomal division 67. Twenty-eight of the annotations provided by Celera Genomics for the 54 annotated proteins seemed unlikely to be entirely correct, as judged from alignment of the protein family and common features of their structures, particularly a short final exon that encodes part of TM7 and that follows a final intron at a conserved location. These revisions were communicated to Swiss-Prot and flybase, and most are incorporated in Release 3.1 of the genome annotations (22).
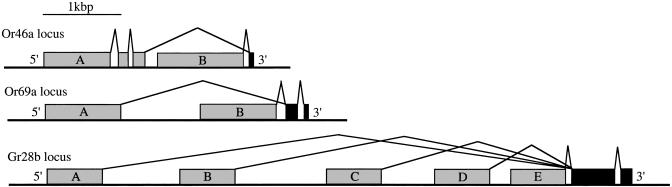
Despite intensive searching, satisfactory C-terminal exons could not be identified for two of the genes, Or46a and Or69b, leading us to hypothesize that they undergo alternative splicing similar to that noted by Clyne et al. (2) for two Drosophila Gr genes and Hill et al. (14) for several Anopheles Gr genes. An appropriate final exon encoding the end of TM7 was not found for Or46a, suggesting that the exons encoding most of Or46a are spliced to the final exon of Or46b. Or69a might similarly be spliced to the final two exons of Or69b (Fig. 1). We have confirmed both of these models by RT-PCR analysis, by using maxillary palp cDNA for Or46a and antennal cDNA for Or69a. These annotation changes require name changes from Or46a and Or46b to Or46aA and Or46aB, and from Or69b and Or69a to Or69aA and Or69aB.
Fig. 1.
Alternative splicing of Or46a, Or69a, and Gr28b. The gray boxes indicate the N-terminal exons that are unique to the differently spliced products, labeled with a letter designating the splice product, whereas the black boxes indicate the shared exons. In the case of Or46a, the encoded proteins share TM7; for Or69a, they share TM6 and -7; and for Gr28b, they share TM5-7. See Clyne et al. (2) for alternative splicing of Gr23a and -39a.
Pseudogenes are rare in the Drosophila genome; however, several are found in the superfamily. In the sequenced Canton-S-derived y; cn bw sp strain, Or85e has suffered a deletion of the 3′ end of the gene relative to an intact cDNA obtained by Vosshall et al. (3) from the Oregon-R strain. In addition, a fragmentary pseudogene was found just upstream of, and in tandem with, Or98a and was named Or98P. Its sequence is comparable to that of Or98a, but it has suffered a ≈1-kbp internal deletion that leaves 138 bp encoding the 46 N-terminal amino acids, the final intron, and the final exon of 69 bp that encodes the 23 C-terminal amino acids. Large deletions like this are thought to be responsible for the paucity of pseudogenes in the Drosophila genome and its small size (e.g., ref. 28).
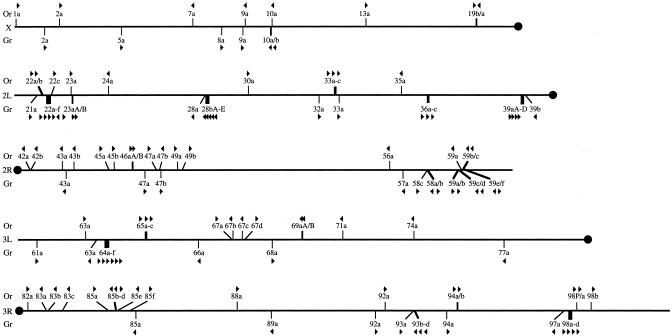
The locations and orientations of these Or genes are shown in Fig. 2. In addition to the recent Or19a/b duplication, a number of them are in short tandem arrays of two or three genes, indicating relatively recent gene duplication, and indeed in some cases the encoded proteins are closely related (Fig. 3; Or22a/b, Or33a-c, Or59b/c, Or65a-c, Or85b-d, and Or94a/b). However, the majority of Or genes are widely spread through the genome, indicating that, in agreement with their high sequence divergence, they are old members of this gene family that have been distributed around the genome by the processes of genome flux (e.g., ref. 29). This genomic distribution of the family is in contrast with the patterns observed with the mammalian olfactory receptors (e.g., refs. 30, 31) and the nematode chemoreceptors in the str, srh, and srj families (32, 33), which commonly are highly clustered on particular chromosomes, in part reflecting the relatively recent expansions of these chemoreceptor families.
Fig. 2.
Genomic locations of the Or and Gr genes. The Or genes are shown above, and the Gr genes below, central lines representing each of the five major chromosome arms drawn to scale following Adams et al. (27). Genes with inferred independent origins are designated by thin lines, whereas clusters of related adjacent genes, or alternatively spliced genes, are shown by thick lines. Orientation of transcription is shown with an arrow; the arrows for the alternatively spliced products are contiguous. The fragmentary Or pseudogene is indicated as 98P. All gene locations and orientations are based on data from Release 3.1 of the genome.
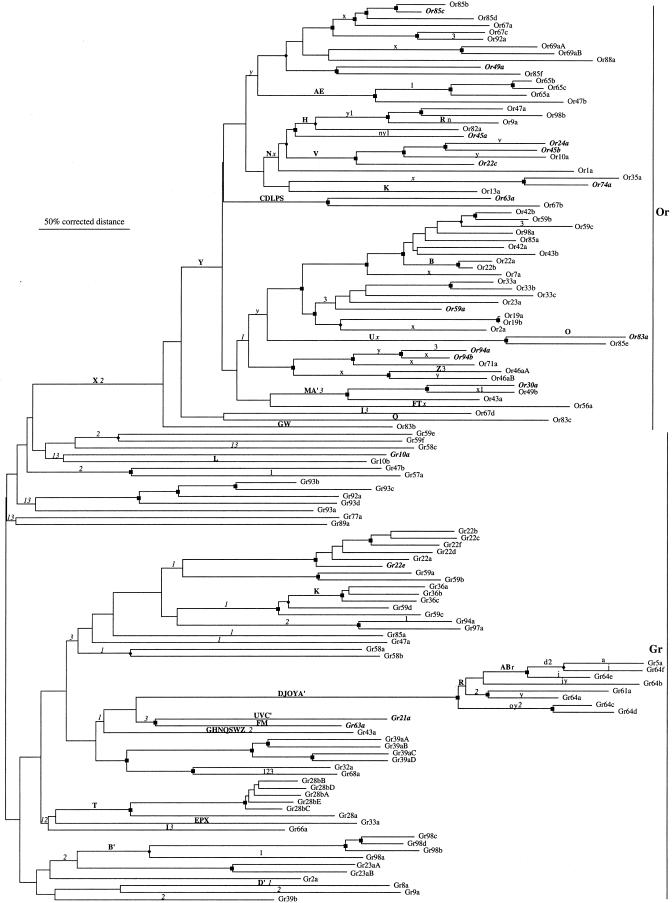
Fig. 3.
Tree of the insect chemoreceptor superfamily. The tree is rooted at the midpoint. The Or and Gr families are indicated on the right, and the scale bar indicates 50% divergence in corrected distances (far larger than the uncorrected distances when comparing distantly related proteins). Branches with 75-100% bootstrap support are indicated with a square and can be considered to be confident, whereas branches with 60-75% bootstrap support are indicated with a diamond and can be considered somewhat confident. Inferred intron gains within the superfamily are indicated above branches in bold uppercase letters, whereas inferred intron losses are shown in lowercase letters (intron losses that are not confidently independent according to the bootstrap support for branches are shown in italics). The Or and Gr families have separate sets of intron letter designations (see Fig. 4), and the putatively ancient ancestral phase-0 C-terminal introns 1-3 are shown as numbers. The Or genes for which no evidence of expression in antenna or maxillary palp has been detected by Vosshall et al. (6) or by us are highlighted in bold italics, as are the four Gr genes that Scott et al. (15) and Dunipace et al. (16) showed to be expressed in the antenna and/or maxillary palp.
The ancestral and idiosyncratic intron locations within the coding regions of the Or genes are shown schematically in Fig. 4, along with those of the Gr genes. Three intron locations appear to be ancestral within the superfamily, as determined by their common location and insertion phase among multiple highly divergent Or and/or Gr lineages. We have named these intron positions 1, 2, and 3 (intron 2 is not present in the Or family, as if it were lost at the base of the Or lineage). There are 27 idiosyncratic Or introns that are not shared in the same location and phase with any Gr lineage but are generally present in one or only a few closely related Or genes. The exceptions are introns x and y, in phases 2 and 0, respectively, which are present in divergent Or lineages and were likely acquired near the base of the Or family tree. It seems likely that at least 25 introns were independently acquired within these single genes or small lineages relatively recently. They are unlikely to be ancient, because then the original Or gene must have been extraordinarily fragmented by introns, and multiple independent losses must have occurred in multiple different Or lineages. Twenty-seven independent losses of introns are inferred on the tree in Fig. 3. Twelve Or genes have lost all but one of their older introns without acquiring any new ones and hence have only one intron within their coding regions; at the other extreme, Or63a and Or67b acquired five new introns in addition to four older ones for a total of nine introns each.
Fig. 4.
Locations and phases of introns in the Or and Gr genes. The intron locations (above the lines) and phases (below the lines) are shown separately for the two families, relative to a scale of the average receptor size in amino acids (determined by excluding the large insertions in some of the receptors). The ancestral phase-0 C-terminal introns 1 and 3 are shared between the two families.
Vosshall et al. (6) detected expression of 40 of the 57 Or genes they examined in sensory neurons of the antenna and maxillary palp by in situ hybridization, and we have been able to detect mRNAs representing 48 of 61 Or transcripts (the analysis did not distinguish between Or19a and -b) in olfactory organs by a combination of in situ hybridization and RT-PCR (ref. 1; C.G.W., unpublished data), leaving 13 transcripts for which there is no evidence of expression in adult olfactory organs. There is no phylogenetic pattern to these 13 Or transcripts in the tree in Fig. 3 (highlighted in bold italics), suggesting that they do not represent a single lineage of genes. It is possible that they have been recruited to expression in other cell types or life stages.
The Gr Family. The combined efforts of Clyne et al. (2), Scott et al. (15), and Dunipace et al. (16) led to the recognition of ≈64 proteins in this family. In addition to the examples described by Clyne et al. (2) of alternatively spliced transcripts from two genes (Gr23a and -39a), together encoding six substantially different proteins, Gr28b encodes five predicted proteins (Fig. 1), which were only partially recognized by Scott et al. (15) and Dunipace et al. (16). We also add two previously undescribed members to the family, Gr9a and -89a, bringing the total to 68 proteins encoded by 60 genes. Most of these were poorly annotated or missed by the automated Celera Genomics annotation. The principal authors of these three papers (2, 15, 16) have coordinated a naming convention for these proteins analogous to that agreed to for the Or proteins; improved annotations for them have been agreed to by all groups and submitted to Swiss-Prot, and most are available in Release 3.1 of the Drosophila annotation (three genes originally designated as members of this family, Gr36d, -43b, and -65a, are no longer considered to be members of the superfamily). Comparison with Gr orthologs in the draft D. pseudoobscura genome indicated that eight of these annotations require further revision; the updated versions have been communicated to flybase and are utilized here (see Or/Gr Proteins, which is published as supporting information on the PNAS web site; see also ClustalX Multiple Sequence Alignment and Table 1 (CG numbers, locations, lengths, and intron numbers), which are published as supporting information on the PNAS web site.
Dunipace et al. (16) noted that Gr22b and -d are pseudogenes because of an in-frame stop codon and a single base-pair deletion in their first exons, respectively. Scott et al. (15) avoided these mutations by starting their annotations downstream, but the resulting proteins do not have TM1. We amplified and sequenced the relevant regions of these two genes from the Oregon-R strain, and both are intact genes in this strain. The apparent TGA stop codon in Gr22b corresponds to a CGA encoding arginine, whereas the single base deletion in Gr22d, located in a string of four Ts, corresponds to a string of five Ts in Oregon-R. Like Or85e, these differences therefore reflect strain polymorphisms also seen in some Anopheles receptors (14); the intact versions are used herein.
The genomic locations of the Gr genes are shown in Fig. 2. Like the Or genes, they are distributed widely around the genome; however, there are some larger clusters (Gr22a-f and Gr64a-f contain six genes each). Some Gr genes appear to have transposed relatively recently: within the Gr22a-f cluster, Gr22e is in the opposite orientation, and Gr22f is ≈28 kb downstream (see also ref. 16). Perhaps the most interesting cases are Gr61a and -5a, which phylogenetically cluster confidently with the Gr64a-f set but are now located elsewhere. Gr5a is the only Gr gene for which a function has been shown; it is required for response to trehalose (17-19), and it is possible that Gr61a and Gr64a-f encode additional sugar receptors. These genes share five idiosyncratic introns and are highly divergent from other receptors within the family. In contrast, the Gr58a-c genes are adjacent to each other, yet only Gr58a/b weakly cluster in the tree, and all three share as little amino acid similarity with each other as many other pairs of Gr proteins (11-16%). We note that Or10a is immediately upstream of and in the same orientation as Gr10a/b, with only ≈350 bp separating the stop codon of Or10a and the start codon of Gr10a. This proximity might be related to the expression of Gr10a in the antenna (15).
Most Gr proteins are extraordinarily divergent, sharing as little as 8% amino acid identity. Multiple alignment and phylogenetic analysis of such highly divergent proteins is difficult. We have used the entire lengths of the proteins, as aligned by clustalx by using slightly relaxed gap penalties, which are particularly capable of achieving alignment of the hydrophobic TMs. In this manner, relationships of the more closely related proteins are well resolved, possibly at the expense of the more distant relationships of the backbone of the tree. As with the Or family, more distant relationships have no bootstrap support (Fig. 3).
The Gr genes contain even more idiosyncratic introns than the Or family. The most recently shared ancestral Gr gene is inferred to have had a long exon encoding TM1-5 followed by three phase 0 introns separating three exons encoding the C-terminal region beyond TM6 (Figs. 3 and 4; see also figure 1 in ref. 2). Mapping of intron losses on the tree is complicated by uncertainty in the backbone of their relationships. However, it seems most likely that there have been 30 idiosyncratic intron gains and at least 21 intron losses, bringing the superfamily totals to 57 gains beyond the ancestral three phase 0 introns and at least 48 losses. Within the Gr family, two genes, Gr68a and -94a, have lost all introns within the coding region. Both of these genes are within the introns of other genes, and Gr68a may have lost its three introns simultaneously through reverse transcription and insertion of a retrocopy. It is possible that these two Gr genes and others have introns in their 5′ UTRs that cannot easily be recognized bioinformatically. In contrast, Gr64e has eight introns.
Discussion
We describe here what may be the full complement of 60 Or and 60 Gr family genes and 130 predicted chemoreceptor proteins that they encode in D. melanogaster. We cannot exclude the possibility of additional highly divergent, or evolutionarily independent, insect chemoreceptors. Scott et al. (15) introduced the notion that these two families of odorant and gustatory receptors are evolutionarily related in an insect chemoreceptor superfamily, and we endorse this view. This superfamily provides a remarkable diversity of receptors that could underlie the entire range of chemoreceptive capabilities of this fly. The Or genes appear to be one lineage, albeit highly expanded in gene number, within the larger Gr family (Fig. 3). To preserve nomenclatural clarity, however, we prefer to retain the Or and Gr designations. In addition to this phylogenetic interrelationship, Scott et al. (15) and Dunipace et al. (16) found that four Gr genes are expressed in subsets of neurons in the antenna and/or maxillary palp (indicated in bold italics in Fig. 3). Neurons expressing Gr21a project axons to glomeruli in the antennal lobe of the brain (15). If these antennal Grs in fact function as odorant receptors, then it would appear that olfactory receptor function has evolved separately several times within the superfamily, perhaps in conjunction with the evolution of terrestrial insects from aquatic arthropod ancestors ≈400 million years ago.
Or83b is extremely divergent from the other Or proteins and is expressed in most olfactory receptor neurons (3). Or83b is also unusual in having an ortholog in A. gambiae that shares 78% amino acid identity, much higher than any other orthologous pair in these two distantly related dipterans (14), as well as 68% identity to CR2 from the moth Heliothis virescens (34). This conservation is sustained throughout the Endopterygota (metamorphosing insects) (ref. 35; G. New, H. Patch, K. Walden, and H.M.R., unpublished results). Dunipace et al. (16) noted that among the Ors, Or83b appears most similar to the Gr family proteins. Our phylogenetic analysis supports this observation (Fig. 3), and although this placement is not supported by bootstrapping, it is obtained regularly when the alignment and tree methodologies are modified considerably. This extraordinary conservation suggests that Or83b serves a function unlike that of other chemoreceptors (14, 35).
The antiquity of this insect chemoreceptor superfamily is supported by several lines of evidence. First, the genes encoding these proteins are roughly evenly spread throughout the genome (Fig. 2). Although there are a few clusters of related proteins that represent recent in situ expansions of gene lineages, the processes of genome flux that led to the current distribution of the genes are clearly evident (for example, the translocation of Gr5a to the X chromosome from the Gr64 cluster on chromosome 3L). This flux is reminiscent of several other ancient gene families in the Drosophila genome, e.g., the tetraspanin superfamily (36).
Second, the amino acid divergences between the Gr and Or proteins, and particularly among the Gr proteins, are extremely high; indeed, Gr proteins commonly share only 8-12% amino acid identity. Some of this divergence could be attributed to an evolving need to adapt to new ecological niches. Nevertheless, the extreme divergence within the family is consistent with an ancient origin. Identification of Or and Gr family members in the moths H. virescens (34) and Manduca sexta (H. Patch, K. Walden, and H.M.R., unpublished results) confirms the antiquity of the families.
Third, the vast majority of introns appear to have been idiosyncratically acquired by limited lineages of genes and commonly single genes (Fig. 3). This pattern of intron evolution is found in other old gene superfamilies (e.g., ref. 36). The ancestral insect chemoreceptor genes appear to have had only three phase 0 introns near their C termini.
Fourth, extended psi-blastp searches initiated with various Grs detected similarities with proteins encoded by five gustatory related (gur) genes, putative seven-TM receptors in the nematode Caenorhabditis elegans (C. Stoetzner and H.M.R., unpublished results). The gur genes are quite distinct from the ≈1,000 candidate chemoreceptors already identified in this nematode (32, 33, 37, 38). The similarity between Gr and GUR proteins suggests that the superfamily predates the arthropod/nematode split.
Note Added in Proof. Bray and Amrein (39) demonstate that Gr68a is expressed in neurons of male-specific contact-chemosensory sensilla on male forelegs and implicate Gr68a in recognition of females in an early step of courtship, when males tap the abdomen of a female with their forelegs, presumably sampling their sex- and species-specific cuticular hydrocarbons. This study provides further support for a gustatory role for most Gr proteins.
Supplementary Material
Acknowledgments
We thank P. Clyne, J. Daniels, J. Kim, E. Moriyama, and A. Ray for assistance with annotation of these genes; and H. Amrein and K. Scott for cooperation in annotation/naming of the Grs. This work was funded by National Science Foundation Grant 9604095 (to H.M.R.) and National Institutes of Health Grants DC04729 and GM63364 (to J.R.C.).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, ”Chemical Communication in a Post-Genomic World,” held January 17-19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviation: TM, transmembrane domain.
References
- 1.Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J. & Carlson, J. (1999) Neuron 22, 327-338. [DOI] [PubMed] [Google Scholar]
- 2.Clyne, P. J., Warr, C. G. & Carlson, J. R. (2000) Science 287, 1830-1834. [DOI] [PubMed] [Google Scholar]
- 3.Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A. & Axel, R. (1999) Cell 96, 725-736. [DOI] [PubMed] [Google Scholar]
- 4.Kim, J., Moriyama, E. N., Warr, C. G., Clyne, P. J. & Carlson, J. R. (2000) Bioinformatics 16, 767-775. [DOI] [PubMed] [Google Scholar]
- 5.Kim, J. & Carlson, J. R. (2002) J. Cell. Sci. 115, 1107-1112. [DOI] [PubMed] [Google Scholar]
- 6.Vosshall, L. B., Wong, A. M. & Axel, R. (2000) Cell 102, 147-159. [DOI] [PubMed] [Google Scholar]
- 7.Gao, Q. & Chess, A. (1999) Genomics 60, 31-39. [DOI] [PubMed] [Google Scholar]
- 8.Drosophila Odorant Receptor Nomenclature Committee (2000) Cell 102, 145-146. [DOI] [PubMed] [Google Scholar]
- 9.Elmore, T. & Smith, D. P. (2001) Insect Biochem. Mol. Biol. 31, 791-798. [DOI] [PubMed] [Google Scholar]
- 10.Dobritsa, A., van der Goes van Naters, W., Warr, C., Steinbrecht, A. & Carlson, J. R. (2003) Neuron 37, 827-841. [DOI] [PubMed] [Google Scholar]
- 11.Wetzel, C. H., Behrendt, H. J., Gisselmann, G., Stortkuhl, K. F., Hovemann, B. & Hatt, H. (2001) Proc. Natl. Acad. Sci. USA. 98, 9377-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stortkuhl, K. F. & Kettler, R. (2001) Proc. Natl. Acad. Sci. USA 98, 9381-9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, A., Pitts, R., Robertson, H., Carlson, J. & Zwiebel, L. (2001) Proc. Natl. Acad. Sci. USA 98, 14693-14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., Cravchik, A., Collins, F. H., Robertson, H. M. & Zwiebel, L. J. (2002) Science 298, 176-178. [DOI] [PubMed] [Google Scholar]
- 15.Scott, K., Brady, R., Jr., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C. & Axel, R. (2001) Cell 104, 661-673. [DOI] [PubMed] [Google Scholar]
- 16.Dunipace, L., Meister, S., McNealy, C. & Amrein, H. (2001) Curr. Biol. 11, 821-835. [DOI] [PubMed] [Google Scholar]
- 17.Dahanukar, A., Foster, K., van der Goes van Naters, W. M. & Carlson, J. R. (2001) Nat. Neurosci. 4, 1182-1186. [DOI] [PubMed] [Google Scholar]
- 18.Ueno, K., Ohta, M., Morita, H., Mikuni, Y., Nakajima, S., Yamamoto, K. & Isono, K. (2001) Curr. Biol. 11, 1451-1455. [DOI] [PubMed] [Google Scholar]
- 19.Chyb, S., Dahanukar, A., Wickens, A. & Carlson, J. R. (2003) Proc. Natl. Acad. Sci. USA 100, 14526-14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., Rapp, B. A. & Wheeler, D. L. (2002) Nucleic Acids Res. 30, 17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, Z., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misra, S., Crosby, M. A., Mungall, C. J., Matthews, B. B., Campbell, K. S., Hradecky, P., Huang, Y., Kaminker, J. S., Millburn, G. H., Prochnik, S. E., et al. (2002) Genome Biol. 3, RESEARCH0083.1-0083.22; Epub 2002 Dec 31. [DOI] [PMC free article] [PubMed]
- 23.Swofford, D. L. (2001) paup*: Phylogenetic Analysis Using Parsimony and Other Methods (Sinauer, New York), Ver. 4.
- 24.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G. & Gibson, T. J. (1998) Trends Biochem. Sci. 23, 403-405. [DOI] [PubMed] [Google Scholar]
- 25.Hall, B. G. (2001) Phylogenetic Trees Made Easy (Sinauer, New York).
- 26.Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. (2002) Bioinformatics 18, 502-504. [DOI] [PubMed] [Google Scholar]
- 27.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 28.Petrov, D. A. (2002) Genetica 115, 81-91. [DOI] [PubMed] [Google Scholar]
- 29.Ranz, J. M., Casals, F. & Ruiz, A. (2001) Genome Res. 11, 230-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glusman, G., Yanai, I., Rubin, I. & Lancet, D. (2001) Genome Res. 11, 685-702. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, X. & Firestein, S. (2002) Nat. Neurosci. 5, 124-133. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, H. M. (2000) Genome Res. 10, 192-203. [DOI] [PubMed] [Google Scholar]
- 33.Robertson, H. M. (2001) Chem. Senses 26, 151-159. [DOI] [PubMed] [Google Scholar]
- 34.Krieger, J., Raming, K., Dewer, Y. M., Bette, S., Conzelmann, S. & Breer, H. (2002) Eur. J. Neurosci. 16, 619-628. [DOI] [PubMed] [Google Scholar]
- 35.Krieger, J., Klink, O., Mohl, C., Raming, K. & Breer, H. (2003) J. Comp. Physiol. A 189, 519-526. [DOI] [PubMed] [Google Scholar]
- 36.Todres, E. Z., Nardi, J. B. & Robertson, H. M. (2000) Insect Mol. Biol. 9, 581-590. [DOI] [PubMed] [Google Scholar]
- 37.Troemel, E. R., Chou, J. H., Dwyer, N. D., Colbert, H. A. & Bargmann, C. I. (1995) Cell 83, 207-218. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta, P., Chou, J. H. & Bargmann, C. I. (1996) Cell 84, 899-909. [DOI] [PubMed] [Google Scholar]
- 39.Bray, S. & Amrein, H. (2003) Neuron 39, 1019-1029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.