Abstract
Glucosinolate profiles differ among Arabidopsis thaliana ecotypes, caused by the composition of alleles at several glucosinolate biosynthetic loci. One of these, GS-Elong, harbors a family of methylthioalkylmalate synthase (MAM) genes that determine the side chain length of aliphatic glucosinolate structures. Fine mapping reveals that GS-Elong constitutes an insect resistance quantitative trait locus, caused by variation in glucosinolate profiles conferred by polymorphism of MAM alleles in this region. A sequence survey of randomly chosen ecotypes indicates that GS-Elong is highly variable among A. thaliana ecotypes: indel polymorphisms are frequent, as well as gene conversion events between gene copies arranged in tandem. Furthermore, statistical methods of molecular population genetics suggest that one of the genes, MAM2, is subject to balancing selection. This may be caused by ecological tradeoffs, i.e., by contrasting physiological effects of glucosinolates on generalist vs. specialist insects.
Resistance to insect herbivores is genetically variable in many plant populations (1). Ecological and evolutionary interactions between host plants and their insect enemies are often mediated by secondary metabolites. Molecular genetics allows us to clone and characterize genes controlling secondary metabolism and insect resistance. When these genes are identified, molecular population genetics provides statistical tests for neutral evolution or natural selection, thus elucidating the evolutionary forces responsible for genetic variation in ecologically important traits. Here we show that a gene family involved in the synthesis of secondary metabolites controls insect resistance, shows complex molecular variation, and maintains excess amino acid polymorphism because of balancing natural selection.
Glucosinolates are amino acid-derived plant secondary compounds present in the Capparales (2, 3). Their biosynthesis occurs in three independent stages, chain elongation of the amino acid, formation of the core structure (consisting of a β-thioglucose moiety and a sulfonated oxime), and side chain modifications. Both side chain elongation and modification contribute to the variation of glucosinolate structures, and >30 different glucosinolates have been identified in the model plant Arabidopsis thaliana (4, 5). However, A. thaliana ecotypes vary extensively both in their glucosinolate composition and quantity (4). Genetically, most of this natural variation can be explained by the combination of alleles at five genetic loci within the A. thaliana genome (4). Among these, GS-Elong has a central role as it controls side chain length of methionine-derived glucosinolates, thereby determining potential for subsequent modification steps (6, 7). GS-Elong consists of a small gene family encoding methylthioalkylmalate synthase (MAM) enzymes responsible for carbon chain elongation in glucosinolate biosynthesis. Leaves of the Columbia (Col-0) ecotype of A. thaliana contain predominantly glucosinolates with four methylene groups (C4) in their basic side chain. In contrast, Landsberg erecta (Ler-0) leaves accumulate primarily glucosinolates with three methylene groups (C3) in their basic side chain. Ecologically, diversification in glucosinolate profiles may represent an adaptation to challenges by microbial pathogens and herbivorous insects.
Materials and Methods
Plant Material. A. thaliana seeds were obtained from the Nottingham Arabidopsis Stock Centre. Ecotypes were chosen without regard to glucosinolate phenotype from throughout the species' native range. Ecotypes were grown as single plants in 5 × 5 cm2 pots filled with a 1:3 vermiculite/standard soil (Einheitserdenwerk, Fröndenberg, Germany) mix under 11.5 h/12.5 h light/dark cycles. For glucosinolate analysis and insect feeding screens, seeds from near-isogenic lines were planted in damp Scotts Redi Earth at a density of 337 plants per m2 in 96-celled flats, covered with clear plastic grow domes and stratified for 48 h at 4°C in the dark. Afterward, flats were moved to ventilated growth rooms with constant air flow and ≈40% humidity and 23°C. Plants were grown at a distance of 30 cm from fluorescent light banks with four bulbs of cool white and four bulbs of wide spectrum lights at a 14 h light/10 h dark photoperiod. Seeds germinated in 2–3 days. Grow domes were removed after 5 days under lights and plants were fertilized once with 1 ml of Scotts Peters Professional Peat Lite Special 20N:10P:20K with trace elements and 1 liter per flat, added to the bottom of the tray. All flats were daily rotated within shelves, between shelves, and end to end to compensate for slight variations in temperature and light intensity in the growth room. For growth rate experiments, five seeds per line were planted in 7.5 × 7.5 cm2 pots, and 18 pots per flat in a random arrangement. Pots were daily rotated between flats. Otherwise, conditions were identical to those for glucosinolate analysis and insect feeding screens.
Herbivory Screens. Diamondback moth (Plutella xylostella) eggs were obtained from New York State Agricultural Experiment Station (Geneva), and a colony was maintained at University of Montana (Missoula). Insects were raised on artificial diet according to published procedures (8). Beet armyworm (Spodoptera exigua) eggs were obtained from Benzon Research (Carlisle, PA), and larvae were raised on an artificial diet from Southland Products (Lake Village, AR). Plants were infested 18 days after transfer to the growth room with one 72-h-old P. xylostella or one 94-h-old S. exigua larva per plant. Plant damage was scored after 48 h.
Growth Rate Measurement. Plants were harvested 16 days after germination. Plants for each pot (i.e., for each line) were pooled, dried in a hot air oven at 45°C, and then weighed. For each line, there were 14 or 15 replicates.
Glucosinolate Analysis. Leaves were harvested 18 days after plants were moved to the growth rooms, i.e., 15–16 days after germination. Glucosinolate extraction and high-pressure liquid chromatography were carried out as described (7).
Molecular Methods. Total leaf DNA was extracted with Qiagen (Hilden, Germany) genomic-tips 100/G following the manufacturer's instructions. Total leaf RNA was isolated with Trizol (Life Technologies). First-strand cDNA was synthesized from 1 μg total RNA following ref. 9. Details on PCRs for MYB37 (=at5g23000), MAM1 (=at5g23010), MAM2, and MAM-L (=at5g23020) genes are given in the supporting information, which is published on the PNAS web site, www.pnas.org.
PCR products were gel purified with QIAquick (Qiagen), and cloned into pCRII TOPO TA, pCR2.1 TOPO TA, or pCR-XL-TOPO vectors (all from Invitrogen). Plasmids were isolated according to standard procedures.
Sequences were obtained either directly from gel-purified PCR products, or from recombinant plasmids. In the latter case, several independent clones were analyzed with vector- and insert-specific primers. Sequencing was done on ABI 377 or 3700 DNA sequencers with Big Dye Terminators (Applied Biosystems). Sequence of the entire Ler-0 GS-Elong region was assembled from two overlapping Ler-0 bacterial artificial chromosomes according to standard methods. Assembly and comparison of DNA sequence data were carried out with DNASTAR (DNASTAR, Madison, WI).
Genotyping of recombinant inbred lines followed ref. 7, including additional markers (see supporting information). Resulting PCR products were purified and analyzed by sequencing, except for those amplified with recMS3f/recMS3r, which were separated on 6% Metaphor (Biozym, Germany) agarose gels.
Statistical Methods. Calculation of Tajima's D, coalescent simulations, and the McDonald–Kreitman test were carried out with DNASP3.84 (10). A neighbor-joining tree (11) of MAM1 and MAM2 alleles was constructed with TREECON (12) following ref. 13. Reliability of the branching order was estimated by boot-strapping (100 replicates; ref. 14).
For measurements of glucosinolates and resistance to S. exigua and P. xylostella, we obtained least square means for each near isogenic line, analyzing a randomized complete blocks design with SYSTAT (SPSS, Chicago). Quantitative trait locus (QTL) mapping with line means used a fixed-effects ANOVA with markers in the GS-Elong interval cross-classified with the AlkOhp marker (15), which was also examined in this experiment (data not shown). Previous studies have shown a QTL influencing glucosinolate concentration in the GS-Elong region (16); hence, in these new experiments, we used a standard P = 0.05 significance threshold based on this a priori expectation.
Results
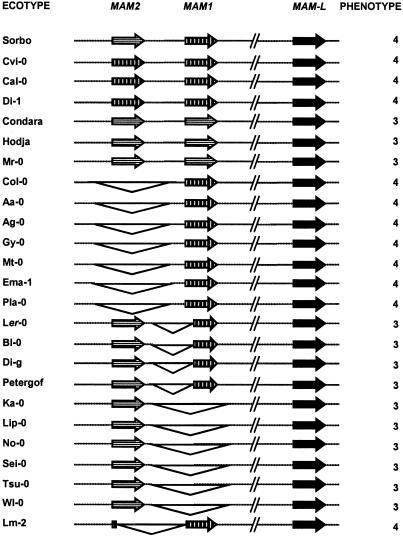
Complex Organization of GS-Elong in A. thaliana Ecotypes. The organization of GS-Elong is highly variable in A. thaliana, and indel polymorphisms of large regions are common (Fig. 1). In addition to a MAM-L (MAM-like) gene present in all ecotypes investigated, GS-Elong may harbor two further loci, MAM1 and MAM2. Some ecotypes contain both loci, whereas others possess either MAM2 or MAM1, or, as the Ler-0 ecotype, a MAM2 in addition to a truncated, nonfunctional MAM1 locus. In the case of the Lm-2 ecotype, the 5′ part of a MAM2-like sequence is fused to the 3′ part of a MAM1-like sequence, which may have been caused by deletion of the intervening region. Reciprocal deletions of MAM2 in Col-0 and MAM1 in Ler-0 have the consequence that the remaining MAM1 and MAM2 genes segregate as alleles of each other, although they are phylogenetically paralogs resulting from a gene duplication event (Fig. 1).
Fig. 1.
GS-Elong region in A. thaliana ecotypes. Maximal levels of divergence between MAM1 and MAM2 (nearly 5%) occur in the Sorbo ecotype, which likely represents the ancestral gene arrangement. Genes are patterned as most similar to Sorbo MAM1 (vertically patterned) or Sorbo MAM2 (horizontally patterned). Triangles indicate large deletions. Predominant glucosinolate side chain length is indicated on the right: glucosinolates with four methylene groups (4, e.g., Col-0) or three methylene groups (3, e.g., Ler-0). Notice that synthesis of C4 glucosinolates is completely associated with occurrence of a full-length Sorbo-like MAM1 gene. Otherwise, C3 glucosinolates are synthesized.
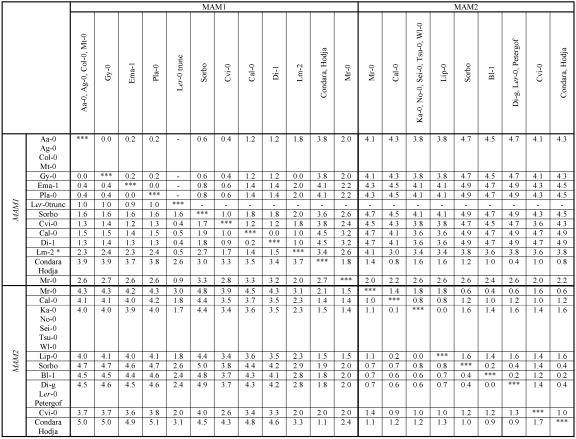
Gene Conversion Between MAM1 and MAM2 Loci. An additional layer of complexity is added by apparent interlocus gene conversion between MAM1 and MAM2. Pairwise comparisons of sequence similarity between MAM genes of ecotypes containing both a MAM2 and a MAM1 locus reveal a maximal divergence between the Sorbo MAM2 and MAM1 genes; both nucleotide and protein sequences differ by ≈5.0% (Table 1). Therefore, the Sorbo ecotype likely represents the basal configuration of MAM genes at GS-Elong in A. thaliana. In contrast, in other ecotypes MAM2 and MAM1 show regions of much greater similarity, with a minimum value of ≈1% divergence between paralogous loci in the Condara and Hodja ecotypes, indicating that genetic information was exchanged between MAM2 and MAM1 in these haplotypes.
Table 1. Pairwise comparisons of amino acids (upper triangle) and nucleotides (lower triangle).
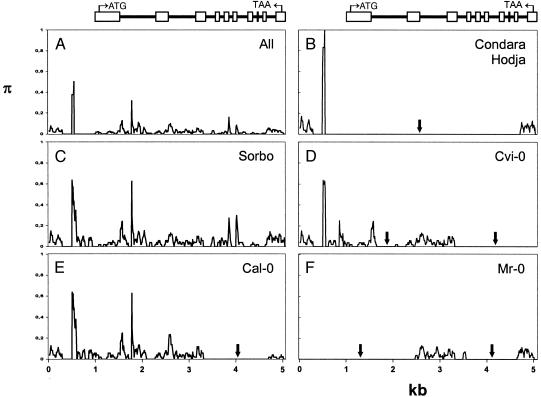
Sliding window analyses of nucleotide polymorphism between MAM1 and MAM2 sequences were performed for those ecotypes harboring both loci, as well as for all MAM1 and MAM2 sequences surveyed (Fig. 2). The nucleotide polymorphism pattern between MAM1 and MAM2 in Sorbo largely reflects the general pattern seen among all MAM1 and MAM2 genes (Fig. 2 A and C), further strengthening the hypothesis that Sorbo reflects the basal configuration of MAM loci at GS-Elong in A. thaliana. In contrast, all other ecotypes harboring two MAM loci, i.e., Cal-0, Condara, Hodja, Cvi-0, and Mr-0, show at least partial sequence identity between the respective genes. However, observed regions of sequence identity differ between ecotypes, suggesting that multiple gene conversion events occurred independently of each other. Genetic information has evidently been transferred in both directions, i.e., in some cases from the MAM1 to the MAM2 locus, and in others from MAM2 to MAM1. For example, in the Condara and Hodja ecotypes most of the MAM1 locus is very similar to the Sorbo MAM2 sequence (Fig. 2B). In contrast, in Cal-0, most of the 3′ part of MAM1 resembles a MAM2-like sequence (Fig. 2E). Finally, in Cvi-0, one central part of MAM2 was converted into a MAM1-like sequence, whereas the 3′ part of MAM1 turned into a MAM2-like sequence (Fig. 2D).
Fig. 2.
Sliding window analysis of nucleotide polymorphism (π) among MAM1 and MAM2 genes. Arrows indicate regions where compared sequences are identical because of one or more presumptive gene conversion events between paralogous genes. The peaks between 500 and 590 and between 1,820 and 1,830 nucleotides stem from complex changes resulting in poor sequence alignment. (A) All sampled MAM1 and MAM2 alleles (except truncated alleles). (B) MAM1 versus MAM2 in the Condara ecotype. Differences between MAM1 and MAM2 are very similar in Hodja. (C) MAM1 versus MAM2 in the Sorbo ecotype. (D) MAM1 versus MAM2 in the Cvi-0 ecotype. (E) MAM1 versus MAM2 in the Cal-0 ecotype. (F) MAM1 versus MAM2 in the Mr-0 ecotype. Window size, 50 nt; step width: 10 nt. Alignment gaps are included in scaling of the horizontal axis. The MAM gene structure is depicted above the panels.
Qualitative Effects Caused by Variation in the Organization of GS-Elong. Ecotypes with a functional MAM1 sequence, like Col-0, accumulate methionine-derived glucosinolates with four methylene groups (C4) in their side chain, whereas those with only a single MAM2 sequence, like Ler-0, sequester predominantly C3 aliphatic glucosinolates (4, 7). Plants with both a MAM1- and a MAM2-like sequence produce primarily C4 glucosinolates, indicating that MAM1 function is dominant over MAM2 function with respect to short-chain glucosinolates. As can be seen from Fig. 1, presence or absence of a full-length Sorbo-like MAM1 gene gives complete prediction of glucosinolate phenotype. Therefore, loss of the MAM1 function, either by deletion of the MAM1 allele or by gene conversion, results in a switch from a C4 to a C3-producing ecotype, provided that MAM2 function is retained.
Quantitative Effects Caused by MAM1/MAM2 Polymorphism. To investigate the impact on plant performance of natural variation between MAM genes, Col-0 was crossed to a recombinant inbred line (17), CL5, which has a Ler-0 MAM2 allele and is predominantly Col-0 elsewhere in the genome (7). A total of 5,000 F2 progeny were screened for recombination in a 210-kb interval containing the GS-Elong region. Prereproductive rosette plants from 58 recombinant, homozygous, near-isogenic F4 lines were assayed for glucosinolates, growth rate, and resistance to larvae of two herbivorous lepidopteran insects, P. xylostella and S. exigua, a crucifer specialist and generalist, respectively (Fig. 3).
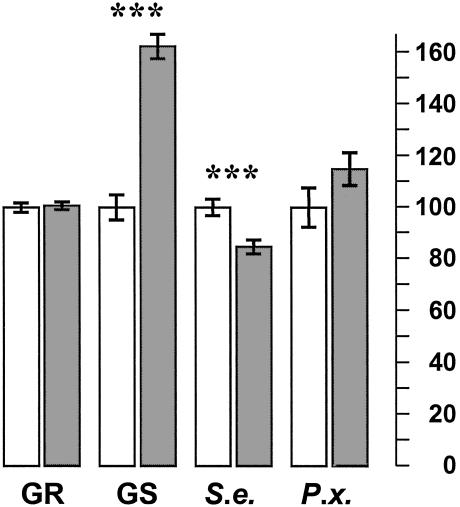
Fig. 3.
Quantitative effects in near-isogenic lines carrying the Col-0 MAM1 (white bars) or the Ler-0 MAM2 allele (gray bars) at GS-Elong. Data are least square means (± standard error) from ANOVA (Col-0 = 100) for growth rate (GR), total aliphatic glucosinolates (GS), damage by S. exigua (S.e.), and by P. xylostella (P.x.). ***, P < 0.001.
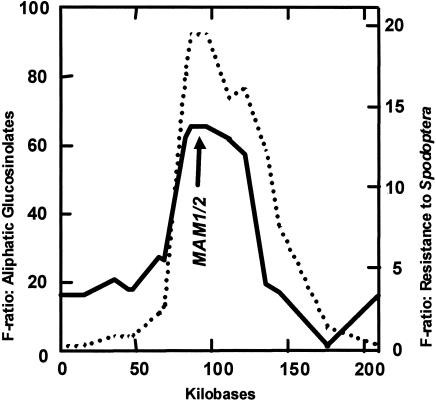
QTLs for total leaf aliphatic glucosinolate concentration and resistance to S. exigua both center at a 15-kb nonrecombinant region containing Col-0 MAM1 or Ler-0 MAM2, but not MAM-L (Fig. 4; F = 92.34, df = 1, 54, P = 0.00001, n = 292 for aliphatic glucosinolates; F = 13.70, df = 1, 54, P = 0.0005, n = 1,212 for S. exigua). These data show that the Ler-0 MAM2 allele at GS-Elong causes increased aliphatic glucosinolate concentration and greater resistance to the generalist herbivore, S. exigua, in comparison to the Col-0 MAM1 allele (Figs. 3 and 4). Herbivory by the specialist insect, P. xylostella, was unaffected by the allelic state at GS-Elong (F = 2.50, df = 1, 54, P = 0.12, n = 1,239). Finally, above-ground biomass was measured in a separate experiment on 3,130 plants in the same 58 mapping lines at 16 days after germination. There was no trace of a significant growth rate QTL at GS-Elong (F = 0.128, df = 1, 54, P = 0.73).
Fig. 4.
QTL mapping of total leaf glucosinolate content (dashed line) and resistance to S. exigua (solid line). Statistical significance is indicated by F ratios.
Nucleotide Polymorphism Patterns. Statistical methods of molecular population genetics were used to test for nonneutral nucleotide polymorphism at GS-Elong (18, 19). Haplotypes for which evidence for gene conversion was found (e.g., Fig. 2) were excluded from significance testing to conform to the statistical assumption of identically and independently distributed point mutations. With the remaining alleles, two independent statistical tests reject the equilibrium neutral hypothesis at MAM2. First, a positive Tajima's D indicates too many intermediate frequency polymorphisms (D = 1.86, P < 0.05 or P < 0.01, based on coalescent simulations without recombination or with free recombination, respectively; Table 2). Second, we found too many amino acid polymorphisms segregating in A. thaliana. When compared with the corresponding gene from closely related Arabidopsis lyrata, the McDonald–Kreitman test (19) shows the ratio of replacement to synonymous nucleotide polymorphisms within A. thaliana is significantly higher than between species (G = 6.098; P = 0.014).
Table 2. Population genetic parameters in the A. thaliana GS-Elong region.
| Gene | MYB37 | MAM2 | MAM1 | MAM-L |
|---|---|---|---|---|
| Coding positions | 721 | 1,518 | 1,518 | 1,509 |
| No. of sequences | 18 | 11 | 8 | 18 |
| No. of haplotypes | 4 | 4 | 3 | 7 |
| πtotal | 0.0014 | 0.0045 | 0.0017 | 0.0034 |
| πsynonymous | 0.0030 | 0.0065 | 0.0039 | 0.0083 |
| πnonsynonymous | 0.0009 | 0.0038 | 0.0010 | 0.0019 |
| Tajima's D | 0.43 (NS) | 1.86* | —1.22 (NS) | 0.40 (NS) |
| McDonald—Kreitman, G; P | 0.003; 0.954 (NS) | 6.098; 0.014* | 0.108; 0.743 (NS) | 0.092; 0.762 (NS) |
All calculations are based on coding regions of genes. Only the third exon of MYB37 was amplified, which comprises > 70% of the entire ORF. Statistical significance of D was investigated by coalescent simulations. Nucleotide diversity (πtotal) at MAM2 is 0.0072 if we include alleles that have experienced gene conversion. NS, not significant.
P < 0.05
Discussion
Molecular Basis of an Insect Resistance QTL. Quantitative analyses demonstrate that GS-Elong constitutes an insect resistance QTL, caused by variation in glucosinolate quantity, quality, or both. Recpirocal deletions of MAM1 and MAM2 have occurred in Ler-0 and Col-0 ecotypes, respectively, so these paralogous loci segregate as alleles in our mapping population. A Ler-0 MAM2 allele at GS-Elong confers higher resistance to the generalist insect herbivore S. exigua than a Col-0 MAM1 allele. Herbivory by a specialist insect, P. xylostella, was unaffected by the allelic state at GS-Elong. This is consistent with studies demonstrating that generalist insects are sensitive toward glucosinolate-based plant defenses, whereas specialists may be able to cope with these compounds (16, 20, 21). Moreover, feeding and oviposition of crucifer specialist insects may be stimulated by glucosinolates and their degradation products (21, 22). Indeed, P. xylostella has evolved a counteradaptation that enables it to circumvent hydrolysis of glucosinolates by myrosinase, and, thus, avoids the formation of toxic breakdown products (23).
This insect resistance QTL displayed complex molecular variation (Fig. 1) with multiple independent mutations causing production of C3 glucosinolates, extensive gene conversion, deletion of large genomic regions, and functionally important genes appearing in Ler-0 that are absent from the A. thaliana Col-0 sequence. Similarly, previous studies in model organisms have shown that gene conversion has important effects on sequence variation in Hsp70 genes in Drosophila (24), and multiple independent deletions at the FRI locus influence life history variation in A. thaliana (25, 26). Conceptually, the GS-Elong region exemplifies complex dynamics predicted for the evolution of gene families (27). In practice, the difficulty of such studies should not be underestimated. Our sustained attempts to characterize allelic variation in the GS-Elong region by using PCR ultimately failed, and we finally sequenced 300 kb of Ler-0 bacterial artificial chromosomes to identify genes which were absent from Col-0.
Evidence for Natural Selection. Two independent statistical tests reject a neutral evolutionary hypothesis at MAM2. We found too many intermediate frequency nucleotide polymorphisms and too many amino acid changes segregating in A. thaliana (P < 0.05 by Tajima's D and McDonald–Kreitman tests; Table 2). Although these findings reject a standard equilibrium neutral model, could these patterns be attributable to nonstandard or nonequilibrium demographic processes rather than to nonneutral evolution? For example, metapopulation structure and population decline can cause positive values of Tajima's D (28, 29). Likewise, relaxation of selection caused by bottlenecks or fixation of deleterious mutations in small populations can cause elevated levels of nonsynonymous polymorphism (30). Because these population processes affect multiple loci throughout the genome, we examined nucleotide polymorphism at three genes immediately flanking MAM2 (Table 2). These results show that sequence polymorphisms adjacent to MAM2 (MYB37, MAM1, and MAML) are compatible with an equilibrium neutral model, based on Tajima's and McDonald–Kreitman analyses (all P > 0.05). Moreover, Haubold et al. (31) examined nucleotide polymorphism at 14 loci in a 170-kb genomic region containing the MAM gene family. They found that genes with contrasting patterns of variation (π and D) are located within a few kilobases of one another. That result corroborates our current finding, where nonneutral variation at MAM2 contrasts sharply with neutral patterns of polymorphism at adjacent loci. Furthermore, this local scale of nucleotide variation appears to be typical for A. thaliana (32).
Molecular variation at MAM2 also contrasts with results from other genes in A. thaliana. Although excess nonsynonymous polymorphism has been observed in comparisons of several A. thaliana genes with congeneric relatives (e.g., refs. 26, 33, and 34), this reflects locus-specific effects that are not found in most genes (34–39). Furthermore, MAM2 displays an excess of intermediate-frequency nucleotide polymorphisms, in contrast to most other A. thaliana genes (26, 33, 34, 37, 39), including data from ≈500 loci sampled throughout the A. thaliana genome (K. Schmid and T.M.-O., unpublished data).
Natural genetic variation at MAM2 shows too much intermediate-frequency nucleotide polymorphism and too many amino acid variants, relative to neutral predictions, suggesting that balancing selection maintains functional diversity at this ecologically important gene. Balancing selection refers to evolutionary mechanisms that maintain more genetic variation than expected under neutrality (40), such as genotype-by-environment interaction, frequency-dependent selection, or trench warfare models of host–enemy coevolution (41). However, MAM2 polymorphism has no impact on glucosinolate identity (Fig. 1), suggesting that nonneutrality at MAM2 is likely caused by selection on glucosinolate quantity and not quality. In support of this interpretation, ecological analyses of natural selection on A. thaliana in the field find stabilizing selection for intermediate glucosinolate concentrations (42).
Allocation Costs or Ecological Tradeoffs? Balancing selection implies that different selective factors favor contrasting phenotypes. For example, genetic variation for insect resistance could be maintained by tradeoffs among components of fitness, if gains in one aspect of fitness were balanced by losses in other fitness components (43). One form of tradeoff, allocation costs, has been proposed to explain genetic polymorphism in plant resistance to insect herbivores. Allocation costs occur when defense mechanisms are energetically expensive, so that genotypes with strong defenses have fewer resources to invest in growth and reproduction (44). To test for possible allocation costs of glucosinolate production, we quantified growth rate (biomass) before bolting. Juvenile biomass is positively correlated with individual fitness in Arabidopsis and Brassica (45, 46). Furthermore, prereproductive vegetative growth rate measures resource availability in the exact environment and growth stage where the Ler-0 MAM2 allele causes increased aliphatic glucosinolate concentration and greater resistance to the generalist herbivore, and allows us to test for allocation costs independently of tolerance (1). However, highly replicated quantification of juvenile growth rate in the fine-scale mapping lines gave no evidence for the existence of allocation costs. In this growth environment and developmental stage, MAM2 provides resistance to generalist insect herbivores without physiological costs. Further experiments will be required to measure fitness consequences in other developmental stages and environments.
An alternative mechanism of tradeoffs is well documented in the Brassica literature. Ecological costs occur when defensive metabolites are toxic to some herbivores but stimulate feeding or oviposition by adapted, specialist insects (43). Indeed, our insect-feeding assays detected enhanced resistance against the generalist herbivore S. exigua, conferred by the MAM2 allele at GS-Elong, whereas herbivory by the crucifer specialist P. xylostella was unaffected by the allelic state of MAM genes at GS-Elong. On the other hand, there is compelling evidence that exposure to glucosinolates, and their degradation products does stimulate feeding, reproduction, or oviposition of a variety of crucifer specialists including butterflies, moths, aphids, beetles, and others, but not by generalists (ref. 22, reviewed in ref. 21). Thus, balancing selection at MAM2 may be explained by ecological tradeoffs caused by contrasting biological effects of glucosinolates on specialist versus generalist herbivores, although we cannot rule out that allocation costs might have additional impact in some environments.
Supplementary Material
Acknowledgments
We thank Deana Pedersen (University of Montana, Missoula) for measuring plant growth rate and Dr. Scott A. McCuine (Department of Soil and Crop Sciences, Texas A&M University, College Station) for providing Ler-0 bacterial artificial chromosomes. T.M.-O. was supported by European Union Contract No. QLRT-2000-01097, the Bundesministerium für Bildung und Forschung, U.S. National Science Foundation Grant DEB-9527725, and the Max-Planck-Gesellschaft. J.K. was supported by the Deutsche Forschungsgemeinschaft and the Max-Planck-Gesellschaft.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Chemical Communication in a Post-Genomic World,” held January 17–19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: MAM, methylthioalkylmalate synthase; Col, Columbia ecotype; Ler, Landsberg erecta; QTL, quantitative trait locus.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ486882–AJ486953).
References
- 1.Rausher, M. D. (2001) Nature 411, 857-864. [DOI] [PubMed] [Google Scholar]
- 2.Halkier, B. A. (1999) Trends Plant Sci. 11, 425-431. [Google Scholar]
- 3.Rask, L., Andreasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B. & Meijer, J. (2000) Plant Mol. Biol. 42, 93-113. [PubMed] [Google Scholar]
- 4.Kliebenstein, D. J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J. & Mitchell-Olds, T. (2001) Plant Physiol. 126, 811-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reichelt, M., Brown, P. D., Schneider, B., Oldham, N. J., Stauber, E., Tokuhisa, J., Kliebenstein, D. J., Mitchell-Olds, T. & Gershenzon, J. (2002) Phytochemistry 59, 663-671. [DOI] [PubMed] [Google Scholar]
- 6.Campos de Quiros, H., Magrath, R., McCallum, D., Kroymann, J., Schnabelrauch, D., Mitchell-Olds, T. & Mithen, R. (2000) Theor. Appl. Genet. 101, 429-437. [Google Scholar]
- 7.Kroymann, J., Textor, S., Tokuhisa, J. G., Falk, K. L., Bertram, S., Gershenzon, J. & Mitchell-Olds, T. (2001) Plant Physiol. 127, 1077-1088. [PMC free article] [PubMed] [Google Scholar]
- 8.Shelton, A. M., Cooley, R. J., Kroening, M. K., Wilsey, W. T. & Eigenbrode, S. D. (1991) J. Entomol. Sci. 26, 17-26. [Google Scholar]
- 9.Frohman, M. A., Dush, M. K. & Martin, G. R. (1988) Proc. Natl. Acad. Sci. USA 85, 8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174-175. [DOI] [PubMed] [Google Scholar]
- 11.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 12.Van de Peer, Y. & De Wachter, R. (1997) Comput. Appl. Biosci. 13, 227-230. [DOI] [PubMed] [Google Scholar]
- 13.Tajima, F. & Nei, M. (1984) Mol. Biol. Evol. 1, 269-285. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. (1985) Evolution (Lawrence, Kans.) 39, 783-791. [DOI] [PubMed] [Google Scholar]
- 15.Kliebenstein, D. J., Lambrix, V. M., Reichelt, M., Gershenzon, J. & Mitchell-Olds, T. (2001) Plant Cell 13, 681-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kliebenstein, D. J., Pedersen, D., Barker, B. & Mitchell-Olds, T. (2002) Genetics 161, 325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister, C. & Dean, C. (1993) Plant J. 4, 745-750. [Google Scholar]
- 18.Tajima, F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald, J. H. & Kreitman, M. (1991) Nature 351, 652-654. [DOI] [PubMed] [Google Scholar]
- 20.Blau, P. A., Feeny, P., Contardo, L. & Robson, D. S. (1978) Science 200, 1296-1298. [DOI] [PubMed] [Google Scholar]
- 21.Raybold, A. F. & Moyes, C. L. (2001) Heredity 87, 383-391. [DOI] [PubMed] [Google Scholar]
- 22.Pivnick, K. A., Jarvis, B. J. & Slater, G. P. (1994) J. Chem. Ecol. 20, 1407-1427. [DOI] [PubMed] [Google Scholar]
- 23.Ratzka, A., Vogel, H., Kliebenstein, D. J., Mitchell-Olds, T. & Kroymann, J. (2002) Proc. Natl. Acad. Sci. USA 99, 11223-11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettencourt, B. R. & Feder, M. E. (2002) J. Mol. Evol. 54, 569-586. [DOI] [PubMed] [Google Scholar]
- 25.Hagenblad, J. & Nordborg, M. (2002) Genetics 161, 289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Corre, V., Roux, F. & Reboud, X. (2002) Mol. Biol. Evol. 19, 1261-1271. [DOI] [PubMed] [Google Scholar]
- 27.Lynch, M., O'Hely, M., Walsh, B. & Force, A. (2001) Genetics 159, 1789-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlesworth, B., Morgan, M. T. & Charlesworth, D. (1993) Genetics 134, 1289-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakeley, J. & Aliacar, N. (2001) Genetics 159, 893-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyre-Walker, A., Keightley, P. D., Smith N. G. & Gaffney, D. (2002) Mol. Biol. Evol. 19, 2142-2149. [DOI] [PubMed] [Google Scholar]
- 31.Haubold, B., Kroymann, J., Ratzka, A., Mitchell-Olds, T. & Wiehe, T. (2002) Genetics 161, 1269-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian, D., Araki, H., Stahl, E., Bergelson, J. & Kreitman, M. (2002) Proc. Natl. Acad. Sci. USA 99, 11525-11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawabe, A., Yamane, K. & Miyashita, N. T. (2000) Genetics 156, 1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen, K. M., Womack, A., Garrett, A. R., Suddith, J. I. & Purugganan, M. D. (2002) Genetics 160, 1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguadé, M. (2001) Mol. Biol. Evol. 18, 1-9. [DOI] [PubMed] [Google Scholar]
- 36.Hauser, M. T., Harr, B. & Schlötterer, C. (2001) Mol. Biol. Evol. 18, 1754-1763. [DOI] [PubMed] [Google Scholar]
- 37.Kawabe, A., Innan, H., Terauchi, R. & Miyashita, N. T. (1997) Mol. Biol. Evol. 14, 1303-1313. [DOI] [PubMed] [Google Scholar]
- 38.Kawabe, A. & Miyashita, N. T. (1999) Genetics 153, 1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuittinen, H. & Aguadé, M. (2000) Genetics 155, 863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nordborg, M. & Innan, H. (2002) Curr. Opin. Plant Biol. 5, 69-73. [DOI] [PubMed] [Google Scholar]
- 41.Bergelson, J., Kreitman, M., Stahl, E. A. & Tian, D. (2001) Science 292, 2281-2285. [DOI] [PubMed] [Google Scholar]
- 42.Mauricio, R. & Rausher, M. D. (1997) Evolution (Lawrence, Kans.) 51, 1435-1444. [DOI] [PubMed] [Google Scholar]
- 43.Purrington, C. B. (2000) Curr. Opin. Plant Biol. 3, 305-308. [DOI] [PubMed] [Google Scholar]
- 44.Tian, D., Traw, M. B., Chen, J. Q., Kreitman, M. & Bergelson, J. (2003) Nature 423, 74-77. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell-Olds, T. (1996) Evolution (Lawrence, Kans.) 50, 140-145. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell-Olds, T. & Bradley, R. D. (1996) Evolution (Lawrence, Kans.) 50, 1859-1865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.