Abstract
Hypoxia is known to favor tumor survival and progression. Numerous studies have shown that hypoxia-inducible factor 1α (HIF-1α), an oxygen-sensitive transcription factor, is overexpressed in various types of human cancers and upregulates a battery of hypoxia-responsive genes for the growth and survival of cancer cells. Although tumor progression involves the acquisition of genetic and/or epigenetic changes that confer additional malignant traits, the underlying mechanisms of these changes remain obscure. We recently identified an alternative mechanism of HIF-1α function by which HIF-1α suppresses DNA repair by counteracting c-Myc transcriptional activity that maintains gene expression. Here we demonstrate that this HIF-α–c-Myc pathway plays an essential role in mediating hypoxic effects on malignant progression via genetic alterations, resulting in formation of malignant tumors with aggressive local invasion and epithelial–mesenchymal transition. We show an absolute requirement of the HIF-α–c-Myc pathway for malignant progression, whereas the canonical transcription function of HIF-1α alone is insufficient and seemingly dispensable. This study indicates that HIF-1α induction of genetic alteration is the underlying cause of tumor progression especially by the hypoxic microenvironment.
Keywords: epithelial–mesenchymal transition, genetic alteration, hypoxia, tumorigenicity, tumor progression
Introduction
Hypoxia—deficiency in the amount of oxygen reaching the tissues—is frequently observed in human cancers. Although the accelerated proliferation of tumor cells contributes to oxygen deficiency by outstripping blood vessel growth, the aberrant vasculature with poor blood flow compounds the issue. As a result, cell death becomes increasingly conspicuous as the stage of the malignancy progresses. On the contrary, overwhelming evidence indicates hypoxia promotes tumor growth, progression, and resistance to therapies (1). Recent studies on the hypoxia-inducible factors, HIFs, have corroborated further the essential role of hypoxia in tumor growth and progression (2, 3).
HIFs are heterodimeric transcription factors consisting of HIF-α and ARNT (4, 5). Whereas both subunits are expressed constitutively at the transcriptional and translational levels, the oxygen-responsive HIF-α that is regulated through ubiquitin-proteasomal degradation has been the focus of intensive investigations (6). In particular, HIF-1α and HIF-2α (also known as EPAS1), the two best-studied members of the HIF-α family, harbor in the oxygen-dependent degradation domain two proline residues that are marked by hydroxylation for the recognition of E3 ubiquitin ligase (7-13). Hypoxic signaling stabilizes HIF-α by inhibiting prolyl hydroxylation and in turn ubiquitin-preteasomal degradation, rendering HIF-α capable of dimerizing with ARNT, binding to the hypoxia-responsive DNA element, and recruiting the transcription coactivator p300/CBP for transcriptional activation of a host of hypoxia-responsive genes (6). Both HIF-1α and HIF-2α are overexpressed frequently in human cancers, and this canonical HIF-α–ARNT pathway accounts for the upregulation of numerous cancer-relevant genes such as VEGFA, PGK1, and LOX for tumor angiogenesis, glycolysis, and metastasis, respectively (2, 14, 15).
Tumor initiation and progression require genetic alterations (16). Both tumor cells and their hypoxic microenvironment exert selective pressure for gene mutations and genetic instability (17, 18). We recently reported that HIF-1α, but not HIF-2α, is responsible for inhibiting DNA repair, leading to genetic instability (19, 20). However, neither HIF-1α DNA binding nor transactivation domain is required for gene repression; rather it is the subregion of HIF-1α's PAS domain, PAS-B, that is both necessary and sufficient to inhibit DNA repair by counteracting c-Myc transcriptional activity that maintains gene expression (21). Although the identification of this HIF-1α–c-Myc pathway has begun to shed light on the role of the hypoxic response in genetic alteration, whether such mechanism is responsible for malignant progression remains to be demonstrated. In this study, we show that the HIF-1α–c-Myc pathway is not only essential to the acquisition of malignant traits by tumor cells but also functions independent of the HIF-α–ARNT pathway. By contrast, the canonical HIF-α–ARNT pathway alone is insufficient to confer malignant traits.
Material and Methods
Cell culture and hypoxic treatment
Human osteosarcoma cell line U-2 OS (HTB-96) and U-118 MG (HTB-15) were obtained from American Type Culture Collection (ATCC). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum at 37°C in a 5% CO2 incubator. For the treatment with short-term hypoxia, cells were incubated in a hypoxic chamber (Innova CO-48, New Brunswick Scientific) maintaining 1% O2 and 5% CO2 overnight. Long-term hypoxia was carried out for 2 weeks in a cycle of 16-h 1% O2 followed by 8-h 21% O2. The choosing of repetitive hypoxia treatment was to ensure sustainable HIF-1α levels that otherwise might have been markedly attenuated during prolonged hypoxic treatment. These cells were then cultured in normal conditions for another 2-4 weeks before further analysis.
Retroviral transduction
An oxygen-insensitive HIF-1α [HIF1α(PP)] (22) with P402A and P564A substitutions was cloned into the retroviral expression vector pBABE-neo (Addgene). Additional mutations were made by site-directed mutagenesis (23) to create HIF1α(PP) mutants VAT [V317L-A321G-T327P (20)] and RFC [R27G (23)-F99L (24)-C800S (25)]. The mutagenic oligonucleotide sequences are listed in Supplemental Table S2. HIF-1α PAS-B and its VAT mutant (20) were fused to a yellow fluorescent protein in the pEYFP-Nuc (Clontech), and the fusion proteins were then cloned into pBABE-neo. Recombinant retroviral particles were generated from the PT67 packaging cell line (ATCC) and harvested for infection with 2–4 consecutive times. Transduced cells were selected with 500 μg/ml of G418 (Sigma) for 2–3 weeks and pooled for further analysis. A short-hairpin RNA targeting HIF1A was based on HIF1A coding sequence 5′-CGTTGTGAGTGGTATTATTCAGCACGACT-3′ and cloned into pSilencer5.1-H1 Retro vector (Ambion). Transduced cells were pooled after selection with 2 μg/ml of puromycin.
Immunofluorescence
Immunofluorescent staining was performed essentially as previously described (20). For the detection of DNA double-strand breaks, cells were incubated with antibodies against γ-H2AX (Millipore) and 53BP1 (Cell Signaling). Images were obtained with a fluorescent and laser-scanning confocal Olympus IX81 microscope. For the detection of epithelial–mesenchymal transition, primary antibodies against β-catenin (Cell Signaling) and E-cadherin and fibronectin (BD Biosciences) were used. For the detection of ZEB2 expression, primary antibodies against SIP1/ZEB2 (Santa Cruz Biotechnology) were used. Secondary antibodies include Texas Red-X goat anti-rabbit (T-6391) and anti-mouse IgG (T-6390), Alexa Fluor 488 anti-rabbit IgG (A-11034), and Marina Blue goat anti-rabbit IgG (M-10992) (Invitrogen). Fluorescent microscopy was performed with an Axiovert 200 fluorescence inverted microscope (Carl Zeiss MicroImaging). Representative images are presented from at least three independent experiments with similar results.
Polymerase chain reaction (PCR)
Conventional reverse transcription (RT)-PCR was performed with primer sequences tabulated in Supplemental Table 2. Genomic DNA was extracted with a DNeasy Blood & Tissue kit (Qiagen) for PCR amplification with primer sequence listed in Supplemental Table 3. For real-time PCR, cDNA was amplified by using the Taqman Universal Polymerase Chain Reaction Master Mix (Applied Biosystems). Human CA9 (6FAM), PGK1 (6FAM), VEGFA (6FAM), MSH2 (6FAM), NBS1 (6FAM), and human ACTB endogenous control (VIC) were purchased from Applied Biosystems.
The Human Cancer PathwayFinder PCR Array (PAHS-033, SuperArray Bioscience) was used for PCR array analysis according to the manufacturer's protocol. Gene-specific real-time PCR products were measured continuously by an ABI PRISM HT7900 Sequence Detection System (Applied Biosystems) during 40 cycles.
Tumorigenesis
Male CD-1 nude mice (up to 42 days old) from Charles River Laboratory were used for bilateral, subcutaneous injections. One million cells in 100 μl of PBS were used. Tumor volume was monitored every 3–4 days beginning 17 days after injection. Mice were euthanized on day 40 according to the protocol approved by the University of Utah Institutional Animal Care and Use Committee. Hematoxylin-eosin staining was performed by the ARUP National Reference Laboratory. Images were examined and photographed with an Axiovert 200 inverted microscope.
Immunoblot
Immunoblot was performed as previously described (26). Specific antibodies include HIF-1α, fibronectin, E-cadherin, and ZEB2 of Santa Cruz Biotechnology; MSH2 of BD Biosciences; NBS1 and β-catenin of Cell Signaling Technology; phospho-histone H2A.X (γ-H2AX) (S139) of Millipore; and β-actin of Sigma. Images were obtained with the Kodak Image Station 4000R. A representative of at least three independent experiments with similar results is shown.
Anchorage-independent growth
Cells (1 × 104) were seeded in six-well plates with a bottom layer of 0.5% Bacto agar in DMEM and a top layer of 0.35% Bacto agar in DMEM. Colonies were counted after two weeks and photographed using a Kodak Image Station 4000R. The total number of colonies per well was determined using Kodak Molecular Imaging software. Results in 6 replicates were presented as mean ± SEM.
Cell proliferation and Matrigel invasion
Cell proliferation was assayed in 96-well plates with the CellTiter-Glo® Luminescent Cell Viability Assay (Promega). At each time point, cell viability in 6 replicates was recorded with an FLx800 microplate fluorescence reader (Bio-Tek) and presented as mean ± SEM. Cell invasion was determined in triplicates with cell suspensions added to the BD BioCoat™ Matrigel™ Invasion Chambers (BD Biosciences) and incubated for 24 h in a humidified tissue culture incubator. Noninvading cells were removed by scrubbing with a cotton-tipped swab. The total number of invading cells was determined according to the manufacturer's protocol and presented as mean ± SEM.
RNA interference
Cells were seeded in 6-well plates and transfected with siRNA duplexes targeting ZEB2 (5′-ACCAUGAAUAGUAAUUUAATT-3′ and 5′-UUAAAUUACUAUUCAUGGUGG -3′) (Qiagen) and control si-GL2 with the Oligofectamine™ reagent (Invitrogen) according to the manufacturer's protocol.
Microsatellite instability
Genomic DNA was isolated from transduced cells and their tumors with a DNeasy Blood & Tissue Kit (Qiagen). Microsatellite instability was analyzed with a panel of 5 markers (BAT25, BAT26, D2S123, D5S346, and D17S250) at the University of Utah Research Core Facility.
Results
Independent functions of the HIF-1α–ARNT pathway and the HIF-1α–c-Myc pathway
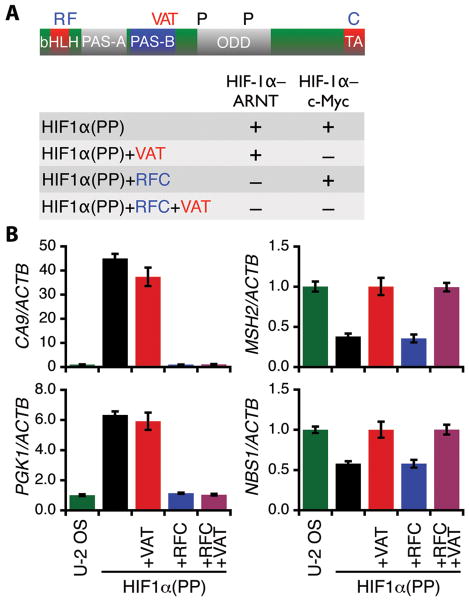
The HIF-1α function via the HIF-1α–ARNT pathway has been a prevalent theory of tumor growth and development by which a large group of hypoxia-responsive genes are transcriptionally upregulated. To demonstrate a critical role of the HIF-1α–c-Myc pathway in cancer biology, we sought to uncouple the activity of the HIF-1α–c-Myc pathway from that of the HIF-1α–ARNT pathway and tested each pathway individually in malignant progression. Hence, the functions of the HIF-1α–ARNT pathway and the HIF-1α–c-Myc pathway were inactivated respectively by site-directed mutagenesis in the context of a stabilized HIF-1α, designated as HIF1α(PP) (Fig. 1A), in which two prolyl hydroxylation sites (P402 and P564) were destroyed to prevent oxygen-dependent proteolysis (11, 12).
Figure 1.
Uncoupling of the HIF-1α–c-Myc and the HIF-1α–ARNT pathways. A, a schematic representation of HIF-1α with functional domains as indicated. bHLH, basic helix-loop-helix; PAS, Per-ARNT-Sim; ODD, oxygen-dependent degradation; TA, transactivation. The HIF-1α–ARNT and the HIF-1α–c-Myc pathways were inactivated (–) individually by site-directed mutagenesis to create the mutants as indicated. B, transduced U-2 OS cells expressing HIF-1α variants as specified were assayed for target gene expression of the HIF-1α–ARNT pathway (left) and the HIF-1α–c-Myc pathway (right) by real-time PCR. Results are representative of two independent experiments in triplicate and presented as mean ± SD.
Subsequently, the HIF-1α–c-Myc pathway was inactivated through simultaneous substitutions of HIF-1α V317, A321, and T327 in the PAS-B (20), yielding the mutant HIF1α(PP)+VAT. Likewise, the HIF-1α–ARNT pathway was mutated at R27 (23), F99 (24), and C800 (25) to create HIF1α(PP)+RFC, a mutant deficient in DNA binding and p300/CBP recruitment. Furthermore, a combinatorial mutant HIF1α(PP)+RFC+VAT was created to inactivate both functions. These HIF-1α mutants were then expressed in various types of tumor cells by retroviral transduction and selection.
As expected, HIF1α(PP) was able to upregulate CA9 and PGK1, two classic targets of the HIF-1α–ARNT pathway (Fig. 1B, left), and downregulate the DNA repair genes MSH2 and NBS1 of the the HIF-1α–c-Myc pathway (Fig. 1B, right; and Supplemental Fig S1, A and B). Likewise, HIF1α(PP)+RFC was unable to activate the HIF-1α–ARNT pathway, yet it remained capable of exerting its activity in the HIF-1α–c-Myc pathway. Furthermore, the mutant failed to activate a HIF-1α–mediated reporter gene that requires DNA binding and p300/CBP recruitment (Supplemental Fig. S1C). By contrast, HIF1α(PP)+VAT was inactive in the HIF-1α–c-Myc pathway but was active in the HIF-1α–ARNT pathway. It is noteworthy that c-Myc protein (Supplemental Fig. S1D) and mRNA (Supplemental Table S1) levels were markedly decreased in the transduced cells with an intact HIF-1α–c-Myc pathway, consistent with HIF-1α counteraction of c-Myc (27, 28). Hence, these HIF-1α mutants by design function independently via the HIF-1α–ARNT pathway and the HIF-1α–c-Myc pathway.
DNA damage and genetic alterations via the HIF-1α–c-Myc pathway
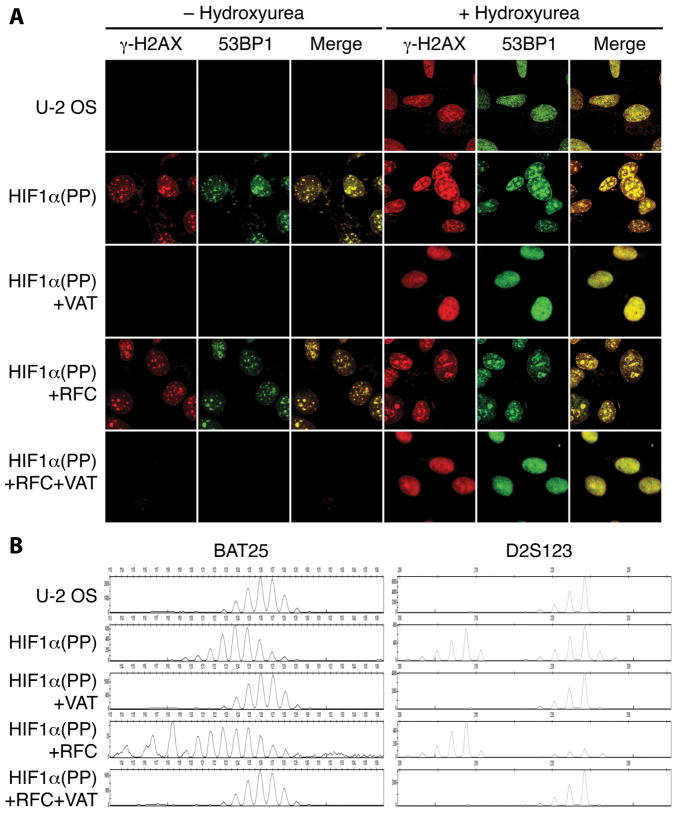
The suppression of DNA repair genes by HIF1α(PP) and HIF1α(PP)+RFC was correlated with conspicuous DNA double-strand breaks, as indicated by a striking increase in γ-H2AX foci, which were colocalized with those of 53BP1 (20) (Fig. 2A). Elevated levels of γ-H2AX and 53BP1 were confirmed by immunoblot analyses (Supplemental Fig. S2A). By contrast, mutants defective in the HIF-1α–c-Myc pathway—HIF1α(PP)+VAT and HIF1α(PP)+RFC+VAT—failed to produce noticeable DNA damage. All of these cells responded to hydroxyurea-induced DNA damage. Similar results were obtained with various tumor cell lines, such as the human glioblastoma cell U-118 MG and the mouse hepatoma cell Hepa 1-6 (data not shown).
Figure 2.
Induction of cumulative DNA damage and microsatellite instability via the HIF-1α–c-Myc pathway. A, DNA double-strand breaks were determined by immunofluorescence for colocalization (Merge) of increased γ-H2AX foci with those of 53BP1 in the transduced cells as indicated. Cells treated with 20 mM hydroxyurea for 24 h are indicated. B, the same transduced cells were analyzed for microsatellite instability with the markers BAT25 and D2S123.
Furthermore, increased microsatellite instability was detected in cells with accumulated DNA damage but not in those without (Fig. 2B and Supplemental Fig. S2B). Chromosomal fragile site instability was also detected in the two most active sites FRA3B and FRA16D, which are harbored within the genomic loci of the tumor suppressor genes FHIT and WWOX, respectively (29), as shown by the loss of FHIT and WWOX expression at mRNA levels (Supplemental Fig. S3A). The lack of gene expression correlated with loss of the exons at the gene locus (Supplemental Fig. 3B) as previously reported (30, 31). In addition, the tumor suppressor genes RB1 and TP53 were markedly suppressed from cells manifesting chronic DNA damage even though no mutations were detected in the coding regions by DNA sequencing (Supplemental Table S1; data not shown). Therefore, the HIF-1α–c-Myc pathway inhibits tumor-suppressing activities, at least in part, via genetic alterations.
Gain of aggressive malignant traits requires the HIF-1α–c-Myc pathway
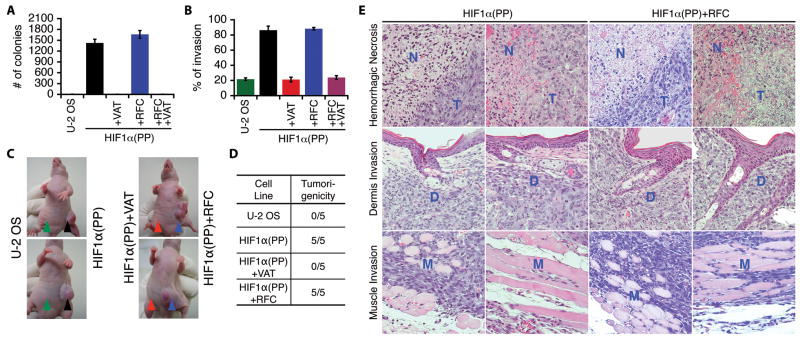
In accordance with diminished tumor-suppressing activities, both HIF1α(PP) and HIF1α(PP)+RFC cells exhibited a remarkable gain of anchorage-independent growth (Fig. 3A and Supplemental Fig. S3C). They also showed a marked increase in cell proliferation and Matrigel invasion, whereas inactivation of the HIF-1α–c-Myc pathway abrogated such increase (Fig. 3B and Supplemental Fig. S3D). Importantly, these cells, in contrast to the parental and HIF1α(PP)+VAT cells, all acquired tumorigenicity by forming rapidly growing tumors within 3 weeks in subcutaneous xenograft mouse models (Fig. 3, C and D). These tumors were highly aggressive in histology, exhibiting massive hemorrhagic necrosis and invasion of neighboring tissues including the dermal layers and skeletal muscles (Fig. 3E).
Figure 3.
Requirement of the HIF-1α–c-Myc pathway for malignant progression and tumorigenicity. A–D, transduced U-2 OS cells as indicated were assayed for anchorage-independent growth (A), Matrigel invasion (B), and tumorigenicity with subcutaneous xenograft in CD-1 nude mice (C). The anchorage-independent colonies and Matrigel invading cells were quantified and presented as mean ± SEM. The injection sites are marked by arrowheads. Summarized result is in (D) with a ratio of 5 mice per group. E, hematoxylin–eosin staining of tumor specimens (T) shows hemorrhagic necrosis (N) and invasion of dermal layers (D) and skeletal muscles (M). Two representative images with 200 × magnification are shown.
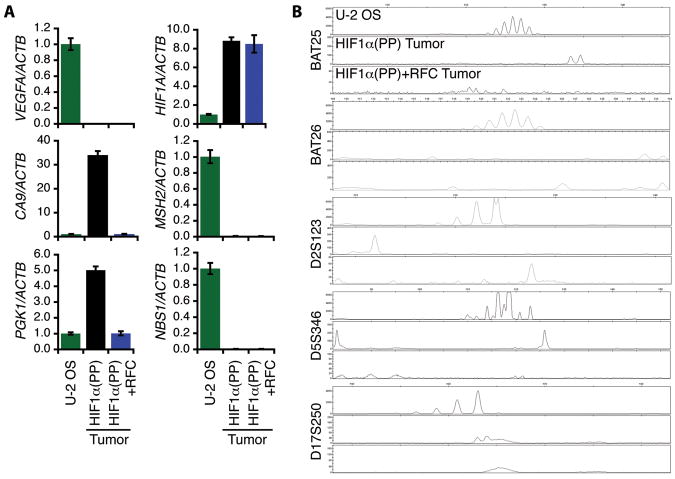
We also performed ex vivo characterization of the HIF1α(PP) and HIF1α(PP)+RFC tumor specimens by analyzing target gene expression of the HIF-1α–ARNT pathway and the HIF-1α–c-Myc pathway in reference to the parental cells. In agreement with the transduced cells, these tumor specimens displayed a similar gene expression pattern, as shown by real-time PCR (Fig. 4A). Interestingly, the angiogenic gene VEGFA was not increased in either type of the tumor specimens and transduced cells (Supplemental Table S1). On the other hand, the tumor specimens showed a further increase in microsatellite instability (Fig. 4B) with reference to the original transduced cells (Fig. 2B), suggesting additional genetic alterations by tumor hypoxia.
Figure 4.
Ex vivo characterization of tumor specimens derived from HIF1α(PP) and HIF1α(PP)+RFC cells. A, total RNA was extracted from the tumors and gene expression as indicated was quantified by real-time PCR in reference to the parental U-2 OS cells. Results are in triplicate and presented as mean ± SD. B, genomic DNA was isolated from the tumors and subjected to microsatellite instability analyses with the 5-marker panel as indicated.
To corroborate the obligatory role of the HIF-1α–c-Myc pathway in acquiring malignant traits, we took advantage of an isolated HIF-1α PAS-B (referred to as PAS1B hereafter), which has been shown previously to be sufficient to activate the HIF-1α–c-Myc pathway (20). We showed that PAS1B but not its mutant VAT yielded identical results as HIF1α(PP) by inducing DNA damage and acquiring malignant traits (Fig. 5, A–C, and Supplemental Fig. S4, A and B). Furthermore, expression of PAS1B in U-2 OS cells was sufficient to confer tumorigenicity in 10 out of 10 nude mice (Supplemental Fig. S4C), and the derived tumors were morphologically as malignant and invasive as those derived from HIF1α(PP) and HIF1α(PP)+RFC cells (Fig. 5D). Likewise, PAS1B expression in U-118 MG cells markedly accelerated tumor growth, leading to the invasion of the mouse dermal layer (Supplemental Fig. S4D). By contrast, the parental U-118 MG and control cells formed only tiny, circumscribed tumors confined within the subcutaneous layer.
Figure 5.
HIF-1α PAS-B is sufficient to confer malignant traits. A, transduced U-2 OS cells expressing EYFP and an EYFP fusion to HIF-1α PAS-B (PAS1B) or to the mutant (VAT) were analyzed for DNA double-strand breaks by immunofluorescent staining for increased γ-H2AX foci that were colocalized with those of 53BP1. Cells were also treated with the DNA-damaging agent doxorubicin (0.5 μM) for 24 h as control. B, transduced cells as above were assayed for anchorage-independent growth. The total number of colonies in 6 replicates was quantified and presented as mean ± SEM. C, Matrigel invasion of transduced cells was assayed in triplicate and presented as mean ± SEM. D, hemorrhagic necrosis (N) and invasion of dermis (D) and skeletal muscle (M) were shown with tumor (T) specimens derived from U-2 OS cells expressing PAS1B. Two representative images are shown in hematoxylin-eosin staining with 200 × magnification.
To ensure the effect of HIF-1α on malignant progression is physiologically relevant, we subjected U-2 OS cells to 1% O2 treatment for different periods. Although a short-term (overnight) hypoxic treatment with or without days of recovery in normoxia failed to induce tumorigenicity, long-term hypoxia (see Materials and Methods) converted the parental cells to 100% tumorigenicity (Supplemental Fig. S5A). However, knockdown of HIF-1α expression (not shown) prior to long-term hypoxia abrogated the tumorigenicity, indicating an essential role of HIF-1α in mediating the hypoxic effect. Moreover, the derived tumors exhibited high degree of malignancy, resembling those of HIF1α(PP) and HIF1α(PP)+RFC cells (Supplemental Fig. S5B).
Induction of ZEB2 expression by HIF-1α for epithelial–mesenchymal transition
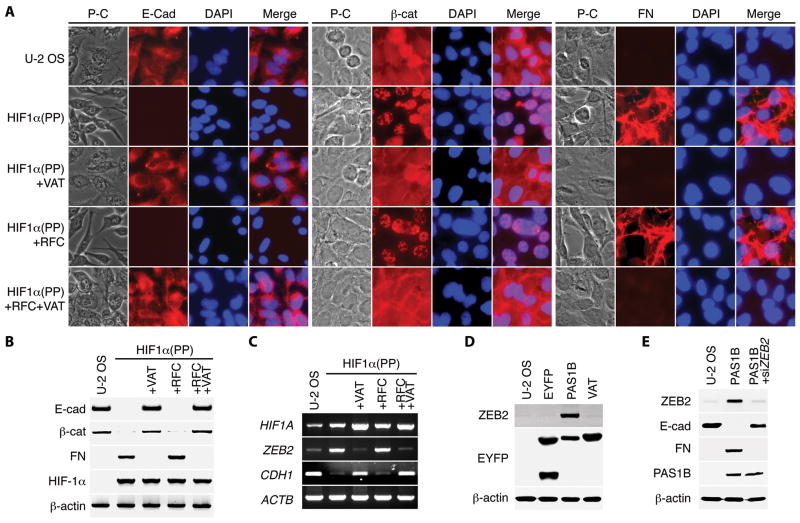
A salient feature observed among the acquired malignant traits was epithelial–mesenchymal transition (EMT), which is characterized by loss of epithelial markers and gain of mesenchymal markers (32, 33). Although the parental U-2 OS cells expressed the epithelial marker E-cadherin, none was detected in cells with an active HIF-1α–c-Myc pathway (Fig. 6, A and B, and Supplemental Fig. S6). Another epithelial marker, β-catenin, became distinctly nuclear in these transduced cells. Furthermore, these cells gained expression of the mesenchymal marker fibronectin. It is worthy to note that no EMT was observed in HIF1α(PP)+VAT cells that were active in the HIF-1α–ARNT pathway, even though several recent studies indicate an important role for HIF-α in EMT (34-37).
Figure 6.
Upregulation of ZEB2 for epithelial–mesenchymal transition by the HIF-1α–c-Myc pathway. A and B, transduced U-2 OS cells as indicated were analyzed for the expression of E-cadherin (E-cad), β-catenin (β-cat), and fibronectin (FN) by immunofluorescent staining (A) and immunoblotting (B). Cell nuclei were visualized with DAPI. P-C, phase-contrast microscopy. C, ZEB2 and CDH1 mRNA levels were determined with conventional RT-PCR in transduced U-2 OS cells as above. D, transduced U-2 OS cells expressing EYFP and an EYFP fusion to HIF-1α PAS-B (PAS1B) or the mutant (VAT) were assayed for ZEB2 expression by immunoblotting. E, transduced cells expressing PAS1B were transfected with small-interfering RNA targeting ZEB2 (siZEB2) and then assayed for specific gene expression with immunoblotting.
Transcriptional repression of CDH1 gene, which encodes E-cadherin, is key to EMT induction (33). Among a panel of candidate CDH1 transcriptional repressors examined, ZEB2/SIP1 (encoding the zinc finger E-box binding homeobox 2) (38) was the one that was markedly upregulated by the HIF-1α–c-Myc pathway (Fig. 6, C and D, and Supplemental Fig. S7A). Knockdown of ZEB2 gene increased E-cadherin but decreased fibronectin levels in these cells (Fig. 6E). However, only partial reversal of EMT was observed in the knockdown cells with immunofluorescent staining (Supplemental Fig. S8), indicating the involvement of additional CDH1 transcriptional repressors, such as Twist1 (35) (Supplemental Table 1). Nevertheless, introduction of the ZEB2 gene in U-2 OS cells was sufficient to decrease E-cadherin while increasing fibronectin levels and to induce EMT (Supplemental Fig. S7, B and C). Taken together, these results indicate that ZEB2 upregulation by HIF-1α is mediated by the HIF-1α–c-Myc pathway and is responsible for maintaining a mesenchymal phenotype of malignant cells.
Discussion
Tumor growth, as judged by size, volume, and weight, has been the gold standard in experimental cancer research. However, such quantitative change often substitutes for tumor progression, a qualitative measure denoting cumulative gain of malignant traits (18). In this study, we have provided a cogent argument for the role of HIF-1α via the HIF-1α–c-Myc pathway in malignant progression; activation of the HIF-1α–c-Myc pathway confers aggressive malignant traits on tumor cells. In particular, we have shown that the HIF-1α–c-Myc pathway is not only necessary to induce rapid tumor formation but more importantly is sufficient to promote malignant behavior including aggressive local invasion. Notably, expression of HIF-1α PAS-B recapitulates the effects of a full-length HIF-1α in this regard. By contrast, activation of the HIF-1α–ARNT pathway is insufficient to do so in the absence of the HIF-1α–c-Myc pathway. Hence, we define the HIF-1α–c-Myc pathway as an underlying mechanism of malignant progression, which is functionally independent of the HIF-1α–ARNT pathway.
The interplay between HIF-1α and c-Myc was first identified as HIF-1α functional counteraction of c-Myc (19, 23, 39), yet additional transcriptional and post-translational mechanisms for c-Myc suppression were also reported for hypoxic adaptation by tumor cells (27, 28). Furthermore, c-Myc protein levels were reduced for survival when tumor cells were located distant from the blood vessels or cultured under low oxygen and glucose-deficient conditions (40). Therefore, lowered c-Myc activity by hypoxia favors tumor survival. However, in Myc-dysregulated tumors HIF-1α has been shown to cooperate with c-Myc to induce angiogenic and glycolytic genes in an inducible MYC lymphoma model (41) and with N-Myc for enhanced aerobic glycolysis in neuroblastomas (42). Therefore, how HIF-1α and Myc interacts seems context dependent (21).
It seems that the HIF-1α–c-Myc pathway is unique in that substitution of HIF-1α V317, A321, and T327 with the corresponding residues of HIF-2α obliterates HIF-1α's ability to confer malignant traits regardless of the status of HIF-1α–ARNT pathway. In keeping with this, HIF-2α does not inhibit DNA repair (20); rather, it stimulates DNA repair and promotes genomic stability in renal clear-cell carcinoma cells (43). In this tumor type, a mutually suppressive interaction between HIF-1α and HIF-2α has been reported along with contrasting properties that HIF-1α retards while HIF-2α enhances tumor growth (44). Moreover, HIF-2α has been shown to cooperate with c-Myc in increasing cell proliferation (39), to override tumor suppression in renal clear-cell carcinomas (45), and to promote an aggressive phenotype in neuroblastomas (46). These findings have led to the notion that HIF-2α is a critical mediator of aggressive phenotypes at least in clear-cell renal carcinoma, neuroblastoma, and non-small cell lung cancer (47). However, an in-depth study of neuroblastomas shows recently that HIF-1α, but not HIF-2α, is preferentially expressed in tumors with poor prognosis (42). In addition, genetic evidence supports a crucial role for HIF-1α in the development of cardiac rhabdomyosarcoma and metastasis (48). Although cell-specific expression of HIF-1α and HIF-2α may explain their apparent discrepancies in tumor progression, further understanding of the mechanisms by which HIF-2α promotes tumor progression is warranted.
Both HIF-1α and HIF-2α have been attributed to the hypoxic induction of EMT, and the HIF-α–ARNT pathway has been shown or implicated for the transcriptional upregulation of CDH1 transcriptional repressors including Twist1, Slug, and ZEB2 (35-37). It remains to be demonstrated, however, that the HIF-α–ARNT pathway is required for ZEB2 regulation (36, 37). Our results indicate that the HIF-1α–c-Myc pathway is essential to the induction of ZEB2 and in turn EMT. This is further substantiated by the sufficiency of HIF-1α PAS-B in this regard. Although how the HIF-1α–c-Myc pathway upregulates ZEB2 expression is yet to be uncovered, recent studies indicate micro RNA (miR-200) plays a key role in regulating ZEB2 expression (49, 50). Therefore, it will be of great interest to determine whether the HIF-1α–c-Myc pathway affects the miR-200s that target ZEB2 transcripts.
The important role of HIF-1α via the HIF-1α–ARNT pathway has been well documented for its essential role in tumor growth by enhancing glycolysis and angiogenesis (2, 3). Although in this study activation of the HIF-1α–ARNT pathway was insufficient and rather seemingly dispensable for conferring malignant traits, tumor growth was retarded from HIF1α(PP) cells that had been depleted of HIF-1α afterwards (data not shown), further supporting the role of HIF-1α in tumor growth. Notably, all the tumor cells used in this study expressed endogenous HIF-1α, which was labile in normoxic culture but presumably activated by tumor hypoxia. In addition, the surrounding mouse tissue consisting of a Hif1a-wild-type microenvironment might have also supported tumor growth. It would be interesting to use a HIF-1α-deficient background of tumor cells and/or of the host mouse to further dissect the individual roles of the HIF-1α–ARNT pathway and the HIF-1α–c-Myc pathway, even though the results may not be straightforward because of the feed-forward relationship between tumor growth and malignant progression via the hypoxic microenvironment (18). In light of previous studies (2, 3), we propose that although HIF-1α plays an important role via the HIF-1α–ARNT pathway in cell proliferation and metabolic adaptation of tumors, the HIF-1α–c-Myc pathway is the underlying mechanism of malignant progression that drives tumors to advanced stages.
Supplementary Material
Acknowledgments
Grant support: Public Health Service grants CA-084563 and CA-131355 from the National Cancer Institute (L.E. Huang).
We thank Kristin Kraus for editorial assistance.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Brown JM. Exploiting the hypoxic cancer cell: mechanisms and therapeutic strategies. Mol Med Today. 2000;6:157–62. doi: 10.1016/s1357-4310(00)01677-4. [DOI] [PubMed] [Google Scholar]
- 2.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giaccia A, Siim BG, Johnson RS. HIF-1 as a target for drug development. Nat Rev Drug Discov. 2003;2:803–11. doi: 10.1038/nrd1199. [DOI] [PubMed] [Google Scholar]
- 4.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 6.Huang LE, Bunn HF. Hypoxia-inducible factor and its biomedical relevance. J Biol Chem. 2003;278:19575–8. doi: 10.1074/jbc.R200030200. [DOI] [PubMed] [Google Scholar]
- 7.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2- dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95:7987–92. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallio PJ, Wilson WJ, O'Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem. 1999;274:6519–25. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 10.Ohh M, Park CW, Ivan M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 11.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 12.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 13.Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci U S A. 2001;98:9630–5. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erler JT, Bennewith KL, Nicolau M, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 15.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 17.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–92. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 18.Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability-a calculated mechanism underlying tumor progression. J Mol Med. 2007;85:139–48. doi: 10.1007/s00109-006-0133-6. [DOI] [PubMed] [Google Scholar]
- 19.Koshiji M, To KK, Hammer S, et al. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 20.To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. Embo J. 2006;25:4784–94. doi: 10.1038/sj.emboj.7601369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang LE. Carrot and stick: HIF-alpha engages c-Myc in hypoxic adaptation. Cell Death Differ. 2008;15:672–7. doi: 10.1038/sj.cdd.4402302. [DOI] [PubMed] [Google Scholar]
- 22.Kageyama Y, Koshiji M, To KK, Tian YM, Ratcliffe PJ, Huang LE. Leu-574 of human HIF-1alpha is a molecular determinant of prolyl hydroxylation. Faseb J. 2004;18:1028–30. doi: 10.1096/fj.03-1233fje. [DOI] [PubMed] [Google Scholar]
- 23.Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. Embo J. 2004;23:1949–56. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Land SC, Tee AR. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J Biol Chem. 2007;282:20534–43. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- 25.Gu J, Milligan J, Huang LE. Molecular mechanism of hypoxia-inducible factor 1alpha -p300 interaction. A leucine-rich interface regulated by a single cysteine. J Biol Chem. 2001;276:3550–4. doi: 10.1074/jbc.M009522200. [DOI] [PubMed] [Google Scholar]
- 26.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. Embo J. 2006;25:1231–41. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Gao P, Fukuda R, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Kaidi A, Williams AC, Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9:210–7. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- 29.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 30.Hinohara S, Satake N, Sekine K, Kaneko Y. Abnormalities of the FHIT transcripts in osteosarcoma and Ewing sarcoma. Jpn J Cancer Res. 1998;89:887–94. doi: 10.1111/j.1349-7006.1998.tb00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners, and functions. J Cell Biochem. 2009;108:737–45. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- 32.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 33.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 34.Esteban MA, Tran MG, Harten SK, et al. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006;66:3567–75. doi: 10.1158/0008-5472.CAN-05-2670. [DOI] [PubMed] [Google Scholar]
- 35.Yang MH, Wu MZ, Chiou SH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 36.Krishnamachary B, Zagzag D, Nagasawa H, et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 2006;66:2725–31. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
- 37.Evans AJ, Russell RC, Roche O, et al. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol Cell Biol. 2007;27:157–69. doi: 10.1128/MCB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comijn J, Berx G, Vermassen P, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 39.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–47. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okuyama H, Endo H, Akashika T, Kato K, Inoue M. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer Res. doi: 10.1158/0008-5472.CAN-10-2720. in press. [DOI] [PubMed] [Google Scholar]
- 41.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. HIF-1 and dysregulated c-Myc cooperatively induces VEGF and metabolic switches, HK2 and PDK1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qing G, Skuli N, Mayes PA, et al. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor (HIF)-1alpha. Cancer Res. 2010;70:10351–61. doi: 10.1158/0008-5472.CAN-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordan JD, Lal P, Dondeti VR, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–46. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raval RR, Lau KW, Tran MG, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–86. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG. Inhibition of HIF is necessary for tumor suppression by the von Hippel- Lindau protein. Cancer Cell. 2002;1:237–46. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 46.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–23. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 47.Qing G, Simon MC. Hypoxia inducible factor-2alpha: a critical mediator of aggressive tumor phenotypes. Curr Opin Genet Dev. 2009;19:60–6. doi: 10.1016/j.gde.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei L, Mason S, Liu D, et al. Hypoxia-inducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel-Lindau protein. Mol Cell Biol. 2008;28:3790–803. doi: 10.1128/MCB.01580-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 50.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.