Abstract
The acquisition and incorporation of genetic material between nonmating species, or horizontal gene transfer (HGT), has been frequently described for phylogenetically related organisms, but far less evidence exists for HGT between highly divergent organisms. Here we report the identification and characterization of a horizontally transferred fragment of the human long interspersed nuclear element L1 to the genome of the strictly human pathogen Neisseria gonorrhoeae. A 685-bp sequence exhibiting 98 to 100% identity to copies of the human L1 element was identified adjacent to the irg4 gene in some N. gonorrhoeae genomes. The L1 fragment was observed in ~11% of the N. gonorrhoeae population sampled but was not detected in Neisseria meningitidis or commensal Neisseria isolates. In addition, N. gonorrhoeae transcripts containing the L1 sequence were detected by reverse transcription-PCR, indicating that an L1-derived gene product may be produced. The high degree of identity between human and gonococcal L1 sequences, together with the absence of L1 sequences from related Neisseria species, indicates that this HGT event occurred relatively recently in evolutionary history. The identification of L1 sequences in N. gonorrhoeae demonstrates that HGT can occur between a mammalian host and a resident bacterium, which has important implications for the coevolution of both humans and their associated microorganisms.
IMPORTANCE
The interactions between microbes and their hosts are relevant to several aspects of biology, including evolution, development, immunity, and disease. Neisseria gonorrhoeae serves as a particularly informative model for this interaction because it has exclusively coevolved with humans and is not known to be found in any other environment. In addition, investigation of the evolutionary relationship between N. gonorrhoeae and humans has practical implications, since gonorrhea is a prevalent sexually transmitted infection worldwide. This study was undertaken to characterize the horizontal transfer of genetic information from humans to N. gonorrhoeae, an event that has been scarcely recognized between any mammalian host and bacterial pathogen. Here we provide evidence that this genetic exchange was the result of a recent evolutionary event that has been propagated within the gonococcal population.
INTRODUCTION
Evolution of asexual organisms can occur by the accumulation and vertical transmission of mutations in an existing genetic repertoire or through the acquisition of novel genetic determinants by horizontal gene transfer (HGT). HGT has the capacity to facilitate the acquisition of complex physiological functions (1) in a single molecular event and has been estimated to account for up to 17% of the total genetic material found in Escherichia coli (2). HGT must therefore be regarded as an important force in the adaptation of species. HGT is particularly well known to have a diverse impact on bacterial physiology, with examples in the literature describing the horizontal acquisition of novel biosynthetic (3), virulence (1), and antibiotic resistance (4) functions by diverse bacterial species. Despite the countless interactions between commensal or pathogenic bacteria and their cognate hosts and the ever-increasing amount of available genomic sequence data, examples of bacterial integration of host genetic information are exceedingly rare. The rarity of HGT from host to bacterium may be due, in part, to potential barriers such as restriction and modification functions, the availability of genetic material, and the fitness cost of deleterious HGT events. However, such an event has the potential to impact the evolution of both the host and the microbe.
Neisseria gonorrhoeae is the causative agent of the sexually transmitted infection gonorrhea. N. gonorrhoeae is naturally competent for transformation and has incorporated DNA from related neisserial species (5, 6) and more divergent bacterial species (7) into its genome. Recombination of exogenous DNA occurs efficiently in N. gonorrhoeae, and it is postulated that this process plays an important role in the microevolution of the organism (8, 9). N. gonorrhoeae has a long and exclusive evolutionary history with its human host, and infection is typically localized to the urogenital tract. Therefore, sources of foreign DNA available to the gonococcus are limited to coinhabiting microorganisms or the host itself. Mammalian genomes contain numerous copies of the long interspersed nuclear element (LINE) L1. Full-length human L1 elements encode a nucleic acid binding protein and a multidomain protein with endonuclease and reverse transcriptase activities (10), which together facilitate mobilization of the element via retrotransposition (11). This work establishes the presence of N. gonorrhoeae genome sequences with strong identity to human LINE L1 (10) and characterizes the results of this HGT event.
RESULTS
The genome sequences of 14 N. gonorrhoeae clinical isolates were recently determined and made publically available by the Neisseria gonorrhoeae group Sequencing Project (http://www.broadinstitute.org/), adding to the existing genome sequences of FA1090 and NCCP11945 (12). Analysis of the unfinished assemblies identified a 685-bp fragment present in strains FA6140 (supercontig 29, positions 23818 to 24502), DGI18 (supercontig 24, positions 24037 to 24721), and PID24 (supercontig 27, positions 23854 to 24538) that exhibited 98 to 100% identity to copies of human retrotransposable element L1. This Neisseria L1 sequence (nL1) also exhibits strong similarity to LINEs from other primates (e.g., Pan troglodytes), but since humans are the only known natural host for N. gonorrhoeae, it is likely that the HGT event that yielded nL1 occurred in humans. The nL1 fragment is identical among the FA6140, DGI18, and PID24 Neisseria strains and occupies the same insertion site (Fig. 1). The nL1 element includes the coding sequence for the first 164 amino acids of the L1 open reading frame 1 (ORF1) nucleic acid binding protein (13) and 192 bases of the L1 5′ untranslated region upstream of ORF1. This region of the ORF1 protein contains a coiled-coil domain that is required for trimerization of the murine ORF1 protein and the first 8 amino acids of a 96-residue RNA recognition motif (14). The insertion site in the gonococcal genome is located 14 bp upstream of a copy of the phage-associated transposase irg4 (15, 16). This genetic region also contains a 24-bp inverted repeat, one of which is interrupted by the nL1 insertion (Fig. 1). Although repeat sequences and mobile genetic elements are often associated with HGT, comparison of nL1-positive and nL1-negative genomes yields no evidence that either the putative Nf4-G4 prophage or the inverted repeat was directly involved in the acquisition of nL1.
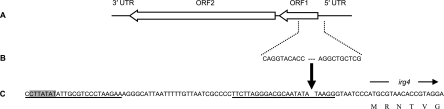
FIG 1 .
Map of the nL1 fragment in N. gonorrhoeae. (A) Schematic of a full-length L1 element. (B) The 10 terminal nucleotides of the 685-bp nL1 insertion are shown in their corresponding locations on the L1 element (GenBank accession no. U09116.1). (C) The nL1 insertion site within the N. gonorrhoeae genome is denoted by the bold arrow and is 14 bp upstream of the irg4 start codon. The specific irg allele was identified using the nomenclature established for the FA1090 annotation (16). The first 6 amino acids of Irg4 are shown. Two 24-bp inverted repeats in the gonococcal genome are underlined, and the highlighted sequence designates one terminus of the Nf4-G4 prophage sequence that includes the irg4 ORF (15). UTR, untranslated region.
Contamination of DNA sequencing samples has previously accounted for the false identification of LINE sequences in human-associated microorganisms (17). To test whether the presence of nL1 was the result of contamination introduced during the sequencing or purification of gonococcal DNA, independently prepared genomic DNA from the sequenced N. gonorrhoeae isolates was assayed for the presence of nL1 by PCR. Multiple reactions containing primer combinations specific to the nL1 sequence and flanking DNA verified the presence of nL1 in the genomic location predicted by the sequences (Fig. 2A to D). Sequencing of nL1-containing PCR products confirmed this result and the composition of nL1 as reported in the genome sequences of all three strains (data not shown). DNA hybridizations using a probe generated from human L1 sequences also confirmed that strains FA6140, DGI18, and PID24 contained nL1 and that this fragment was not located at an alternative site in the genomes of the remaining 12 strains from Fig. 2 (data not shown). Together, these analyses conclusively demonstrate that the presence of L1 DNA in strains PID24, DGI18, and FA6140 is attributable to HGT and is not an artifact of genome sequencing.
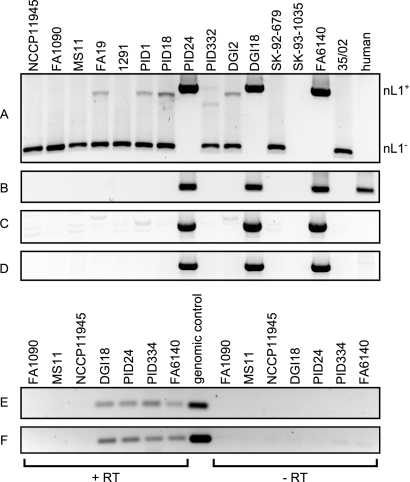
FIG 2 .
PCR amplification of the nL1 fragment in N. gonorrhoeae isolates and nL1 transcript detection by RT-PCR. Purified genomic DNA from sequenced gonococcal isolates and human chromosomal DNA were used as templates for PCR amplification of the nL1 fragment and L1. (A) Primers IS1106for and IRGrev anneal to sequences flanking the nL1 insertion site and yield products of 1,090 and 405 bp for nL1-positive and nL1-negative alleles, respectively. Neither product was detected in DNA from strain SK-93-1035, but this isolate is not predicted to harbor the nL1 fragment. (B) Reactions using primers LINEfor and LINErev, which anneal to sequences internal to the human L1 element and the nL1 fragment. Combinations of flanking and internal primers LINErev and IS1106for (C) or LINEfor and IRGrev (D) were used to confirm the genomic location of nL1. The background bands visible in panels A and C are not due to nL1 sequence amplification, since all of the strains shown here, except PID24, DGI18, and FA6140, were also determined to be nL1 negative by DNA hybridization. Total RNA from nL1-containing isolates (DGI18, PID24, PID334, and FA6140) was used as the template in RT reaction mixtures containing (+RT) or lacking (−RT) reverse transcriptase. The resulting cDNA was amplified with internal nL1 primers L1for and L1rev. Isolates FA1090, MS11, and NCCP11945 lack the L1 fragment and were included as negative controls. Purified genomic DNA from isolate PID334 was used as a positive control. Products were obtained with cDNA generated from positive-strand (E) and negative-strand (F) RNA transcripts.
To assess the frequency of nL1 in the population, a diverse collection of Neisseria strains, isolated at different times and locations, were screened for the presence of nL1 using a combination of molecular and bioinformatic techniques (see Table S1 in the supplemental material). Four additional nL1-positive N. gonorrhoeae strains were identified by this analysis (97G181, ATCC 49226, PID334, and WHO I) and determined to harbor a 685-bp sequence identical to that reported for the sequenced nL1-positive strains. Therefore, the nL1 allele occurred at a frequency of ~11% among the 62 N. gonorrhoeae strains that were included in this study. Interestingly, the different disease manifestations of N. gonorrhoeae infection (uncomplicated infection, pelvic inflammatory disease, disseminated gonococcal infection) were all represented among the nL1-positive strains identified here. Although closely related members of the genus Neisseria are also exclusively associated with human infection, examination of 212 Neisseria meningitidis isolates and 19 commensal Neisseria isolates produced no evidence of the nL1 fragment (see Table S1 in the supplemental material). Assuming that the frequency of occurrence is similar in N. meningitidis and N. gonorrhoeae, the probability of observing zero nL1 events in the N. meningitidis population tested is 1.9E−11, since the number should follow a binomial distribution with n = 212 and P = 0.11. The lack of nL1 sequences in the highly genetically related species N. meningitidis suggests that the acquisition of nL1 by N. gonorrhoeae was a recent evolutionary event that occurred subsequent to the divergence of N. meningitidis and N. gonorrhoeae. The identification of nL1 in gonococci but not meningococci may reflect differences in the species-specific interactions between bacteria and host cells, the availability of host DNA during infection with N. gonorrhoeae versus N. meningitidis, or most likely an event that occurred after the two species diverged.
The absolute conservation of the nL1 sequence among the seven positive isolates is striking and suggests that this locus is under selective pressure or has not had sufficient time to degenerate. Reverse transcription (RT)-PCR using total RNA and internal L1 primers detected transcripts containing the nL1 sequence originating from both strands of DNA (Fig. 2E and F). Therefore, the HGT event that yielded nL1 has the potential to produce a novel gene product and may contribute to the observed sequence conservation. However, because the frequency of nL1 occurrence in the tested population is modest, any selective force conferred by nL1 may be small and difficult to identify experimentally. During laboratory culture, no consistent gross phenotypic differences between nL1-positive and nL1-negative strains were observed.
In addition to selective pressure, clonal expansion could also account for the observed lack of variation of the nL1 allele among N. gonorrhoeae isolates. Multilocus sequencing typing (MLST) (18) was used to assess the relatedness of four nL1-positive isolates compared to 12 confirmed L1-negative isolates for which allele sequences were available. Strains DGI18 and PID24 have identical MLST profiles belonging to sequence type 8418 and may represent clonal expansion of the nL1 allele (Table 1). However, strains FA6140 and PID334 have sequence types divergent from both sequence type 8418 and each other. Since three distinct sequence types are represented among the four nL1 strains analyzed, either the presence of the nL1 fragment in these strains was the result of multiple identical HGT events or, more likely, an initial acquisition of nL1 from humans was subsequently transferred horizontally between gonococcal strains.
TABLE 1 .
MLST profiles of the N. gonorrhoeae strains used in this study
| Strain | MLST allele |
Sequence type | ||||||
|---|---|---|---|---|---|---|---|---|
| abcZ | adk | aroE | fumC | gdh | pdhC | pgm | ||
| NCCP11945 | 109 | 39 | 170 | 111 | 148 | 153 | 65 | 1901 |
| FA19 | 59 | 39 | 67 | 156 | 188 | 71 | 133 | 1892 |
| MS11 | 126 | 39 | 67 | 78 | 146 | 153 | 133 | 6959 |
| 1291 | 126 | 39 | 170 | 158 | 149 | 531 | 65 | 8422 |
| 35/02 | 59 | 39 | 67 | 78 | 148 | 153 | 65 | 7363 |
| FA6140 | 126 | 39 | 67 | 78 | 149 | 71 | 65 | 1927 |
| PID1 | 126 | 39 | 67 | 156 | 149 | 71 | 133 | 8417 |
| PID18 | 59 | 113 | 67 | 158 | 148 | 154 | 133 | 1926 |
| PID24 | 109 | 39 | 170 | 78 | 228 | 71 | 65 | 8418 |
| PID332 | 109 | 39 | 67 | 156 | 152 | 71 | 65 | 1594 |
| PID334 | 126 | 39 | 67 | 156 | 146 | 71 | 65 | 8423 |
| FA1090 | 109 | 39 | 67 | 190 | 147 | 71 | 65 | 1899 |
| SK-92-679 | 59 | 39 | 67 | 111 | 149 | 153 | 65 | 6715 |
| SK-93-1035 | 129 | 39 | 67 | 156 | 149 | 153 | 65 | 1595 |
| DGI2 | 59 | 113 | 67 | 158 | 147 | 530 | 133 | 8421 |
| DGI18 | 109 | 39 | 170 | 78 | 228 | 71 | 65 | 8418 |
DISCUSSION
The high level of identity between human L1 and nL1 sequences, combined with its absence in closely related Neisseria species, suggests that this HGT event happened relatively recently in evolutionary history and/or that the nL1 region is under strong selective pressure. In addition, the low penetrance of nL1 within the gonococcal population is consistent with the hypothesis that nL1 was acquired recently from humans. Since numerous different L1 copies exist in any one human genome, including ones that contain sequences identical to nL1 (GenBank accession no. AC013546), analysis based on the accumulation of synonymous and nonsynonymous substitutions relative to a donor sequence is not feasible. N. gonorrhoeae can be found intracellularly and extracellularly associated with neutrophils and epithelial cells and thus has the opportunity to encounter host DNA during infection (19, 20). The natural competence of N. gonorrhoeae provides the bacterium with the means to acquire exogenous DNA, and the fact that the signature Neisseria DNA uptake sequence is not absolutely required for internalization of exogenous DNA (21) increases the probability of this event’s occurrence. Although N. gonorrhoeae can invade and replicate within host cells, there is no evidence for nuclear association, making interaction with host DNA in live cells unlikely. A potential scenario in which HGT between humans and intracellular N. gonorrhoeae could more readily occur is after an apoptotic (22) or necrotic event in which the host DNA is fragmented or disrupted within the dying cell. Alternatively, the active secretion of neutrophil extracellular traps (23) provides a means by which host DNA could become accessible to extracellular N. gonorrhoeae.
The mechanism by which this foreign genetic material was incorporated into the N. gonorrhoeae genome is a matter of speculation. Comparison of the insertion site between nL1-positive and nL1-negative isolates demonstrates that the recombined fragment is the result of a simple insertion with no loss or gain of flanking chromosomal information (Fig. 1). Mobilization of L1 elements within the nuclei of host cells occurs via target-primed RT (11), which requires the function of ORF1p and ORF2p and results in target site duplication. No such signature duplication is present near the nL1 site, and retrotransposition is unlikely to account for the insertion of nL1 into the N. gonorrhoeae genome. There is no homology between the gonococcal insertion site and the L1 sequences flanking the nL1 fragment, suggesting that direct recombination after transformation is also an unlikely mechanism for the integration of nL1. Therefore, we postulate that nonhomologous end joining (NHEJ) could have occurred between a fragmented copy of L1 and the recipient region of the N. gonorrhoeae genome. Although N. gonorrhoeae lacks the NHEJ machinery possessed by some bacteria, alternative pathways of low-frequency NHEJ that have the potential to mediate HGT of exogenous sequences have been proposed (24).
Regardless of the mechanism by which HGT occurred, it is possible that nL1 has an effect on the physiology of N. gonorrhoeae by way of residual activity of the truncated ORF1 protein (if produced) or via polar effects of the insertion itself. More significantly, though, the identification of human sequences within the N. gonorrhoeae genome has important implications for modeling of the generation of genetic diversity and evolution of this and other obligate human pathogens and establishes that HGT can occur between a mammalian host and an associated microorganism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Neisseria isolates used in this study and their sources are listed in Table S1 in the supplemental material. N. gonorrhoeae and commensal Neisseria species were grown on gonococcal medium base (Difco) containing Kellogg supplements as described previously (25). N. meningitidis strains were cultivated on gonococcal medium supplemented with 1% IsoVitaleX (Becton Dickinson). Bacteria used for the preparation of RNA were grown to exponential phase as described previously (25) in GCBL broth (1.5% proteose peptone no. 3 [Difco], 0.4% K2HPO4, 0.1% KH2PO4, 0.1% NaCl) supplemented with 1% IsoVitaleX.
Detection of the nL1 allele.
All of the oligonucleotide primers used in this study are listed in Table S2 in the supplemental material. Genomic DNA was purified on QIAamp columns (Qiagen) and used for PCR and slot blot hybridizations. The nL1 genotype of the N. gonorrhoeae strains listed in Fig. 2 was established using the following primer combinations: IS1106for and IRGrev, LINEfor and LINErev, IS1106for and LINErev, and LINEfor and IRGrev. All other Neisseria isolates screened by PCR as reported in Table S1 in the supplemental material were screened by the LINEfor and LINErev primer pair at minimum. PCR products were sequenced by the Northwestern University Genomics Core Facility. A sampling of isolates of each Neisseria species screened was analyzed by slot blot hybridization using a 541-bp probe PCR amplified from human chromosomal DNA using primers LINEfor and LINErev. The product was labeled with digoxigenin-dUTP via the random-primed method (Roche), and hybridizations were performed using standard techniques. The binomial distribution was used to determine the probability of observing zero nL1 events in N. meningitidis based on the N. gonorrhoeae frequency and assuming equivalent rates. The Northwestern University Biostatistics Collaboration Center assisted with the statistical analysis.
RT-PCR.
Total RNA was purified from N. gonorrhoeae cultures using the RNeasy kit (Qiagen) and treated with RQ1 DNase (Promega) to remove genomic contamination. The LINEfor and LINErev oligonucleotides were used to prime cDNA synthesis using Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s recommendations. Internal nL1 primers L1for and L1rev were used to PCR amplify cDNA products.
MLST.
Seven housekeeping genes; abcZ, adk, aroE, fumC, gdh, pdhC, and pgm, were used to establish the sequence types of N. gonorrhoeae strains as described previously (18), with the exception of the oligonucleotide primers that were used for amplification and sequencing of MSLT alleles. Allele numbers were assigned by querying the Neisseria MLST database (18) using existing genomic sequences when available. The MLST primers listed in Table S2 in the supplemental material were used to PCR amplify and sequence (both strands) all alleles from strain PID334 and any other allele not previously established in the database. The profiles of all of the strains listed in Table 1 have been deposited in the Neisseria MLST database.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
We acknowledge M. Apicella, J. Dillard, K. Lee, R. Nicholas, P. Rice, W. Shafer, D. Trees, and W. Whittington for contributing Neisseria strains; J. Bodily for providing preparations of human genomic DNA; K. Clark for performing the bioinformatics screen that identified the nL1 sequence; D. Ward and M. Giovanni for facilitating the genomic sequencing of N. gonorrhoeae strains; M. Fitzgerald for providing genomic assemblies; M. Maiden for providing access to N. meningitidis genome sequences; K.-Y. Kim for assisting with the statistical analysis; and A. Chen and A. Hauser for critical reading of the manuscript.
This work was supported by NIH grants R01 AI044239 and R37 AI033493 to H.S.S. M.T.A. was partially supported by NIH fellowship F32AI080083.
Footnotes
Citation Anderson, M. T., and H. S. Seifert. 2011. Opportunity and means: horizontal gene transfer from the human host to a bacterial pathogen. mBio 2(1):e00005-11. doi:10.1128/mBio.00005-11.
REFERENCES
- 1. McDaniel T. K., Kaper J. B. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399–407 [DOI] [PubMed] [Google Scholar]
- 2. Lawrence J. G., Ochman H. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383–397 [DOI] [PubMed] [Google Scholar]
- 3. Goldman B. S., Kranz R. G. 1998. Evolution and horizontal transfer of an entire biosynthetic pathway for cytochrome c biogenesis: Helicobacter, Deinococcus, Archae and more. Mol. Microbiol. 27:871–873 [DOI] [PubMed] [Google Scholar]
- 4. Davies J., Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74:417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feil E., Zhou J., Smith J. Maynard, Spratt B. G. 1996. A comparison of the nucleotide sequences of the adk and recA genes of pathogenic and commensal Neisseria species: evidence for extensive interspecies recombination within adk. J. Mol. Evol. 43:631–640 [DOI] [PubMed] [Google Scholar]
- 6. Spratt B. G., Bowler L. D., Zhang Q. Y., Zhou J., Smith J. M. 1992. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J. Mol. Evol. 34:115–125 [DOI] [PubMed] [Google Scholar]
- 7. Kroll J. S., Wilks K. E., Farrant J. L., Langford P. R. 1998. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. U. S. A. 95:12381–12385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fudyk T. C., Maclean I. W., Simonsen J. N., Njagi E. N., Kimani J., Brunham R. C., Plummer F. A. 1999. Genetic diversity and mosaicism at the por locus of Neisseria gonorrhoeae. J. Bacteriol. 181:5591–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hobbs M. M., Alcorn T. M., Davis R. H., Fischer W., Thomas J. C., Martin I., Ison C., Sparling P. F., Cohen M. S. 1999. Molecular typing of Neisseria gonorrhoeae causing repeated infections: evolution of porin during passage within a community. J. Infect. Dis. 179:371–381 [DOI] [PubMed] [Google Scholar]
- 10. Scott A. F., Schmeckpeper B. J., Abdelrazik M., Comey C. T., O’Hara B., Rossiter J. P., Cooley T., Heath P., Smith K. D., Margolet L. 1987. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cost G. J., Feng Q., Jacquier A., Boeke J. D. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21:5899–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung G. T., Yoo J. S., Oh H. B., Lee Y. S., Cha S. H., Kim S. J., Yoo C. K. 2008. Complete genome sequence of Neisseria gonorrhoeae NCCP11945. J. Bacteriol. 190:6035–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin S. L., Bushman F. D. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21:467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khazina E., Weichenrieder O. 2009. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl. Acad. Sci. U. S. A. 106:731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawai M., Uchiyama I., Kobayashi I. 2005. Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Res. 12:389–401 [DOI] [PubMed] [Google Scholar]
- 16. Skaar E. P., Lecuyer B., Lenich A. G., Lazio M. P., Perkins-Balding D., Seifert H. S., Karls A. C. 2005. Analysis of the Piv recombinase-related gene family of Neisseria gonorrhoeae. J. Bacteriol. 187:1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deitsch K. W., Carlton J. M., Wootton J. C., Wellems T. E. 2001. Host sequences in Plasmodium falciparum and Plasmodium vivax genomic DNA: horizontal transfer or contamination artifact? FEBS Lett. 491:164–165 [DOI] [PubMed] [Google Scholar]
- 18. Bennett J. S., Jolley K. A., Sparling P. F., Saunders N. J., Hart C. A., Feavers I. M., Maiden M. C. 2007. Species status of Neisseria gonorrhoeae: evolutionary and epidemiological inferences from multilocus sequence typing. BMC Biol. 5:35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Apicella M. A., Ketterer M., Lee F. K., Zhou D., Rice P. A., Blake M. S. 1996. The pathogenesis of gonococcal urethritis in men: confocal and immunoelectron microscopic analysis of urethral exudates from men infected with Neisseria gonorrhoeae. J. Infect. Dis. 173:636–646 [DOI] [PubMed] [Google Scholar]
- 20. Ovcinnikov N. M., Delektorskij V. V. 1971. Electron microscope studies of gonococci in the urethral secretions of patients with gonorrhoea. Br. J. Vener. Dis. 47:419–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duffin P. M., Seifert H. S. 2010. DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J. Bacteriol. 192:4436–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walker P. R., Sikorska M. 1994. Endonuclease activities, chromatin structure, and DNA degradation in apoptosis. Biochem. Cell Biol. 72:615–623 [DOI] [PubMed] [Google Scholar]
- 23. Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y., Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532–1535 [DOI] [PubMed] [Google Scholar]
- 24. Chayot R., Montagne B., Mazel D., Ricchetti M. 2010. An end-joining repair mechanism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:2141–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stohl E. A., Criss A. K., Seifert H. S. 2005. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58:520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.