Abstract
A DNA strand-transfer reaction is an early step in the transposition of phage Mu. It has been shown that an efficient reaction in vitro requires, in addition to buffer and salt, only the Mu A protein, Mu B protein, host protein HU, ATP, and Mg2+. We have determined that, of the three protein factors involved, only the Mu B protein has an ATPase activity. The Mu B ATPase is stimulated by Mu A protein and DNA but not by either of these factors alone. Double-stranded DNA is a much better cofactor than single-stranded DNA, but there is no apparent sequence specificity. In the absence of the Mu B protein and/or ATP, the intermolecular Mu DNA strand-transfer reaction is extremely inefficient, and the strand-transfer products are predominantly the result of an intramolecular reaction. This contrasts with the efficient intermolecular reaction that occurs if Mu B protein and ATP are provided. The Mu B protein, in the presence of Mu A protein and protein HU, therefore, seems to facilitate interactions between potential DNA target sites and pairs of Mu DNA ends.
Full text
PDF
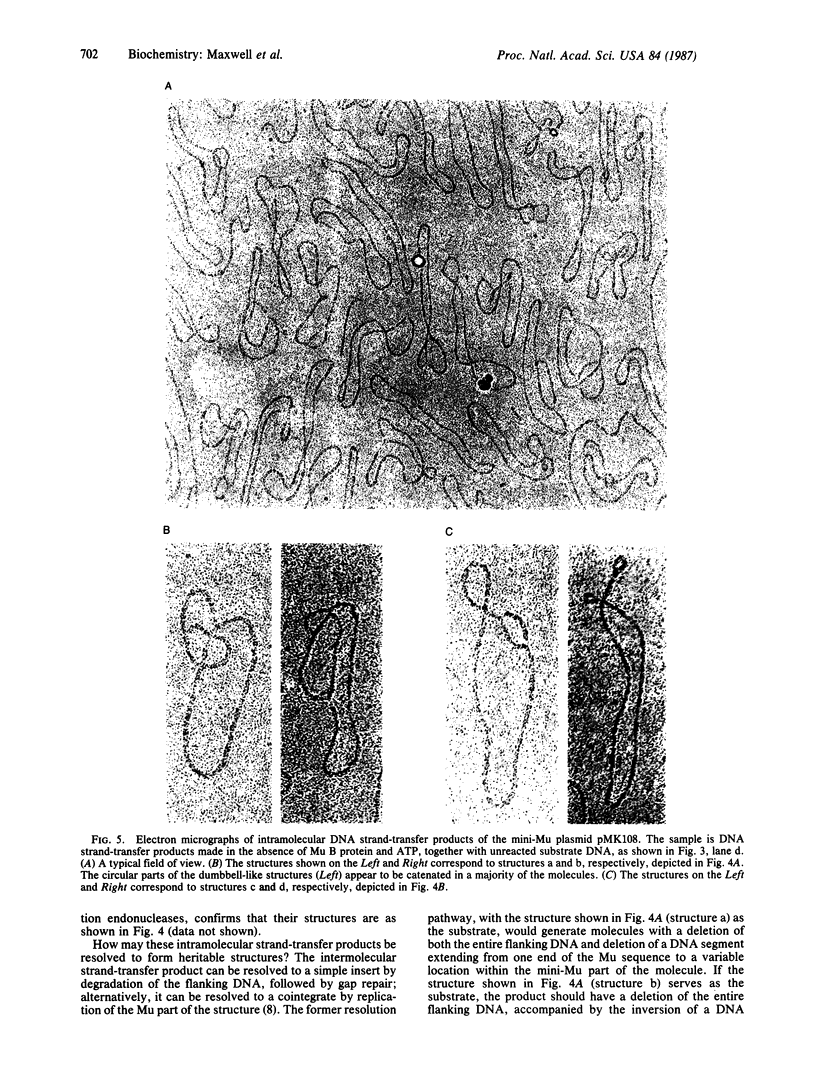

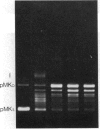
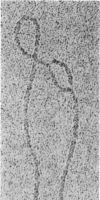
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chaconas G., Gloor G., Miller J. L. Amplification and purification of the bacteriophage Mu encoded B transposition protein. J Biol Chem. 1985 Mar 10;260(5):2662–2669. [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Sarvetnick N., Bukhari A. I. Predominant end-products of prophage Mu DNA transposition during the lytic cycle are replicon fusions. J Mol Biol. 1981 Aug 15;150(3):341–359. doi: 10.1016/0022-2836(81)90551-9. [DOI] [PubMed] [Google Scholar]
- Chen Y., Maxwell A., Westerhoff H. V. Co-operativity and enzymatic activity in polymer-activated enzymes. A one-dimensional piggy-back binding model and its application to the DNA-dependent ATPase of DNA gyrase. J Mol Biol. 1986 Jul 20;190(2):201–214. doi: 10.1016/0022-2836(86)90293-7. [DOI] [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin D. J., Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Cloning of the A gene of bacteriophage Mu and purification of its product, the Mu transposase. J Biol Chem. 1985 Feb 10;260(3):1832–1835. [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985 Jul;41(3):867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell. 1986 Jun 20;45(6):793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Faelen M., Huisman O., Toussaint A. Involvement of phage Mu-1 early functions in Mu-mediated chromosomal rearrangements. Nature. 1978 Feb 9;271(5645):580–582. doi: 10.1038/271580a0. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Groenen M. A., Timmers E., van de Putte P. DNA sequences at the ends of the genome of bacteriophage Mu essential for transposition. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2087–2091. doi: 10.1073/pnas.82.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey R. M. Transposition without duplication of infecting bacteriophage Mu DNA. Nature. 1984 Oct 11;311(5986):580–581. doi: 10.1038/311580a0. [DOI] [PubMed] [Google Scholar]
- Liebart J. C., Ghelardini P., Paolozzi L. Conservative integration of bacteriophage Mu DNA into pBR322 plasmid. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4362–4366. doi: 10.1073/pnas.79.14.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. The DNA dependence of the ATPase activity of DNA gyrase. J Biol Chem. 1984 Dec 10;259(23):14472–14480. [PubMed] [Google Scholar]
- Mizuuchi K., Craigie R. Mechanism of bacteriophage mu transposition. Annu Rev Genet. 1986;20:385–429. doi: 10.1146/annurev.ge.20.120186.002125. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983 Dec;35(3 Pt 2):785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Mechanism of transposition of bacteriophage Mu: polarity of the strand transfer reaction at the initiation of transposition. Cell. 1984 Dec;39(2 Pt 1):395–404. doi: 10.1016/0092-8674(84)90018-7. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Lotterman B. Kinetics of Mu DNA synthesis. Mol Gen Genet. 1977 Mar 7;151(2):169–174. doi: 10.1007/BF00338691. [DOI] [PubMed] [Google Scholar]