Abstract
Voltage-gated sodium and potassium channels underlie electrical activity of neurons, and are dynamically regulated by diverse cell signaling pathways that ultimately exert their effects by altering the phosphorylation state of channel subunits. Recent mass spectrometric-based studies have led to a new appreciation of the extent and nature of phosphorylation of these ion channels in mammalian brain. This has allowed for new insights into how neurons dynamically regulate the localization, activity and expression through multisite ion channel phosphorylation.
Keywords: Mass spectrometry, Phosphoproteomics, VGSC (Nav channel), VGKC (Kv channel), Brain ion channel
1. Introduction
Ion fluxes through voltage-dependent ion channels that are selectively permeable to sodium and potassium ions underlie much of the electrical signaling in neurons. As such, voltage-gated sodium channel (VGSC) and potassium channel (VGKC) activity is highly regulated in both excitable and non-excitable cells. These ion channels exist as supramolecular protein complexes composed of pore-forming transmembrane principal or α subunits, auxiliary or regulatory β subunits [1], and a diverse array of interacting proteins [2]. VGSCs mediate the rapid influx of Na+ that underlies action potential initiation and propagation, dendritic excitability, and many other aspects of neuronal excitability. These channels consist of a highly glycosylated 24 transmembrane segment α subunit of ≈260 kDa, associated with smaller auxiliary single pass transmembrane Navβ subunits [3]. VGKCs are composed of four pore-forming and voltage-sensing α subunits, each of which has six transmembrane segments S1–S6, extensive cytoplasmic N- and C-termini, and intracellular linkers between transmembrane segments S2–S3 and S4–S5 [4]. VGKCs typically contain additional single-pass transmembrane or cytoplasmic auxiliary subunits [1].
Diverse posttranslational events acting on components of VGSC and VGKC complexes can dynamically regulate the expression, localization and function of these ion channels. While numerous non-covalent mechanisms such as ligand binding, sensing of transmembrane voltage, and interaction with other proteins can play prominent roles in regulating VGSCs and VGKCs, direct covalent modification of components of these multiprotein ion channel complexes represents a widely used and potent mechanism for neurons to achieve dynamic and reversible changes in VGSC and VGKC function, and impact their contribution to neuronal signaling.
Phosphorylation constitutes the most common covalent post-translational modification in eukaryotes [5], with (as of early 2009) up to 25,000 described phosphorylation sites (phosphosites) on 7,000 human proteins, out of an estimated 500,000 potential phosphosites that exist in a cellular proteome {Lemeer, 2009 #109}. In neurons, reversible activity-dependent phosphorylation represents a major mechanism of dynamic regulation of synaptic development [6], potentiation, depression and homeostatic plasticity [7], through phosphorylation of a large number of synaptic proteins including ligand-gated ion channels [8]. Neurons also exhibit cellular plasticity at the level of intrinsic excitability, achieved through phosphorylation of components of VGSC [9] and VGKC [10] channel complexes, which localize in distinct neuronal compartments [9, 11, 12]. As opposed to the classical approaches of in vivo or in vitro radiolabeling with 32P, peptide mapping and/or sequencing, and site-directed mutagenesis [e.g., [13, 14], mass spectrometry (MS)-based phosphoproteomic techniques have recently emerged as the primary tool for the identification phosphorylation on VGSC and VGKC subunits, and have revealed unanticipated extent of multisite phosphorylation on both VGSCs and VGKCs. Here we review the current state of how applying such approaches has impacted our view of the extent and nature of phosphorylation of mammalian neuronal VGSC and VGKC, and perspectives for future research into these important determinants of neuronal signaling.
2. Strategies to identify phosphorylation on brain VGSCs and VGKCs
2.1. Complexity and dynamics of brain proteome
Recent innovations in proteomics and bioinformatics have expanded our knowledge of the mammalian brain proteome to include ≈10,000 different proteins [15]. It remains a major challenge to overcome the high complexity of the brain proteome, and the dynamic nature of reversible phosphorylation, to define the VGSC and VGKC phosphoproteome. To understand how the signaling networks present in neuronal cells impinge on the expression, localization and function of these ion channels, the need for comprehensive information on VGSC and VGKC phosphosites is paramount. MS-based proteomic approaches represent a powerful approach to define the VGSC and VGKC phosphoproteome, a critical first step in understanding how dynamic changes in phosphorylation at specific sites impacts dynamic plasticity in neuronal electrical activity. Proteomic analyses of a variety of VGSCs and VGKCs, whose polypeptide sequences have been deduced from molecular cloning and genomic studies, have led to the identification of a large and rapidly expanding set of identified in vivo phosphosites. Here, we review these studies and discuss practical aspects and implication of phosphosite identification of brain VGSC and VGKC proteins.
2.2. Sample preparation for phosphoproteomic analysis of brain ion channels
2.2.1. Antibody-dependent approaches
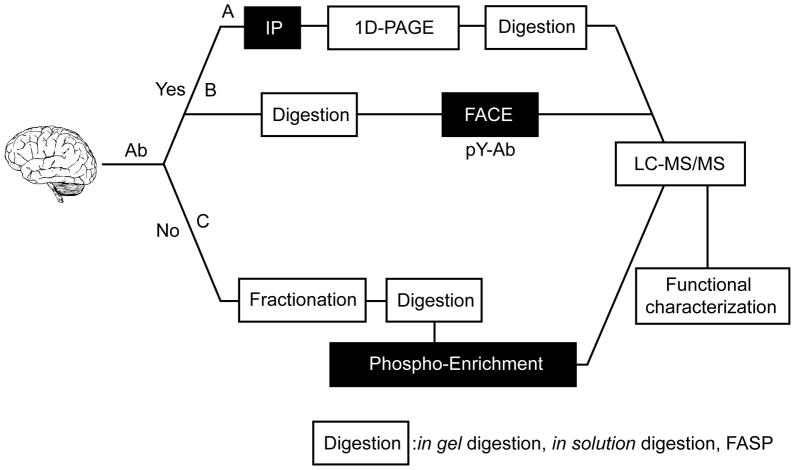
Given the high complexity of the brain proteome, and highly variable expression levels of expression of different VGSCs and VGKCs, enrichment for specific channel subunits can greatly benefit attempts at a comprehensive analysis of the extent and nature of their phosphorylation. Antibody based approaches represent a powerful tool to directly isolate, as target antigens, specific ion channel α subunits, auxiliary subunits or interacting proteins for subsequent analysis of phosphosites. A suitable antibody can allow for the simple yet specific immunopurification of the target VGSC or VGKC subunit directly from samples with a high degree of proteomic complexity (e.g., crude brain preparations) as shown in Figure 1 (Path A). Following further fractionation by preparative one-dimensional SDS-PAGE, VGSC or VGKC subunits can then be subjected to MS analysis in a sample with a limited proteomic complexity. When excised bands of immunopurified proteins are digested in gel slices, unmodified peptides, as well as less abundant and more difficult to detect phosphopeptides, present in the sample in femtomole levels, can be detected in conventional nano-liquid chromatography MS (LC-MS) without any enrichment steps [16, 17]. Further enrichment of phosphopeptides by simple methods such as Immobilized Metal Affinity Chromatography (IMAC) can lead to identification of additional phosphosites not detected in unenriched samples [16].
Figure 1.
Flowchart for identification/characterization of phosphorylation in VGSCs and VGKCs. Antibody-dependent (A, B) and -independent (C) approaches are shown (see text for details). A. Conventional antibody-dependent method for purification of target proteins from a complex sample. B. Enrichment of phosphopeptides from a complex sample using a phosphospecific antibody (Filter aided antibody capturing and elution: FACE). C. High-throughput strategy for phosphorylation identification from complex samples in the absence of an antibody (FASP: filter aided sample preparation)
The antibody-dependent approach has been used in a number of recent studies employing MS-based approaches to identify phosphosites on brain VGSC [16] and VGKC [17–19] α subunits. This approach has also been applied to identification of phosphosites on co-purifying interacting proteins, such as in the MS-based identification of phosphosites on the auxiliary Kvβ2 subunit, following its copurification from brain with antibodies directed against the Kv1.2 α subunit [20].
An alternative antibody-based approach is to use antibodies that recognize an entire family of VGSC or VGKC proteins. A particularly useful set of such “pan” antibodies, targeting VGSCs, has been raised by immunizing animals with the cytoplasmic linker region between domains III and IV (i.e., the ID III-IV linker) that is absolutely conserved among all vertebrate Nav channels. Polyclonal [21] and monoclonal [22] antibodies raised against this segment have proven extremely useful in labeling VGSC α subunits on immunoblots and by immunocytochemistry. The pan-VGSC α subunit monoclonal antibody K58/35 was recently used in concert with a Nav1.2-specific monoclonal antibody in immunopurification of mammalian brain VGSCs for MS-based studies of in vivo phosphorylation sites [16].
An even broader antibody-based approach uses anti-phosphotyrosine (anti-pTyr) antibodies to isolate and identify pTyr-containing proteins, or from tryptic digests, pTyr-containing phosphopeptides. Enrichment can be performed on material that has already been enriched by other means (e.g., subcellular fractionation, immunopurification with specific antibodies, etc.), on membrane fractions, or on whole brain extracts (Path B in Figure 1) in large-scale proteomic studies. The latter approach was used to begin to define the extent and nature of mouse brain tyrosine phosphorylation, and led to identification of 414 unique pTyr phosphosites, including four sites on VGSC and VGKC subunits. Together, antibody-dependent approaches have yielded valuable insights into the brain VGSC and VGKC phosphoproteome.
2.2.2. Antibody-independent approaches
High throughput, antibody-independent approaches represent a powerful approach to obtain important information on VGSC and VGKC phosphorylation in the absence of suitable antibodies, or when one aims to obtain a more global view of phosphorylation that does not focus on individual channel subunits. A number of recent studies have effectively used antibody-independent approaches that incorporate various fractionation procedures, as well as phosphoprotein/phosphopeptide enrichment methods, to reduce the overall complexity of the sample. This has allowed for detection of relatively low abundant VGSC and VGKC phosphopeptides. Such strategies are outlined in Figure 1 as Path C.
A number of key recent studies have provided important new information on the VGSC and VGKC phosphoproteome, without having this as a specific focus per se. A global phosphoproteomic analysis of mouse brain proteins present in synaptic membranes and/or synpatosomes yielded 1,367 unique in vivo phosphosites, including a number of VGSCs and VGKCs [23]. A more recent global analysis of the phosphoproteome of a mouse brain extract yielded over 12,000 phosphosites on ≈4,600 brain phosphoproteins, using an antibody-independent approach for pSer and pThr sites, and an antibody-dependent approach for less abundant pTyr sites [24]. The complexity of whole brain proteins and peptides was reduced by multiple fractionation steps (Path C in Figure 1), including strong anion exchange (SAX), strong cation exchange (SCX), size exclusion chromatography (SEC), and immobilized metal affinity chromatography (IMAC) and TiO2 phospho-peptide enrichment. One item of note from these studies is that the overall ratio obtained for pSer:pThr:pTyr phosphosites in these mouse brain samples was 85:14:1 [23] and 83:15:2 [24], comparable to those (86:12:2) obtained from previous studies on a human cell line [25].
Together, the recent application of broader antibody-independent high-throughput approaches, in combination with a more focused antibody-dependent approach, has proved advantageous in identifying an unexpected wealth of phosphosites on brain VGSCs and VGKCs.
3. Proteomic analyses of brain ion channel phosphorylation
In this section we review available data on phosphorylation of brain VGSCs and VGKCs, focusing on data obtained in recent antibody-dependent and independent MS-based studies. Note that this synopsis is assuredly not comprehensive, as there are likely other high-throughput studies that have yielded data leading to identification of phosphosites that we have not included in our analyses, which focuses on data from five recent antibody-independent studies [23, 24, 26–28], and data obtained from recent antibody-dependent approaches.
3.1. Voltage-gated Sodium Channels (VGSCs)
Antibodies recognizing specific VGSC α subunits have been used to define the expression and localization Nav1.1, Nav1.2, Nav1.3 and Nav1.6 in mammalian brain [29]. Two distinct monoclonal antibodies, one specific for Nav1.2, and one with pan-VGSC specificity, were used in parallel immunopurification and MS analyses of rat brain VGSC phosphorylation [16]. These studies identified fifteen phosphosites on Nav1.2, and three on Nav1.1 (Supplemental Table). Note that two sites (S468, S610) were only observed after additional IMAC phosphopeptide enrichment following immunopurification [16]. Phosphorylation on the cytoplasmic ID I-II linker (11 sites identified) is known to modulate Nav1.2 gating [9]. The C-terminus (three sites identified) is the site of G protein binding, which also impacts Nav1.2 gating [30], however, whether phosphorylation affects G protein binding has not been investigated. The Nav1.1 phosphosites, while in peptides unique to Nav1.1, are in sequence alignments in positions identical to three of the phosphosites found on the Nav1.2 ID I-II linker [16], suggesting a conservation of VGSC modulation through phosphorylation at these sites.
A recent antibody-independent global phosphoproteomics study on mouse brain [24] identified 45 phosphosites on VGSC α subunits, and 3 phosphosites on Navβ subunits, as shown in Supplemental Table. Eight phosphosites were identified on the Nav1.1 ID I-II linker, including two of the sites (S551 and S607) identified in the antibody-dependent study of rat brain Nav1.1 [16], suggesting conservation of phosphorylation between mouse and rat Nav1.1. A total of twenty sites were identified on Nav1.2, including nine of the twelve sites within the ID I-II linker identified in the antibody-dependent study of rat brain [16], again suggesting strong conservation of phosphorylation between mouse and rat Nav1.2 orthologs. Note that previous biochemical studies [reviewed in [9]] had also demonstrated phosphorylation on the Nav1.2 ID I-II linker at S579. Together, these antibody-dependent and –independent studies together yielded a wealth of new phosphosites on Nav1.1 (11 sites) and Nav1.2 (29 sites).
MS-based phosphorylation site analysis has not been performed on other VGSC α subunits purified from brain. However, recent antibody-independent analysis of the mouse brain phosphoproteome employing FASP sample preparation [24] yielded a total of five Nav1.6 phosphosites, all in the ID I-II linker, and four sites on Nav1.9, two in the ID I-II linker, and two in the ID II-III linker. This study also yielded the first data on phosphorylation of auxiliary Navβ subunits. These data represent a huge advance in our understanding of molecular determinants of VGSC modulation, and future studies are likely to yield important information on the role of phosphorylation at these sites in regulating neuronal excitability.
3.2. Voltage-gated potassium channels (VGKCs)
Phosphoproteomic analysis of VGKCs has also been accomplished using both antibody-dependent [17–19, 31] and antibody-independent [23, 24, 26, 28] approaches. Kv1 channels are predominantly localized in axons, with certain subunits present at high levels in presynaptic terminals, the juxtaparanodes of nodes of Ranvier in myelinated axons, and axon initial segments [29]. With the exception of Kv1.2, antibody-dependent approaches have not been used to address the extent and nature of phosphorylation of brain Kv1 channel subunits. MS analysis of Kv1.2 channels immunopurified from rat, mouse and human brain identified four phosphosites (S434, S440, S441, and S449) in heterologous expression systems [19]. Functional characterization of these residues demonstrated that phosphorylation at S440/S441 is specifically associated with cell surface Kv1.2, while phosphorylation at S449 is associated with newly synthesized, intracellular Kv1.2 [19]. These sites have also identified in antibody-independent phosphoproteome analyses from mouse brain, which identified additional phosphosites T421, Y429, T433, and S447 [23, 24, 26, 32]. Together, these studies have provided a total of eight phosphosites on brain Kv1.2, all on the cytoplasmic C-terminal tail. The role of many of these sites is not known, although mutation of the S440, S441 and S449 phosphosites negatively impacts biosynthetic trafficking of Kv1.2 to the plasma membrane [19]. The location of these C-terminal phosphosites in the Kv1.2 structure is also not known, as this domain is lacking in the fragment of Kv1.2 used in crystallographic studies [33].
While antibody-dependent approaches have not been used in MS-based studies to characterize phosphorylation on other Kv1 family α subunits, recent antibody-independent phosphoproteomics analyses [23, 24, 26, 28] have provided some initial data on their phosphorylation status. These studies have identified N-terminal phosphosites on Kv1.1 (1 site), Kv1.4 (3 sites), and Kv1.6 (2 sites), and C-terminal phosphorylation of Kv1.5 (1 site). It is intriguing that unlike Kv1.2, so many other Kv1 family members exhibit phosphorylation on the N-terminus. The phosphosites identified on Kv1.4 are all located on the so-called “chain” segment involved in the “ball and chain” mechanism of N-type inactivation, and one of these sites (S122) is required for the CamKII-dependent modulation of Kv1.4 inactivation kinetics [34] that mediates firing frequency-dependent inactivation of Kv1.4-containing channels in presynaptic terminals [35]. The N-terminal phosphorylation of Kv1.6 is also intriguing as it occurs within the so-called NIP domain that prevents N-type inactivation in Kv1.6-containing Kv1 channels [36], suggesting that, as in Kv1.4-containing channels, N-terminal phosphorylation of Kv1.6 may regulate channel inactivation. The Kv1 family associated auxiliary Kvβ1 and Kvβ2 subunits have also been found phosphorylated in vivo in mouse brain in a number of high throughput phosphoproteomic studies [23, 24, 26, 28, 32, 37].
The delayed rectifier Kv channel Kv2.1 plays a highly conditional role in determining the firing of neuronal action potentials [38, 39]. Kv2.1 channels are found in large plasma membrane clusters in somatodendritic regions of neurons, with a minor population on axon initial segments [29]. Early studies suggested that Kv2.1 is present in a highly phosphorylated state in mammalian brain and that phosphorylation determines the voltage-dependence of channel activity [40]. More recent studies revealed that changes in neuronal activity during in vivo epileptic and hypoxic episodes, and upon glutamate stimulation of cultured neurons, trigger Kv2.1 dephosphorylation [39, 41–43]. Dephosphorylation is mediated by the phosphatase calcineurin, and leads to dispersion of Kv2.1 clusters, and hyperpolarizing shifts in voltage-dependent activation and inactivation of Kv2.1 channels [39, 41–43]. Similar phosphorylation-dependent modulation is seen for recombinant Kv2.1 expressed in heterologous cells [40, 44, 45].
An antibody-dependent approach was used to purify recombinant Kv2.1 protein from heterologous cells, and endogenous Kv2.1 protein from rat brain [18, 46]. Subsequent MS analysis led to the initial identification of 21 Ser and two Thr residues (Table 1) as in vivo phosphosites [18]. All phosphosites except two were located on the cytoplasmic C-terminus. Stable Isotope Labeling with Amino acids in cell Culture (SILAC) allowed for quantitative MS analysis of the subset of sites (9/16 sites identified in the original analysis) targeted during calcineurin-mediated dephosphorylation [18]. Subsequent high throughput analyses on mouse brain [23, 24, 26, 28] have yielded seven additional Kv2.1 phosphosites (one N-terminal and six C-terminal), as well as providing corroborating evidence for five of the sixteen sites identified in the antibody-dependent analysis of rat brain Kv2.1 (Supplemental Table). The recent antibody independent analysis of mouse brain phosphoproteome revealed seven phosphosites on the Kv2.2 C-terminus; the functional significance of these new phosphosites has not yet been determined. Two Kv2.2 phosphosites (S488 and S507) are present in sequence alignments in positions identical to two of the phosphosites (S484 and S503) that regulate Kv2.1 gating.
Table I.
Phosphorylation sites identified on VGSC and VGKC α subunits by MS.
| α Subunit | Total | N-terminal | Linker | C-terminal |
|---|---|---|---|---|
| Nav1.1/Scn1a | 11 | 0 | 10 | 1 |
| Nav1.2/Scn2a1 | 29 | 1 | 24 | 4 |
| Nav1.6/Scn8a | 5 | 0 | 5 | 0 |
| Nav1.9/Scn9a | 4 | 0 | 4 | 0 |
| Kv1.1/Kcna1 | 1 | 1 | 0 | 0 |
| Kv1.2/Kcna2 | 8 | 0 | 0 | 8 |
| Kv1.4/Kcna4 | 3 | 3 | 0 | 0 |
| Kv1.5/Kcna5 | 1 | 0 | 0 | 1 |
| Kv1.6/Kcna6 | 2 | 2 | 0 | 0 |
| Kv2.1/Kcnb1 | 23 | 2 | 0 | 21 |
| Kv2.2/Kcnb2 | 7 | 0 | 0 | 7 |
| Kv3.1/Kcnc1 | 5 | 2 | 0 | 3 |
| Kv3.2/Kcnc2 | 4 | 0 | 0 | 4 |
| Kv3.3/Kcnc3 | 5 | 0 | 0 | 5 |
| Kv3.4/Kcnc4 | 1 | 0 | 0 | 1 |
| Kv4.1/Kcnd1 | 2 | 0 | 0 | 2 |
| Kv4.2/Kcnd2 | 5 | 1 | 0 | 4 |
| Kv4.3/Kcnd3 | 1 | 1 | 0 | 0 |
| Kv7.2/Kcnq2 | 16 | 1 | 1 | 14 |
| Kv7.3/Kcnq3 | 6 | 1 | 1 | 4 |
| Kv7.5/Kcnq5 | 2 | 0 | 0 | 2 |
| Slo1 BK/Kcnma1 | 37 | 2 | 2 | 33 |
KCNQ/Kv7 α subunits form the VGKCs corresponding to the M-type current. These channels localize in axon initial segments and nodes of Ranvier nodes throughout the nervous system [29], modulate neuronal excitability, bursting, and neurotransmitter release, and are subject to Gq/11 modulation [47]. One prominent form of these channels that is also highly regulated by phosphorylation is formed by Kv7.2/Kv7.3 heteromers [48–50]. Phosphoproteomic analysis of recombinant human Kv7.2 and Kv7.3 expressed in heterologous cells revealed phosphorylation T217 (in Kv7.2) and T246 (in Kv7.3), both localized within the cytoplasmic S4–S5 linker, and S579 in the C-terminus of Kv7.3 [31]. High throughput phosphoproteomic studies have identified phosphorylation at a site corresponding to human T579 on the mouse ortholog of Kv7.3 [24, 28]. These studies and others [23, 26] identified numerous other sites (Supplemental Table), primarily on the Kv7.2 and Kv7.3 C-termini. Interestingly, one of these sites (i.e. S52) had been previously identified as crucial to PKA-mediated activation of Kv7.2 channels in heterologous cells [51]. A number of Kv7.2 phosphosites cluster near the C-terminal region proposed to mediate PIP2 binding [52], which is required for Kv7 channel function [47]. How phosphorylation at these sites on Kv7.2, and the others identified on Kv7 subunits in mouse brain, affect PIP2 binding and other aspects of Kv7 function, is not known.
BK channels are composed of tetramers of a large α subunit that is encoded by a single gene, termed Slo1, whose transcripts are subjected to extensive alternative splicing to generate numerous BK channel isoforms [53]. BK channels are unique among VGKCs in that under physiological conditions they require both membrane depolarization and increased [Ca2+]i to activate. The extended C-terminal region, a substrate for phosphorylation, is a major determinant of BK channel function, as both a direct regulator of gating via calcium binding, and as an organizing center for numerous protein-protein interactions that influence BK channel function [54, 55]. A recent study employing immunopurification of rat brain BK channels, followed by analysis of in vivo phosphosites using MS analysis, identified 39 Ser/Thr phosphosites [17]. Mutation of phosphosite S891 in the 1209 a.a. rat BK sequence Swiss Prot Q62976-2 causes a hyperpolarizing shift in the voltage-dependent activation curve of the channel, suggesting a role for phosphorylation at this site in modulating BK channel gating. Interestingly, phosphorylation at this same site (the S924 residue in mouse Q08460-1 and the S885 position in mouse Q08460-2) was also identified in analyses of the synaptosomal [37] and global [24] phosphoproteomes of mouse brain. Importantly, different BK channel phosphosites can make stoichiometrically distinct contribution to function, with certain sites exhibiting dominant and other recessive effects [56]. As such multisite BK channel phosphorylation can have diverse effects on function. Moreover, the effects of phosphorylation on BK channels can be impacted by changes in BK channel sequences outside of the region containing the phosphosites themselves, through alternative splicing of BK channel; mRNAs to alternative isoforms with different sensitivities to phosphorylation-dependent modulation [57]. Recent high throughput proteomic analyses have allowed the identification of a small number of additional phosphosites in BK channels [26, 28, 37]. However, changes in the in vivo phosphorylation state of BK channels at specific sites during neuronal signaling events that regulated BK channel expression, localization and function, have not been observed and remain an important topic for future experiments.
4. Useful phosphoproteomic technologies
The sections above detail recent studies that, through their increased coverage of VGSC and VGKC sequences, and enhanced sensitivity, have allowed for identification of a wealth of novel phosphosites. Advances in LC/MS-based phosphoproteomic technologies, and in sample preparation, phosphopeptide enrichment, chemical tagging, etc. have and will continue to impact studies of the VGSC and VGKC phosphoproteome. Here, we discuss aspects of these methods and their impact on phosphoproteome analysis.
Filter-aided sample preparation (FASP), combines the advantages of in-gel and in-solution protein digestion, and was developed specifically for MS-based proteomics [58]. Analyses of the mouse brain phosphoproteome employing FASP led to the remarkable identification of ≈5,000 phosphosites on ≈2,000 proteins in a single LC-MS experiment on one mg of total mouse brain protein, representing an unprecedented depth of phosphoproteome coverage [24]. Much of the data reviewed above came from this single study in which the FASP technique was applied to analyses of the mouse brain phosphoproteome.
Based on the success of this study, FASP-based analyses will likely form the core of many future phosphoproteome analyses, especially those targeting membrane proteins such as VGSCs and VGKCs. Based on the strong chelating properties of these metals with negatively charged phosphopeptides, immobilizing Fe3+ (in immobilized metal affinity chromatography or IMAC), or TiO2 on beads allows for effective enrichment of phosphopeptides from high complexity mixtures of digested peptides, and these methods have achieved wide use for phospho-protein/peptide enrichment [59]. Hydrophilic interaction chromatography (HILIC) can also be very advantageous for phosphopeptide enrichment, due to the hydrophilic nature phosphopeptides. When HILIC is used prior to IMAC, the selectivity of the metal affinity resin for phosphopeptides versus other acidic peptides is improved to greater than 95% [60, 61]. High-resolution MS instruments, such as Orbitrap and LTQ-FT mass spectrometers, can help reduce false positives in phosphopeptide identification, due to the low part-per-million (ppm) precursor ion mass accuracy. As a tagging method for quantitative analysis, iTRAQ and mTRAQ (Isobaric and multiple tags for relative and absolute quantification) are useful tools [62] for quantifying changes in phosphorylation at specific VGSC and VGKC phosphosites, and how this relates to their dynamic regulation in mammalian brain.
5. What’s next?
Generation of antibodies against many of the principal and auxiliary subunits of VGSCs and VGKCs has allowed not only for their detection on immunoblots and brain sections in analyses of their expression and localization, but has also greatly facilitated their purification. This provides a major opportunity for obtaining VGSCs and VGKCs in sufficient quantities and purity for comprehensive MS-based phosphoproteomic studies. However, validated antibodies suitable for purification are not available for every VGSC and VGKC subunit expressed in brain. Moreover, each purification of the dozens of VGSC and VGKC subunits represents a large undertaking. Improvements in high-throughput proteomic methods are likely to allow for increased insights into the extent and nature the brain VGSC and VGKC phosphoproteome; whether they are able to replace analyses based on target protein purification and in themselves allow for a comprehensive data set remains to be seen.
Since changes in phosphorylation status are dynamic and highly regulated processes, the determination of different mechanisms of VGSC and VGKC phosphorylation related to neurodegenerative diseases (e.g. Alzheimer’s Disease, Parkinson’s Disease and Multiple Sclerosis) and psychiatric/psychological disorders (e.g. addiction, depression, autism and schizophrenia) remain a major focus of ongoing research. Generation of phosphospecific antibodies against specific brain VGSC and VGKC phosphosites may provide effective tools for understanding the role of phosphorylation in the pathophysiology of these diseases, and in some cases, as diagnostic tools for the diseases themselves. In this context, improvements in generation of specific antibodies useful for VGSC and VGKC purification, and in general membrane protein purification methods as they are applied to high-throughput MS approaches are fundamental to increasing our knowledge of the VGSC and VGKC phosphoproteome. Further proteomic method development may allow for quantitative comparisons of VGSCs and VGKCs in small amounts of material from normal and diseased brain, and allow the determination of specific changes associated with disease. The need for normal and diseased brain tissue and detailed clinical histories underscores the importance of high quality tissue banks, and highlights the importance of a close working relationship between basic and clinical researchers. Another major focus is determining the role of phosphorylation at these newly identified sites in regulating the expression, localization and function of neuronal VGSCs and VGKCs, by careful analyses of channel isoforms with phosphorylation-eliminating or –mimicking mutations at specific sites.
The power of sensitive antibody-dependent and independent MS-based analyses of the VGSC and VGKC phosphoproteome represents a new world of possibilities for integrative studies in neuroscience. It is important to remember that the biological significance of any identified phosphosites still relies on careful analyses through classical methodologies such as immunohistochemistry, biochemistry, site-directed mutagenesis, and electrophysiology, and the generation of animal models expressing VGSC and VGKC subunits with altered phosphorylation state.
Supplementary Material
Table II.
Phosphorylation sites identified on VGSC and VGKC auxiliary subunits by MS.
| Auxiliary Subunit | Total | N-terminal | Core | C-terminal |
|---|---|---|---|---|
| Navβ2/Scn2b | 2 | 0 | 0 | 2 |
| Navβ3/Scn3b | 1 | 0 | 0 | 1 |
| Kvβ1/Kcnab1 | 2 | 0 | 1 | 1 |
| Kvβ2/Kcnab2 | 8 | 5 | 2 | 1 |
| KChIP2/Kcnip2 | 3 | 3 | 0 | 0 |
| KChIP3/Kcnip3 | 1 | 1 | 0 | 0 |
Acknowledgments
We thank to Ms. Ashleigh Evans for critical comments on the manuscript, and Dr. Ry Tweedie-Cullen for providing data sets. The work in our laboratory described here was supported by NIH grants NS38343, NS42225, NS64428 and NS64957.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pongs O, Schwarz JR. Ancillary subunits associated with voltage-dependent K+ channels. Physiol Rev. 2010;90:755–96. doi: 10.1152/physrev.00020.2009. [DOI] [PubMed] [Google Scholar]
- 2.Dai S, Hall DD, Hell JW. Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol Rev. 2009;89:411–52. doi: 10.1152/physrev.00029.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 4.Swartz KJ. Towards a structural view of gating in potassium channels. Nat Rev Neurosci. 2004;5:905–16. doi: 10.1038/nrn1559. [DOI] [PubMed] [Google Scholar]
- 5.Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem. 2001;268:5001–10. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 6.Saneyoshi T, Fortin DA, Soderling TR. Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr Opin Neurobiol. 2010;20:108–15. doi: 10.1016/j.conb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–35. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins MO, Grant SG. Supramolecular signalling complexes in the nervous system. Subcell Biochem. 2007;43:185–207. doi: 10.1007/978-1-4020-5943-8_9. [DOI] [PubMed] [Google Scholar]
- 9.Cantrell AR, Catterall WA. Neuromodulation of Na+ channels: an unexpected form of cellular plasticity. Nat Rev Neurosci. 2001;2:397–407. doi: 10.1038/35077553. [DOI] [PubMed] [Google Scholar]
- 10.Schulz DJ, Temporal S, Barry DM, Garcia ML. Mechanisms of voltage-gated ion channel regulation: from gene expression to localization. Cell Mol Life Sci. 2008;65:2215–31. doi: 10.1007/s00018-008-8060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah MM, Hammond RS, Hoffman DA. Dendritic ion channel trafficking and plasticity. Trends Neurosci. 2010;33:307–16. doi: 10.1016/j.tins.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerda O, Trimmer JS. Analysis and functional implications of phosphorylation of neuronal voltage-gated potassium channels. Neurosci Lett. 2010 doi: 10.1016/j.neulet.2010.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa MR, Casnellie JE, Catterall WA. Selective phosphorylation of the alpha subunit of the sodium channel by cAMP-dependent protein kinase. J Biol Chem. 1982;257:7918–21. [PubMed] [Google Scholar]
- 14.Costa MR, Catterall WA. Phosphorylation of the alpha subunit of the sodium channel by protein kinase C. Cell Mol Neurobiol. 1984;4:291–7. doi: 10.1007/BF00733592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, et al. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J Proteome Res. 2006;5:361–9. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berendt FJ, Park KS, Trimmer JS. Multisite phosphorylation of voltage-gated sodium channel alpha subunits from rat brain. J Proteome Res. 2010;9:1976–84. doi: 10.1021/pr901171q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan J, Olsen JV, Park KS, Li W, Bildl W, Schulte U, et al. Profiling the phospho-status of the BKCa channel alpha subunit in rat brain reveals unexpected patterns and complexity. Mol Cell Proteomics. 2008;7:2188–98. doi: 10.1074/mcp.M800063-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park KS, Mohapatra DP, Misonou H, Trimmer JS. Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science. 2006;313:976–9. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- 19.Yang JW, Vacher H, Park KS, Clark E, Trimmer JS. Trafficking-dependent phosphorylation of Kv1.2 regulates voltage-gated potassium channel cell surface expression. Proc Natl Acad Sci U S A. 2007;104:20055–60. doi: 10.1073/pnas.0708574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vacher H, Clark E, Park KS, Yang JW, Trimmer JS. Neuroscience Meeting Planner. Vol. 44.13. San Diego, CA: Society for Neuroscience; 2007. Regulation of Kv1 channel axonal targeting by phosphorylation of Kvbeta2; p. I5. Online. [Google Scholar]
- 21.Dugandzija-Novakovic S, Koszowski AG, Levinson SR, Shrager P. Clustering of Na+ channels and node of Ranvier formation in remyelinating axons. J Neurosci. 1995;15:492–503. doi: 10.1523/JNEUROSCI.15-01-00492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasband MN, Peles E, Trimmer JS, Levinson SR, Lux SE, Shrager P. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J Neurosci. 1999;19:7516–28. doi: 10.1523/JNEUROSCI.19-17-07516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tweedie-Cullen RY, Reck JM, Mansuy IM. Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J Proteome Res. 2009;8:4966–82. doi: 10.1021/pr9003739. [DOI] [PubMed] [Google Scholar]
- 24.Wisniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J Proteome Res. 2010;9:3280–9. doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- 25.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, et al. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol Cell Proteomics. 2007;6:283–93. doi: 10.1074/mcp.M600046-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol Cell Proteomics. 2006;5:914–22. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, et al. Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics. 2008;7:684–96. doi: 10.1074/mcp.M700170-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88:1407–47. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantegazza M, Yu FH, Powell AJ, Clare JJ, Catterall WA, Scheuer T. Molecular determinants for modulation of persistent sodium current by G-protein betagamma subunits. J Neurosci. 2005;25:3341–9. doi: 10.1523/JNEUROSCI.0104-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surti TS, Huang L, Jan YN, Jan LY, Cooper EC. Identification by mass spectrometry and functional characterization of two phosphorylation sites of KCNQ2/KCNQ3 channels. Proc Natl Acad Sci U S A. 2005;102:17828–33. doi: 10.1073/pnas.0509122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballif BA, Carey GR, Sunyaev SR, Gygi SP. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J Proteome Res. 2008;7:311–8. doi: 10.1021/pr0701254. [DOI] [PubMed] [Google Scholar]
- 33.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 34.Roeper J, Lorra C, Pongs O. Frequency-dependent inactivation of mammalian A-type K+ channel KV1.4 regulated by Ca2+/calmodulin-dependent protein kinase. J Neurosci. 1997;17:3379–91. doi: 10.1523/JNEUROSCI.17-10-03379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiger JR, Jonas P. Dynamic control of presynaptic Ca(2+) inflow by fast-inactivating K(+) channels in hippocampal mossy fiber boutons. Neuron. 2000;28:927–39. doi: 10.1016/s0896-6273(00)00164-1. [DOI] [PubMed] [Google Scholar]
- 36.Roeper J, Sewing S, Zhang Y, Sommer T, Wanner SG, Pongs O. NIP domain prevents N-type inactivation in voltage-gated potassium channels. Nature. 1998;391:390–3. doi: 10.1038/34916. [DOI] [PubMed] [Google Scholar]
- 37.Collins MO, Yu L, Coba MP, Husi H, Campuzano I, Blackstock WP, et al. Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem. 2005;280:5972–82. doi: 10.1074/jbc.M411220200. [DOI] [PubMed] [Google Scholar]
- 38.Du J, Haak LL, Phillips-Tansey E, Russell JT, McBain CJ. Frequency-dependent regulation of rat hippocampal somato-dendritic excitability by the K+ channel subunit Kv2.1. J Physiol. 2000;522(Pt 1):19–31. doi: 10.1111/j.1469-7793.2000.t01-2-00019.xm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohapatra DP, Misonou H, Pan SJ, Held JE, Surmeier DJ, Trimmer JS. Regulation of intrinsic excitability in hippocampal neurons by activity-dependent modulation of the KV2.1 potassium channel. Channels (Austin) 2009;3:46–56. doi: 10.4161/chan.3.1.7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakoshi H, Shi G, Scannevin RH, Trimmer JS. Phosphorylation of the Kv2.1 K+ channel alters voltage-dependent activation. Mol Pharmacol. 1997;52:821–8. doi: 10.1124/mol.52.5.821. [DOI] [PubMed] [Google Scholar]
- 41.Misonou H, Menegola M, Mohapatra DP, Guy LK, Park KS, Trimmer JS. Bidirectional activity-dependent regulation of neuronal ion channel phosphorylation. J Neurosci. 2006;26:13505–14. doi: 10.1523/JNEUROSCI.3970-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misonou H, Mohapatra DP, Menegola M, Trimmer JS. Calcium- and metabolic state-dependent modulation of the voltage-dependent Kv2.1 channel regulates neuronal excitability in response to ischemia. J Neurosci. 2005;25:11184–93. doi: 10.1523/JNEUROSCI.3370-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misonou H, Mohapatra DP, Park EW, Leung V, Zhen D, Misonou K, et al. Regulation of ion channel localization and phosphorylation by neuronal activity. Nat Neurosci. 2004;7:711–8. doi: 10.1038/nn1260. [DOI] [PubMed] [Google Scholar]
- 44.Mohapatra DP, Trimmer JS. The Kv2.1 C terminus can autonomously transfer Kv2.1-like phosphorylation-dependent localization, voltage-dependent gating, and muscarinic modulation to diverse Kv channels. J Neurosci. 2006;26:685–95. doi: 10.1523/JNEUROSCI.4620-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohapatra DP, Siino DF, Trimmer JS. Interdomain cytoplasmic interactions govern the intracellular trafficking, gating, and modulation of the kv2.1 channel. J Neurosci. 2008;28:4982–94. doi: 10.1523/JNEUROSCI.0186-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park KS, Mohapatra DP, Trimmer JS. Proteomic analyses of Kv2.1 channel phosphorylation sites determining cell background specific differences in function. Channels (Austin) 2007;1:59–61. doi: 10.4161/chan.4388. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez CC, Zaika O, Tolstykh GP, Shapiro MS. Regulation of neural KCNQ channels: signalling pathways, structural motifs and functional implications. J Physiol. 2008;586:1811–21. doi: 10.1113/jphysiol.2007.148304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoshi N, Zhang JS, Omaki M, Takeuchi T, Yokoyama S, Wanaverbecq N, et al. AKAP150 signaling complex promotes suppression of the M-current by muscarinic agonists. Nat Neurosci. 2003;6:564–71. doi: 10.1038/nn1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajo K, Kubo Y. Protein kinase C shifts the voltage dependence of KCNQ/M channels expressed in Xenopus oocytes. J Physiol. 2005;569:59–74. doi: 10.1113/jphysiol.2005.094995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gamper N, Stockand JD, Shapiro MS. Subunit-specific modulation of KCNQ potassium channels by Src tyrosine kinase. J Neurosci. 2003;23:84–95. doi: 10.1523/JNEUROSCI.23-01-00084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder BC, Kubisch C, Stein V, Jentsch TJ. Moderate loss of function of cyclic-AMP-modulated KCNQ2/KCNQ3 K+ channels causes epilepsy. Nature. 1998;396:687–90. doi: 10.1038/25367. [DOI] [PubMed] [Google Scholar]
- 52.Hernandez CC, Zaika O, Shapiro MS. A carboxy-terminal inter-helix linker as the site of phosphatidylinositol 4,5-bisphosphate action on Kv7 (M-type) K+ channels. J Gen Physiol. 2008;132:361–81. doi: 10.1085/jgp.200810007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fury M, Marx SO, Marks AR. Molecular BKology: the study of splicing and dicing. Sci STKE. 2002;2002:PE12. doi: 10.1126/stke.2002.123.pe12. [DOI] [PubMed] [Google Scholar]
- 54.Lu R, Alioua A, Kumar Y, Eghbali M, Stefani E, Toro L. MaxiK channel partners: physiological impact. J Physiol. 2006;570:65–72. doi: 10.1113/jphysiol.2005.098913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levitan IB. Signaling protein complexes associated with neuronal ion channels. Nat Neurosci. 2006;9:305–10. doi: 10.1038/nn1647. [DOI] [PubMed] [Google Scholar]
- 56.Tian L, Coghill LS, McClafferty H, MacDonald SH, Antoni FA, Ruth P, et al. Distinct stoichiometry of BKCa channel tetramer phosphorylation specifies channel activation and inhibition by cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 2004;101:11897–902. doi: 10.1073/pnas.0402590101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L, Tian L, MacDonald SH, McClafferty H, Hammond MS, Huibant JM, et al. Functionally diverse complement of large conductance calcium- and voltage-activated potassium channel (BK) alpha-subunits generated from a single site of splicing. J Biol Chem. 2005;280:33599–609. doi: 10.1074/jbc.M505383200. [DOI] [PubMed] [Google Scholar]
- 58.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Y, Jensen ON. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9:4632–41. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNulty DE, Annan RS. Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol Cell Proteomics. 2008;7:971–80. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Tolstikov VV. Metabolic analysis. Methods Mol Biol. 2009;544:343–53. doi: 10.1007/978-1-59745-483-4_22. [DOI] [PubMed] [Google Scholar]
- 62.White FM. Quantitative phosphoproteomic analysis of signaling network dynamics. Curr Opin Biotechnol. 2008;19:404–9. doi: 10.1016/j.copbio.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.