Abstract
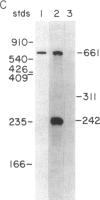
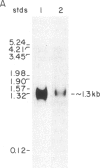
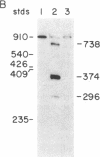
Synthetic oligonucleotides were used to screen a rat striatal cDNA library for sequences corresponding to the tachykinin peptides substance P and neurokinin A. The cDNA library was constructed from RNA isolated from the rostral portion of the rat corpus striatum, the site of striatonigral cell bodies. Two types of cDNAs were isolated and defined by restriction enzyme analysis and DNA sequencing to encode both substance P and neurokinin A. The two predicted preprotachykinin protein precursors (130 and 115 amino acids in length) differ from each other by a pentadecapeptide sequence between the two tachykinin sequences, and both precursors possess appropriate processing signals for substance P and neurokinin A production. The presence of a third preprotachykinin mRNA of minor abundance in rat striatum was established by S1 nuclease protection experiments. This mRNA encodes a preprotachykinin of 112 amino acids containing substance P but not neurokinin A. These three mRNAs are derived from one rat gene as a result of differential RNA processing; thus, this RNA processing pattern further increases the diversity of products that can be generated from the preprotachykinin gene.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A. F., Finnie M. D., Smyth D. G. Mechanism of C-terminal amide formation by pituitary enzymes. Nature. 1982 Aug 12;298(5875):686–688. doi: 10.1038/298686a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Douglass J., Civelli O., Herbert E. Polyprotein gene expression: generation of diversity of neuroendocrine peptides. Annu Rev Biochem. 1984;53:665–715. doi: 10.1146/annurev.bi.53.070184.003313. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Itakura K., Rossi J. J., Wallace R. B. Synthesis and use of synthetic oligonucleotides. Annu Rev Biochem. 1984;53:323–356. doi: 10.1146/annurev.bi.53.070184.001543. [DOI] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J. E., Reiner A. J., Advis J. P., McKelvy J. F. In vivo biosynthesis of [35S]- and [3H]substance P in the striatum of the rat and their axonal transport to the substantia nigra. J Neurosci. 1984 Mar;4(3):775–785. doi: 10.1523/JNEUROSCI.04-03-00775.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Nawa H., Hirose T., Takashima H., Inayama S., Nakanishi S. Nucleotide sequences of cloned cDNAs for two types of bovine brain substance P precursor. Nature. 1983 Nov 3;306(5938):32–36. doi: 10.1038/306032a0. [DOI] [PubMed] [Google Scholar]
- Nawa H., Kotani H., Nakanishi S. Tissue-specific generation of two preprotachykinin mRNAs from one gene by alternative RNA splicing. Nature. 1984 Dec 20;312(5996):729–734. doi: 10.1038/312729a0. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Nicoll R. A., Schenker C., Leeman S. E. Substance P as a transmitter candidate. Annu Rev Neurosci. 1980;3:227–268. doi: 10.1146/annurev.ne.03.030180.001303. [DOI] [PubMed] [Google Scholar]
- Peacock S. L., McIver C. M., Monahan J. J. Transformation of E. coli using homopolymer-linked plasmid chimeras. Biochim Biophys Acta. 1981 Sep 28;655(2):243–250. doi: 10.1016/0005-2787(81)90014-9. [DOI] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. G., Massey D. E. A new glycopeptide in pig, ox and sheep pituitary. Biochem Biophys Res Commun. 1979 Apr 27;87(4):1006–1010. doi: 10.1016/s0006-291x(79)80007-8. [DOI] [PubMed] [Google Scholar]
- Tatemoto K., Lundberg J. M., Jörnvall H., Mutt V. Neuropeptide K: isolation, structure and biological activities of a novel brain tachykinin. Biochem Biophys Res Commun. 1985 Apr 30;128(2):947–953. doi: 10.1016/0006-291x(85)90138-x. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]