Abstract
Serotyping of Salmonella has been an invaluable subtyping method for epidemiologic studies for more than 70 years. The technical difficulties of serotyping, primarily in antiserum production and quality control, can be overcome with modern molecular methods. We developed a DNA-based assay targeting the genes encoding the flagellar antigens (fliC and fljB) of the Kauffmann-White serotyping scheme. Fifteen H antigens (H:a, -b, -c, -d, -d/j, -e,h, -i, -k, -r, -y, -z, -z10, -z29, -z35, and -z6), 5 complex major antigens (H:G, -EN, -Z4, -1, and -L) and 16 complex secondary antigens (H:2, -5, -6, -7, -f, -m/g,m, -m/m,t, -p, -s, -t/m,t, -v, -x, -z15, -z24, -z28, and -z51) were targeted in the assay. DNA probes targeting these antigens were designed and evaluated on 500 isolates tested in parallel with traditional serotyping methods. The assay correctly identified 461 (92.2%) isolates based on the 36 antigens detected in the assay. Among the isolates considered correctly identified, 47 (9.4%) were partially serotyped because probes corresponding to some antigens in the strains were not in the assay, and 13 (2.6%) were monophasic or nonmotile strains that possessed flagellar antigen genes that were not expressed but were detected in the assay. The 39 (7.8%) strains that were not correctly identified possessed an antigen that should have been detected by the assay but was not. Apparent false-negative results may be attributed to allelic divergence. The molecular assay provided results that paralleled traditional methods with a much greater throughput, while maintaining the integrity of the Kauffmann-White serotyping scheme, thus providing backwards-compatible epidemiologic data. This assay should greatly enhance the ability of clinical and public health laboratories to serotype Salmonella.
In the United States, Salmonella serotypes are the basis for the National Salmonella Surveillance System. The identification of serotype for over 30,000 culture-confirmed cases of Salmonella infection each year is the cornerstone of our understanding of Salmonella epidemiology in the United States. (3).
The genus Salmonella is divided into two species, Salmonella enterica and Salmonella bongori (33). S. enterica is further divided into seven subspecies that can be abbreviated by Roman numerals, I, II, IIIa, IIIb, IV, VI, and VII. Subspecies VII was described by multilocus enzyme electrophoresis (MLEE) and by phylogenetic analysis of housekeeping genes (2, 25). Subspecies V is now recognized as the separate species S. bongori (29).
Serotyping further divides the Salmonella subspecies into subtypes, or serovars, by immunologic characterization of two surface structures, O-polysaccharide (O antigen) and flagellin protein (H antigen) (16, 28). Serotyping was initially proposed by Fritz Kauffmann and P. Bruce White in 1934 as a classification scheme for Salmonella (27). The Kauffmann-White serotyping scheme has expanded over the years to include the currently recognized 2,587 serotypes (5, 9, 10). A serotype is represented by an antigenic formula (e.g., I 4,5,12:i:1,2) indicating the subspecies and the O, H1, and H2 antigens. Serotypes in subspecies I are also given names (e.g., Salmonella enterica serotype Typhimurium).
The H antigens of Salmonella have been well described and are primarily encoded by one of two genes, fliC and fljB, which express the phase 1 H antigen and the phase 2 H antigen, respectively (11, 14, 15, 20, 22, 35, 37). The fliC gene is located in one of the flagellar biosynthesis operons; this gene is present in all salmonellae and has homologues in other enteric bacteria (21). The fljB gene is located in a region of the genome that is unique to Salmonella enterica and is present in four of the S. enterica subspecies (subspecies I, II, IIIb, and VI). The two flagellin loci fliC and fljB are coordinately regulated so that only one antigen is expressed at a time in a single cell via a phase variation mechanism (31). Serotypes which express two flagellin types are called diphasic; those with only one flagellar antigen type are considered monophasic. Subspecies IIIa, IV, VII and S. bongori do not contain the fljB operon and are historically monophasic. In rare instances, Salmonella isolates express a third flagellar antigen and the serotype is referred to as triphasic (1, 32).
Currently, there are 114 recognized H antigen types described in the Kauffmann-White scheme (26). Sixty-nine flagellar antigens are characterized by a single epitope which typically shows little or no immunologic relatedness with other H antigens (e.g., H:a, -b, -c, and -d). The remaining H antigens have been described as having multiple epitopes and are grouped into antigenically related complexes. Antigen complexes account for 45 H antigens in the serotyping scheme and consist of a major common epitope plus one or more secondary epitopes. The antigen complexes are named based on the major immunologic epitope (e.g., the 1 complex, the EN complex, the G complex, the L complex, and the Z4 complex). For example, the 1 complex is identified by the major antigen H:1 and further subdivided by the secondary epitopes H:2, -5, -6, and -7. Therefore, an antigen could be represented as H:1,2, -1,5, -1,6, or -1,7 as well as combinations of these single factors, such as H:1,5,7 (17). Eight antigens are recognized in the 1 complex (e.g., H:1,2, -1,5…), 4 in the Z4 complex (e.g., H:z4,z23, -z4,z24…), 3 in the EN complex (e.g., H:e,n,x, -e,n,z15, and -e,n,x,z15), 6 in the L complex (e.g., H:l,v, -l,w, -l,z13…), and 21 in the G complex (H:f,g,t, -g,m…).
Alignment of the fliC and fljB gene sequences shows that the 5′ and 3′ ends of the genes from different antigenic types are highly conserved. The central region sequences of these genes can be highly variable between immunologically distinct antigen types and are presumed to be the basis for the antigenic differences of flagellar antigens (26, 36). The variable regions of the fliC and fljB alleles of immunologically related antigens, however, are highly similar (26). Differences which correlate to these antigen protein sequences can differ in as few as one amino acid residue (26). However, these changes have not been identified as the immunological epitope.
Traditional H antigen serotyping is technologically simple; however, the production and quality control of the hundreds of antisera required to generate the >2,500 serotypes are difficult and time-consuming. Many isolates require 3 to 5 days or more for full determination of the serotype, delaying information submitted into the public health data information systems.
To avoid the difficulties of traditional serotyping methods, we developed a system for determination of serotype based on DNA markers within genes encoding H antigens. By using DNA targets from sequences derived from the H antigen-encoding genes, we form the critical bridge of the molecular data to the surveillance data by mirroring the traditional Kaufmann-White serotyping scheme in a high-throughput DNA-based assay. A similar assay has already been described to detect common O groups of Salmonella (6). The H antigen assay described here, when used in conjunction with the O-group assay, should provide a useful tool for determination of serotype in Salmonella.
MATERIALS AND METHODS
Isolates.
All isolates were from the CDC collection. Isolates were referred to the National Salmonella Reference Laboratory for identification or confirmation of serotype. Serotypes were designated according to the Kauffmann-White scheme (9, 10).
DNA sequences.
The fliC and fljB sequences utilized for the assay design were from reference 26. DNA sequencing of additional fliC and fljB alleles was performed as described previously (26).
DNA alignment and analysis were performed using MEGA 4.1 (18). Neighbor-joining phylogenetic trees were generated using MEGA 4.1. Analysis was performed with 1,000 bootstraps using the Kumar two-parameter model with Jukes-Cantor correction.
Genomic DNA extraction.
Isolates were grown overnight on Trypticase soy agar or in 1 ml Luria broth (LB) (BD Diagnostics, Franklin Lakes, NJ). DNA was extracted with a QIAamp DNeasy kit (Qiagen, Valencia, CA) or Instagene Matrix (Bio-Rad, La Jolla, CA) in accordance with the manufacturer's protocols, except that incubation times were reduced to 4 min at 55°C followed by 4 min at 96 to 100°C. All genomic DNA preparations were quantified by A260/280 spectrophotometry and diluted to 200 ng per μl before use.
Primer and probe design.
Visual OMP software (DNAsoftware, Inc., Ann Arbor, MI) was used for probe and primer design and for experimental simulation to evaluate cross-reactivity, probe binding specificity, and hairpin formation in silico.
Assay for H-antigen determination.
The assay described here uses the final assay conditions developed as described in Results.
(i) Multiplex PCR.
Nine forward primers (5′) and 11 reverse primers (3′) were used to amplify the central variable regions of the fliC and fljB genes (Table 1). The 5′ ends of the reverse primers were biotinylated for use in the hybridization step of the assay. The biotinylated labeled primer was added at a 5-fold excess concentration to the forward primer. Primers were designed to anneal to sequences conserved between different antigens, for example, two forward primers and one reverse primer for amplification of all antigens from the 1 complex, the EN complex, and the L complex.
Table 1.
Primer sequences for the H-antigen PCR assay
| Primer name | Concn (μmol) | Sequence |
|---|---|---|
| 1EN_F1 | 1 | CGCTGAACGTGCAGAAAGAGTATGATGT |
| 1EN_F2 | 1 | AACGTGCAGAAAAAATATGATGT |
| ri_F | 1 | GTGCAACAAAAATATAAGGTCAGC |
| bd_F | 1 | GCGAACGACGGTGAAACTAT |
| G_F | 1 | TCCAGCTTCAAGAATGTTACGGG |
| z51_F | 1 | ATCTAATTTCAAAAACGTTACTGG |
| z29_F | 1 | AAAAAGCCTTGGAATGGATGG |
| Z4_F | 1 | AGCTGGGCTTAGATAAATTAGATGT |
| L_F | 1 | GCAGAAAAAATATGATGTGAAGAGC |
| 1ENL_R | 5 | AACTGCCTTCAATTGTCTTACC |
| ri_R | 5 | CACCCGCTGCTGTCAATG |
| b_R | 5 | CGGTCACCTCAACGAAGTAG |
| G_R | 5 | TATAAACATTTTTGCTTGATTGTAAGG |
| mt_R | 5 | TAGCAGTATATTCAGCTCCCATT |
| c_R | 5 | ATTTATTCGTCAGCAGTTTTTTC |
| y_R | 5 | AACCGCCTTCAATTGTTTTACC |
| eh_R | 5 | CGTCAATAGATTTTCCATTTTTATC |
| z29_R | 5 | CCGCGTTAACAAATGACAGC |
| Z4_R | 5 | GGCAGATTCAAAACGGTTC |
| d_R | 5 | GCATAGCCACCATCAATAACC |
The PCR cycling conditions were described by Fitzgerald et al. (6). The PCRs and hybridizations were performed with a DNA Engine thermocycler (Bio-Rad, Hercules, CA). The amplification parameters were as follows: a preheat step of 95°C for 15 min, followed by 30 cycles of 94°C for 30 s, 48°C for 60 s, and 72°C for 90 s. This was followed by a finishing cycle at 72°C for 10 min.
(ii) Coupling of oligonucleotide probes to beads.
DNA oligonucleotide probes were synthesized with a 5′ amino-linked 6-carbon spacer arm. These were coupled to the carboxylated microsphere polystyrene microspheres (Luminex Corporation, Austin, TX) in accordance with the procedure described in the work of Fitzgerald et al. (6). In brief, 2.5 × 106 microspheres were carbodiimide coupled with 200 pmol of oligonucleotide probe by use of 30 μg/ml freshly prepared N-(3-dimethylaminodipropyl)-N-ethylcarbodiimide (EDC) (Pierce Chemicals, IL). The mixtures were incubated twice for 30 min at room temperature, with 1 μl of freshly prepared EDC added at each 30-min interval. Following coupling, the microspheres were centrifuged for 1 min at 9,000 × g and washed with 1.0 ml of 0.02% Tween 20 (Sigma-Aldrich Corporation, St. Louis, MO), followed by a second wash with 0.5 ml of 0.1% sodium dodecyl sulfate (Sigma-Aldrich Corporation, St. Louis, MO). The microspheres were then suspended in 50 μl of TE buffer (0.01 M Tris-EDTA, pH 8.0; Sigma-Aldrich Corporation, St. Louis, MO) and stored at 4°C in the dark. Individual microspheres coupled to probes were stored in TE at a concentration of approximately 250 microspheres/μl. Labeled beads were pooled into the appropriate microsphere sets corresponding to the complement of antigens desired to be detected.
(iii) Hybridization and detection assay.
A hybridization mixture of microspheres in 1.5× TMAC buffer (1.5 M tetramethylammonium chloride, 75 mM Tris, 6 mM EDTA, and 0.15% Sarkosyl [pH 8.0]) was prepared fresh daily. Ten microliters of microsphere solution per antigen, corresponding to approximately 2,500 coupled microspheres, was added to give a final volume of 1 ml of 1.5× TMAC buffer. For the H-antigen assay, 33 μl of this hybridization mixture was added to 5 μl of H-antigen PCR product and 12 μl of TE (Tris-EDTA, pH 8.0; Sigma-Aldrich, St. Louis, MO) in a low-profile 96-well plate (Bio-Rad, Hercules CA). This reaction mixture was mixed and incubated first at 94°C for 5 min for DNA denaturation, followed by 30 min at 52°C. Following the hybridization reaction, the 96-well plate containing the samples was moved to the brass heating plate in a BioPlex instrument preheated to 52°C.
Seventy-five microliters of detection buffer (R-phycoerythrin-conjugated streptavidin [SAPE] [Invitrogen, Bethesda, MD] diluted to 4 μg/ml in 1× TMAC) was mixed directly into the hybridization reaction mixture. Samples were incubated for 10 min at 52°C in the analyzer and then read by a BioPlex instrument (Bio-Rad, Hercules, CA). The instrument was set to high sensitivity (RP1) for the Cal2 calibration, which produces fluorescence signal relative to the background.
Each probe was assessed based on the signal strength (expressed in median fluorescence intensity [MFI] value) and evaluated relative to the background MFI value. Machine-to-machine and run-to-run variations were normalized by calculating the ratio of the sample MFI value to the negative control (P/N ratio). Acceptable signal strength for each probe was approximately 1,000 or higher with a relative background on high calibration setting of approximately 100 (P/N ratio of 10). However, allowing for variability of both signal and background, we have set the cutoff for what is considered a positive result at a P/N value of 6.
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank under accession numbers HM141979 to HM142068, HQ405742 to HQ405751, and HQ540511 to HQ540513.
RESULTS
DNA Sequence analysis of fliC and fljB alleles.
Most of the fliC and fljB DNA sequences used in target identification and primer and probe design were reported previously; however, 74 additional alleles of fliC and 16 additional alleles of fljB were sequenced in order to characterize antigens whose alleles were noted to be more divergent and to help ensure the specificity of probes that were designed. The alleles sequenced were fliC H:1,5,7 (3 strains), -1,7 (2 strains), -a (16 strains), -b (6 strains), -c (1 strain), -f,g,t (1 strain), -g,m (1 strain), -g,z51 (5 strains), -k (2 strains), -m,t (3 strains), -z27 (4 strains), -z29 (11 strains), -z35 (1 strain), -z4,z23 (4 strains), -z4,z23,z32 (1 strain), -z4,z24 (9 strains), and -z4,z32 (4 strains) and fljB H:1,2 (1 strain), -1,5 (8 strain), -e,n,x (2 strains), -e,n,z15 (2 strains), -e,n,x,z15 (1 strain), and -k (2 strains).
H:a and H:1,5 were the most-divergent antigens; six distinct lineages among 15 H:a serotypes sequenced and five distinct lineages among 26 H:1,5 serotypes were identified (see Fig. S1 and S2 in the supplemental material). Alleles encoding H:t, a secondary antigen within the G complex, fell into three lineages (see Fig. S3 in the supplemental material).
Probe design and evaluation.
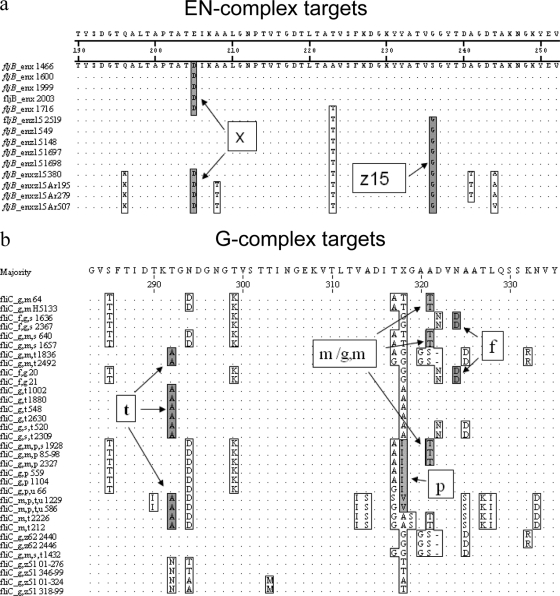
Alignment of closely related fliC and fljB allele sequences and deduced protein sequences identified conserved nucleotide substitutions or amino acid residues corresponding to the antigen type (Fig. 1a and b). The probes were designed to detect a common DNA sequence for all antigens of that type (e.g., H:x) (Fig. 1a) within the variable region of the coding sequence that was distinct from the corresponding sequences for the other antigen types.
Fig. 1.
Location of H-antigen complex secondary antigen target sequences. (a) Conserved EN-complex amino acid sequences targeted by probes to detect single factors H:x and -z15. (b) G-complex amino acid sequences targeted by probes to detect single factors H:f, -m/g,m, -p, and -t.
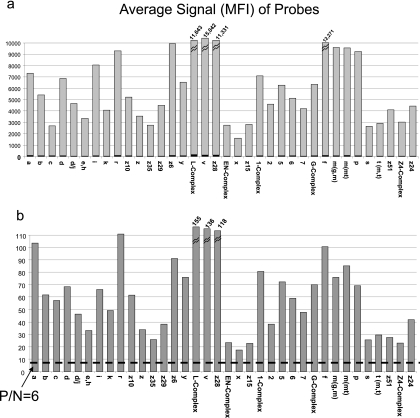
Typically, four probes for each antigen were designed and preliminarily evaluated. The probes were tested with five strains that possessed the antigen that the probe was designed to detect and a panel of 10 to 30 strains possessing different antigens. More strains were used for antigens such as the antigenic complexes where cross-reactivity with immunologically related antigens might be encountered. Probes that demonstrated a P/N ratio of greater than 6.0 when hybridized to the allele of the respective antigen type (Fig. 2) and did not cross-react with closely related antigen types were further evaluated. The second test panel consisted of approximately 10 strains that possessed the antigen of interest from a variety of additional subspecies and serotypes and 20 to 30 appropriate negative controls. Those probes that again met the criteria for a P/N ratio of ≥6 without cross-reacting with other antigen types were next tested on a panel of strains representing the top 100 serotypes. As necessary, additional probes were designed and evaluated in order to obtain probes with the desired specificity. In total, approximately 600 probes were designed and tested in order to identify the 38 probes used in the assay.
Fig. 2.
Signal strength of H antigen probes. (a) Raw data averages for five isolates run in the H antigen assay. The signal strength of each H antigen probe is expressed in median fluorescence units, as indicated by the gray bar. Negative-control signal strength is indicated in black on each bar. (b) Average ratios of positive/negative values for five isolates. A positive reaction is represented by a P/N ratio greater than 6.0, as indicated by a dashed line.
The probes for the final optimized H-antigen assay are listed in Table 2. Thirty-eight probes were used to identify 36 antigens; 15 single antigens (H:a, -b, -c, -d, -j, -e,h, -i, -k, -r, -y, -z, -z10, -z29, -z35, and -z6), the 5 complex major antigens (G, EN, Z4, 1, and L), and 16 secondary epitopes within the complexes (H:2, -5, -6, -7, -f, -m/g,m, -m/m,t, -p, -s, -t, -v, -x, -z15, -z24, -z28, and -z51).
Table 2.
Probe sequences
| H antigen | Sequence |
|---|---|
| a-1a | CCTTCGGCTACATTAAGCACT |
| a-2a | TCTACAACTGATACTGGTTC |
| b | TGCCTATACGCCAAAAGGTACC |
| c | CTGGCGCAGCTAGCTTGAAAG |
| d | ACTACAAAGAAAGTTAATATTGATAC |
| j | ACTGGCGCCGATAAGGAC |
| e,h | GGCGATTCCTTGTCTGCTACG |
| i | TTGATAAGACGAACGGTGAGGT |
| k | ATGGTTTCCTTAAAGTTGACGTTAATAC |
| r | GGCACACCAACAGGACCAAT |
| z10 | AACCAACGAATGCAGTTGAA |
| z | CAATTGGGGCCTCGACTACTA |
| z35 | TGCTAAGAGCGGTTACTATAA |
| z29 | TGGCGCGCACAAAGCAAC |
| z6 | TTACAGACCCAGAAATTGCTG |
| y | AAGCACTACGATGCCTACTGC |
| L-comp | AAAAGGCCAATTAGTTACGAT |
| vc | AATGCCGACAACCACtGAAAG |
| z28 | AACCGCCGCGAAAGTGACA |
| EN-comp-1a | CACTGTAAGTGGTTATAC |
| EN-comp-2a | CACTGTAGGTGGTTATACCGATGC |
| xc | ATATTATTCCACTGTAAGTGtGTTATACC |
| z15c | CTGTAGGTGtGTTATACCGATGC |
| 1-comp | CAATAATGGTACTACACTGGATGTAT |
| 2b,c | GTGtGTACGAATGGTACtGGC |
| 5-1a | GTGGTACGACTGGTACGGC |
| 6 | GTACGCTTGGCACGGCTTCTGTAA |
| 7 | CGAATGGTGCACCTAGTGTAACAGGTA |
| G-comp | AAGCGACAGTGGGTGATCTGA |
| fc | CGGtCGAATGTTGATGtCTGCTAC |
| m/g,m | TATTGCCACTGGCGCGAC |
| m/m,t | GTTTATACTTCCGTTGTAAGCGGTC |
| p | TGATATTGCCATTGGCGCTGGCG |
| s | CTGCGGTTAACCTATTCAtAGACGACTA |
| t-1 | GCTCCAACTGTTCCTGATAAAGTATACGTA |
| z51 | TATTAATTCTGGAGCAGTAACTGATGA |
| Z4-comp | ACCGAGCTGGGCTTAGATAAAT |
| z24 | GCTACCGTAGATAATAGTACTGGG |
Antigens that required more than probe to capture all or most alleles.
The probe contains locked nucleic acid nucleotides (shown in bold).
The probe contains an additional thymine nucleotide(s) indicated as a lowercase t, required for improvement of specificity.
Two probes were needed to detect all H:a and -EN alleles; the two probes for each antigen are coupled to a single microsphere. The H:5 probe, 5-1, which detects the most common lineage of H:1,5, detected most but not all H:1,5 alleles. The H:t probe, t-1, detects one of the three H:t antigen alleles, commonly found in Salmonella enterica serotype Oranienburg. The H:x and -z15 probes did not detect H:e,n,x,z15 alleles, and the H:z28 probes did not detect H:l,z13,z28 alleles. Additional characteristics of the probes are presented in Table 3.
Table 3.
Interpretation of reactivity with specific probes
| Reaction | Interpretation | Comment |
|---|---|---|
| d+ only | H:d | H:d from a Salmonella serotype other than Typhi |
| d+, j+ | H:d | H:d from Salmonella serotype Typhi |
| j+ only | H:j | H:j from Salmonella serotype Typhi |
| a+, 1-comp+, 5+ | Possibly Salmonella serotype Paratyphi A | Salmonella serotype Paratyphi A strains have an inactive H:1,5 allele; confirmed by O-group determination |
| 2+, 5-1 weak | H:2 | H:2 and 5 differ by a single nucleotide in the target region; weak cross-reaction (<500) sometimes seen |
| 5-1+, 2 weak | H:5 | H:2 and 5 differ by a single nucleotide in the target region; weak cross-reaction (<500) sometimes seen |
| x+, z15 weak | H:x | H:x and z15 differ by a single nucleotide in the target region; weak cross-reaction (<500) sometimes seen |
| L-comp+ only | May be H:l,w, -l,z13, -l,z13,z28, or -l,z40 | The secondary antigens in these H antigens are not detected in the assay; confirmed by traditional methods |
| 1-comp+ only | Likely H:1,5 | The H:5 probe in the assay detects the most-common lineages of H:1,5; confirmed by traditional serotyping |
| EN-comp+ only | May be H:e,n,x,z15 | H:e,n,x,z15 alleles do not react with the H:x or H:z15 probes; confirmed by traditional serotyping |
| G-comp+ m/m,t+ | Likely H:m,t | Some H:m,t alleles do not react with the H:t-1 probe, confirmed by traditional serotyping |
| G-comp+ t-1+ | Likely H:g,t | H:g,t is a very rare antigen; confirmed by traditional serotyping |
| G-comp+ f+ | H:f,g or -f,g,t | Some H:f,g,t alleles may not react with the H:t-1 probe; confirmed by traditional serotyping |
| G-comp+ s+ | Likely H:g,s,t | H:g,s,t alleles do not react with the H:t-1 probe, including Salmonella serotype Senftenburg; confirmed by traditional serotyping |
| Z4-comp+ only | May be H:z4z23, -z4,z32, -z4,z23,z32, -z36, -z38, or -z36,z38 | Only secondary antigen H:z24 is detected in the assay; H:z36 and z38 are genetically related to the Z4 complex and are detected with the Z4-comp probe (24) |
For closely related alleles, different methods were employed to increase specificity while maintaining high signal strength. To reduce cross-reaction within a specific region of an oligonucleotide probe, a thymine base was introduced within 4 bases of the target substitution (e.g., H:2, -f, -m/g,m, -p, -s, -v, -x, and -z15). Locked nucleic acid (LNA) bases (Exiqon, Vedbaek, and Denmark) were incorporated into probes (e.g., H:2 and -m/m,t) to increase the hybridization efficiency of the probe with a smaller number of nucleotides in an oligonucleotide probe (Table 2).
The average MFI values ranged from a low of 1,596 (H:x) to a high of 15,042 (H:v). The average signal strength of each probe from five isolates expressing the immunologically defined antigens is reported in Fig. 2a. The corresponding average P/N ratio data for each of the probes are shown in Fig. 2b. The average background signal level in the probes for this assay ranged from 46 to 148.
Evaluation of the assay.
The complete H-antigen assay described here was evaluated for 500 isolates in parallel with the serotype determined by traditional methods (Fig. 3 and Table 4). The molecular-serotyping results matched the results for traditional H-antigen serotype completely for 402 (80%) of the isolates. Forty-six isolates (9.2%) possessed an antigen that was not detected in the assay. In all instances, the partial serotype determined for that isolate matched that determined by traditional serotyping. Nine isolates (1.0%) were determined to be monophasic by traditional methods, but two H antigens were identified in the molecular assay. Four of these were Salmonella enterica serotype Paratyphi A strains, which are known to encode an H:1,5 allele that is not expressed (23), and the other strains were variants of typically diphasic serotypes. Four isolates (1.2%) were nonmotile, and thus, H antigens could not be detected by traditional methods; the flagellar antigens determined by molecular methods for all four strains were consistent with common serotypes.
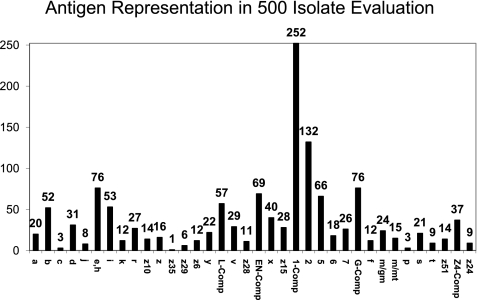
Fig. 3.
Frequency of H antigens among the 500 isolates in the evaluation panel.
Table 4.
Evaluation of the H antigen assay on 500 isolates
| Description | Explanation | No. of isolates | % of total |
|---|---|---|---|
| Identified as expected | |||
| Matched traditional serotyping | Molecular results matched traditional results completely | 402 | 80.4 |
| No probe for one or more antigens | Possessed an H antigen not detected by the probes currently in the assay, resulting in partial serotype determination | 46 | 9.2 |
| Monophasic isolates | Monophasic by traditional methods; two H antigens detected by molecular method; includes 4 serotype Paratyphi A strains | 9 | 1.8 |
| Nonmotile isolates | Nonmotile variant; H antigen detected in molecular assay matched a common serotype. | 4 | 0.8 |
| Subtotal | 461 | 92.2 | |
| Not identified as expected | |||
| H:m,t and -g,s,t | Did not react with t-1 probe; allelic diversity suspected | 19 | 3.8 |
| H:1,5 | Did not react with 5-1 probe; allelic diversity suspected | 6 | 1.2 |
| H:z | All are subspecies IIIb serotypes | 6 | 1.2 |
| H:m,t | Salmonella serotype Pensacola; did not react with m/m,t probe | 2 | 0.4 |
| H:l,v | Salmonella serotype I 9,12:l,v:−; did not react with v probe | 2 | 0.4 |
| H:d | Salmonella serotype Putten; shown to have a divergent H:d allele | 1 | 0.2 |
| H:1,5,7 | Did not react with 5-1 or 7 probe; subspecies II strain | 1 | 0.2 |
| H:e,n,x,z15 | Did not react with x probe; subspecies II strain | 1 | 0.2 |
| H:z35 | Subspecies IIIb strain | 1 | 0.2 |
| Subtotal | 39 | 7.8 | |
| Total | 500 |
Thirty-nine isolates (7.8%) had no reaction with the probes that they were expected to react with based on traditional serotyping, resulting in a partial serotype determined by molecular methods. The specific antigens that were not detected are listed in Table 4. Lack of reactivity with either the 5-1 or the t-1 probe may be attributed to known allelic diversity for these antigens (see Fig. S2 and S3 in the supplemental material). One isolate, a Salmonella enterica serotype Putten (formula, I 13,23:d:l,w) isolate, did not react with the H:d probe. Three additional isolates of Salmonella serotype Putten also resulted in negative a reaction for the H:d probe. Sequence analysis of the H:d allele of Salmonella serotype Putten identified 3-bp differences at the H:d probe target site (see Fig. S4 in the supplemental material). Lack of reactivity in the other strains was not further examined but in some instance may be attributed to the fact that the strains belonged to other subspecies of Salmonella, where sequence divergence may be present.
DISCUSSION
For over 70 years, the Kauffmann-White Salmonella serotyping scheme has served as the primary method of subtyping Salmonella and acted as an international language for microbiologists and public health scientists. Traditional methods for identifying serotype can be problematic because production and quality control of the hundreds of antisera required to identify the >2,500 serotypes are difficult and time-consuming, and many isolates can require 3 to 5 days or more for full determination of the serotype. New methods are being developed using DNA-based assays for serotype identification to replace traditional serotyping (4, 12, 19). Ideally, these new methods should correlate with the Kauffmann-White serotyping scheme in order to preserve the continuity of the epidemiologic information based on serotype and the ability to communicate with laboratories still using traditional methods. Molecular methods based on other surrogate targets may not group the isolates similarly, making it difficult to compare results to historical surveillance data.
In order to preserve the link to serotype determined by traditional methods, we have developed a molecular serotyping assay for the H antigens by targeting the genes encoding the immunologically recognized antigen. In developing the assay, we focused on the 100 most common serotypes among isolates reported from humans in the United States (see Table S1 in the supplemental material). In the United States, more than 161,000 isolates, representing approximately 1,000 different serotypes, were reported to the CDC from 2002 to 2006; however, the 100 most common serotypes accounted for about 98% of the isolates.
The H-antigen assay described here used a 20-primer multiplex PCR to amplify the variable region of the flagellin genes of Salmonella, followed by a hybridization of the PCR amplicons with 38 antigen-specific probes coupled to 36 different microspheres. The H-antigen PCR and hybridization conditions were optimized to match the conditions reported for the Salmonella O group assay (6), which allows the O- and H-antigen assays to be run simultaneously, with only the primer and probe combinations being different.
Signal strength, reported in MFI values, was quite variable between probes. Each probe generally has a characteristic MFI range as well as a typical MFI value for the background. It is unknown why some probes produced stronger MFI values than others (e.g., H:v mean MFI value of 15,042 and H:x mean MFI value of 1,596). Factors such as G+C content, length of oligonucleotide, location of probe target within the amplicon, balance of G+C across the probe sequence, and location of substitutions within the probe have been considered; however, none of these appeared to correlate directly with signal strength. Increased strength of hairpin formation predicted in simulation by probe design software correlated with decreased signal in preliminary tests; therefore, efforts were made to reduce the likelihood of hairpin formation in the design of the probes.
PCR product length has also been suggested to impact signal strength. Preliminary assays with longer PCR amplicons (1,100 to 1,300 bp) produced MFI values in 2,000-to-4,000 MFI value range. However, the levels for five probes (H:2, -m/g,m, -v, -y, and -z29) were unacceptably low (e.g., MFI values of <1,000). To increase the overall MFI values, we designed the 20-primer multiplex PCR described here, which resulted in a PCR amplicon length of 300 to 600 bp. Primers were designed to amplify conserved sequences within multiple antigen groups when possible (e.g., two forward primers and one reverse primer amplify all 1-complex, EN-complex, and L-complex antigen alleles). This strategy maximized MFI signal while minimizing the need for different PCR primers to amplify both fliC and fljB alleles.
In the evaluation of the assay on 500 strains, all probes were specific for the antigen targets for which they were intended; no cross-reactions were observed (Table 4). About 10% of the isolates in the evaluation were not fully serotyped, because they possessed an H antigen not detected in the assay. This is higher than what might be expected during routine use because the National Salmonella Reference Laboratory tends to receive atypical strains for serotype identification or confirmation. Only 381 (76%) of the 500 strains in the evaluation panel were a “top 100” serotype. Among the strains that did not react as expected in the assay (Table 4), the majority possessed the H:m,t, -g,s,t, or -1,5 flagellar antigen, each of which is known to exhibit allelic diversity (see Fig. S2 and S3 in the supplemental material). Several other strains that did not react as expected belonged to subspecies II or IIIb; flagellin alleles in non-subspecies I serotypes were shown to be genetically distinct for H:k and -i (26). Allelic diversity may also be a factor for the flagellar antigen not detected here (H:z in subspecies IIIb and H:z35 and -1,5,7 in subspecies II). Two isolates, serotype I 9,12:l,v:−, did not react with the H:v probe. Sequencing of the fliC and fljB genes from isolates that did not react as expected is needed in order to determine why these strains did not react with the probes as expected.
DNA sequencing of fliC and fljB revealed multiple alleles for some flagellar antigen types. In particular, genes encoding antigens H:a, H:1,5, and H:t consisted of multiple genetic lineages and posed challenges in probe design (see Fig. S1 to S3 in the supplemental material). When a single probe sequence that detected all alleles of a particular antigenic type could not be found, more than one probe, representing the different alleles, was used. All H:a and EN-complex alleles were detected using two probes each. The H:1,5 antigen is encoded by at least five different alleles (see Fig. S2 in the supplemental material). The H:5 probe in the assay detected the most-common H:1,5 lineages. In the evaluation of the assay, six H:1,5 isolates did not react with the 5-1 probe. These were all rare serotypes and may possess one of the H:1,5 alleles not yet detected in the assay.
The G-complex secondary antigens H:m and H:t also had more than one genetic lineage, based on DNA sequence alignment and phylogenetic analysis (J. McQuiston, unpublished observations). The antigenic diversity of H:m is well recognized, and commercially available antiserum is commonly a pool of two different antisera for detection of both H:m types. The different lineages of H:t have been less problematic for traditional serotyping, as they are typically detected with a single, polyclonal rabbit antiserum. Kauffmann described antigenic diversity for H:t that correlates with the allele distribution observed here (17); however, the specifics of this observation have been lost in the literature over the years. In the evaluation, 19 isolates possessing an H:t allele reacted with the G-complex probe but did not react with the t-1 probe, including all Salmonella enterica serotype Senftenberg isolates. This result was expected based on sequences of fliC in these serotypes (see Fig. S3 in the supplemental material); additional probes detecting these alleles are currently being evaluated.
A less common antigen in the EN complex, H:e,n,x,z15, is genetically distinct from the more-common antigens H:e,n,x and H:e,n,z15; however, the secondary complex antigens H:x and H:z15 of H:e,n,x,z15 react with standard H:x and -z15 antisera. H:e,n,x,z15 alleles reacted with the EN-complex probe, but they did not react with the H:x and H:z15 probes; thus, full characterization of H:e,n,x,z15 strains will require additional probes. Similarly, the secondary antigens H:z13 and H:z28 of antigen H:l,z13,z28 are genetically distinct from both H:l,z13 and H:l,z28. H:l,z13,z28 strains reacted only with the L-complex probes and will require additional probes for full characterization as well.
Deletion of a 261-base segment of fliC has been shown to convert flagellar antigen H:d to H:j in Salmonella enterica serotype Typhi (7). The deleted region is flanked by direct repeat sequences, suggesting that the deletion occurs via recombination. Two probes were designed to differentiate H:d and H:j alleles. The H:d probe corresponded to a sequence within the 261-base deletion and detected most H:d alleles. The H:j probe corresponds to a region of fliC that appears to be unique to the H:d allele found in all Salmonella serotype Typhi isolates characterized to date. Thus, reactivity with only the H:d probe would be indicative of H:d in Salmonella serotypes other than Typhi, reactivity with only the H:j probe would be indicative of an H:j variant allele likely in Salmonella serotype Typhi, and reactivity with both would be indicative of H:d alleles likely in Salmonella serotype Typhi (Table 3).
Salmonella has been shown to be unique among the Enterobacteriaceae in that it possesses two different flagellar antigens that are coordinately expressed via a phase variation mechanism (30). However, many Salmonella serotypes possess only one flagellar antigen (e.g., Salmonella enterica serotypes Enteritidis and Typhi); these are commonly referred to as monophasic serotypes. Monophasic variants of typically diphasic serotypes have also been described, where expression of one flagellar antigen in a typically diphasic serotype has been lost (e.g., Salmonella enterica serotype I 4,5,12:i:−) (8). Monophasic variants result from loss or inactivation of the flagellin gene itself, from loss of expression of a flagellar antigen gene, or from a mutation in the phase inversion mechanism which prevents switching between the two flagellar types. Loss or inactivation of a flagellar gene can occur through deletion of all or part of the gene and has been described only for fljB (24). Nonmotile variants arise through a variety of different mutations that result in the inability to produce functional flagella; the flagellin genes are almost always still intact in these isolates.
In the evaluation, nine variants identified as monophasic by traditional methods were identified as diphasic by molecular methods. Depending on the genetic basis for a monophasic serotype or serotype variant, the isolate may or may not type the same by traditional and molecular methods. When fljB is absent, as with most commonly recognized monophasic serotypes and many monophasic variants, the isolate will type the same by both traditional and molecular methods. Isolates that still possess a particular flagellar antigen gene but do not express the antigen will be identified by molecular methods, provided that the antigen is represented in the assay. For example, Salmonella serotype Paratyphi A (antigenic formula, I 2,12:a:−) contains a fljB H:1,5 allele but commonly does not express it (23). This allele is detected in the molecular assay. Isolates that become monophasic through the deletion of part of fljB react differently in the molecular method, depending on the nature of the fljB deletion. If the portion of fljB that is homologous to a particular probe is still present enough so that the primers will amplify the target, this portion will react with that probe even though the antigen is not expressed and cannot be detected by traditional methods. Since nonmotile isolates typically still have at least one intact flagellin gene, they can usually be typed as the serotype or partial serotype from which they were derived in the molecular assay. In the evaluation, four isolates that were nonmotile by traditional methods were typed as common serotypes in the molecular method (Table 4).
Additional DNA targets are being planned for future versions of the assay in order to improve the accuracy and completeness of results. A PCR-probe combination that detects the conserved regions of fljB will indicate that fljB is present when the specific flagellar antigen type encoded by fljB is not detected in the assay or indicate that fljB is absent in monophasic strains that have lost the entire gene. A PCR-probe combination targeting sdf (34), shown to be specific for Salmonella serotype Enteritidis, will differentiate Salmonella serotype Enteritidis from Salmonella enterica serotypes Gallinarum and Pullorum. A PCR-probe combination for detection of viaB, encoding the Vi antigen, will help confirm the identification of Salmonella serotype Typhi, and a three-gene assay will differentiate the Salmonella enterica biovars Paratyphi B and Paratyphi B var. tartrate+ (formerly Salmonella enterica serovar Java).
Use of the liquid microsphere array format allows for flexibility in assay development. Additional probes for antigens not yet covered by the assay can be added one by one as they become available without disruption or redesign of the assay. At this time, this H-antigen assay cannot completely replace traditional methods, since it does not detect all flagellar antigen types. However, it will greatly reduce the time needed to subtype common serotypes, therefore enhancing surveillance activities for Salmonella, and dramatically increase the speed for complete serotyping, as phase reversal would no longer be required. The greatest impact of this assay will result from its use in combination with the O-group assay of Fitzgerald et al. (6). We have found it relatively easy to perform 100 or more O- and H-antigen assays in a single working day. During outbreaks, this assay has been extremely helpful in ruling out nonoutbreak isolates within hours after they are obtained, greatly reducing the workload for traditional methods. The combined O- and H-antigen assay should greatly enhance the serotyping ability of public health laboratories throughout the world.
Supplementary Material
ACKNOWLEDGMENTS
We thank the DFBMD Genomics Unit for assistance with DNA sequencing.
Use of trade names is for identification only and does not imply endorsement by the Public Health Service or by the U.S. Department of Health and Human Services.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 15 December 2010.
REFERENCES
- 1. Baker S., et al. 2007. A novel linear plasmid mediates flagellar variation in Salmonella Typhi. PLoS Pathog. 3:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyd E. F., Wang F. S., Whittam T. S., Selander R. K. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Department of Health and Human Services 2006. Salmonella: annual summary 2006. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ncidod/dbmd/phlisdata/salmtab/2006/SalmonellaAnnualSummary2006.pdf [Google Scholar]
- 4. Echeita M. A., Herrera S., Garaizar J., Usera M. A. 2002. Multiplex PCR-based detection and identification of the most common Salmonella second-phase flagellar antigens. Res. Microbiol. 153:107–113 [DOI] [PubMed] [Google Scholar]
- 5. Ewing W. H. 1972. The nomenclature of Salmonella, its usage, and definitions for the three species. Can. J. Microbiol. 18:1629–1637 [DOI] [PubMed] [Google Scholar]
- 6. Fitzgerald C., et al. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 45:3323–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frankel G. 1989. Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi. EMBO J. 8:3149–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garaizar J., et al. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grimont P. A. D., Weill F.-X. (ed.). 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Centre for Reference and Research on Salmonella, Paris, France [Google Scholar]
- 10. Guibourdenche M., et al. 2010. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 161:26–29 [DOI] [PubMed] [Google Scholar]
- 11. He X. S., Rivkina M., Stocker B. A., Robinson W. S. 1994. Hypervariable region IV of Salmonella gene fliCd encodes a dominant surface epitope and a stabilizing factor for functional flagella. J. Bacteriol. 176:2406–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herrera-Leon S., et al. 2004. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J. Clin. Microbiol. 42:2581–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reference deleted.
- 14. Joys T. M. 1985. The covalent structure of the phase-1 flagellar filament protein of Salmonella typhimurium and its comparison with other flagellins. J. Biol. Chem. 260:15758–15761 [PubMed] [Google Scholar]
- 15. Joys T. M. 1969. Recombination in H1, the gene determining the flagellar antigen-i of Salmonella typhimurium; mapping of H1 and fla mutations. J. Gen. Microbiol. 58:267–275 [DOI] [PubMed] [Google Scholar]
- 16. Judicial Commission of the International Committee on Systematics of Prokaryotes 2005. The type species of the genus Salmonella Lignieres 1900 is Salmonella enterica (ex Kauffmann and Edwards 1952) Le Minor and Popoff 1987, with the type strain LT2T, and conservation of the epithet enterica in Salmonella enterica over all earlier epithets that may be applied to this species. Opinion 80. Int. J. Syst. Evol. Microbiol. 55:519–520 [DOI] [PubMed] [Google Scholar]
- 17. Kauffmann F. 1966. The bacteriology of Enterobacteriaceae, 1st ed., vol. 1, p. 122–124 The Williams and Wilkins Company, Baltimore, MD [Google Scholar]
- 18. Kumar S., Tamura K., Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 19. Leader B. T., Frye J. G., Hu J., Fedorka-Cray P. J., Boyle D. S. 2009. High-throughput molecular determination of Salmonella enterica serovars by use of multiplex PCR and capillary electrophoresis analysis. J. Clin. Microbiol. 47:1290–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MacNab R. M. 1996. Flagella and motility, p. 123–145 In Neidhardt F. C. (ed.), Escherichia coli and Salmonella cellular and molecular biology, 2nd ed., vol. 1 ASM Press, Washington, DC [Google Scholar]
- 21. Macnab R. M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131–158 [DOI] [PubMed] [Google Scholar]
- 22. Masten B. J., Joys T. M. 1993. Molecular analyses of the Salmonella g… flagellar antigen complex. J. Bacteriol. 175:5359–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McClelland M., et al. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268–1274 [DOI] [PubMed] [Google Scholar]
- 24. McQuiston J. R., Fields P. I., Tauxe R. V., Logsdon J. M., Jr 2008. Do Salmonella carry spare tyres? Trends Microbiol. 16:142–148 [DOI] [PubMed] [Google Scholar]
- 25. McQuiston J. R., et al. 2008. Molecular phylogeny of the salmonellae: relationships among Salmonella species and subspecies determined from four housekeeping genes and evidence of lateral gene transfer events. J. Bacteriol. 190:7060–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McQuiston J. R., et al. 2004. Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. J. Clin. Microbiol. 42:1923–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salmonella Subcommittee of the International Society of Microbiology 1934. The genus Salmonella Lignieres, 1900. J. Hyg. (Lond.) 22:333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Popoff M. Y., Bockemuhl J., Gheesling L. L. 2004. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 155:568–570 [DOI] [PubMed] [Google Scholar]
- 29. Reeves M. W., Evins G. M., Heiba A. A., Plikaytis B. D., Farmer J. J., III 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27:313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Silverman M., Simon M. 1980. Phase variation: genetic analysis of switching mutants. Cell 19:845–854 [DOI] [PubMed] [Google Scholar]
- 31. Silverman M., Zieg J., Hilmen M., Simon M. 1979. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc. Natl. Acad. Sci. U. S. A. 76:391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith N. H., Selander R. K. 1991. Molecular genetic basis for complex flagellar antigen expression in a triphasic serovar of Salmonella. Proc. Natl. Acad. Sci. U. S. A. 88:956–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tindall B. J., Grimont P. A., Garrity G. M., Euzeby J. P. 2005. Nomenclature and taxonomy of the genus Salmonella. Int. J. Syst. Evol. Microbiol. 55:521–524 [DOI] [PubMed] [Google Scholar]
- 34. Trafny E. A., Kozlowska K., Szpakowska M. 2006. A novel multiplex PCR assay for the detection of Salmonella enterica serovar Enteritidis in human faeces. Lett. Appl. Microbiol. 43:673–679 [DOI] [PubMed] [Google Scholar]
- 35. Vanegas R. A., Joys T. M. 1995. Molecular analyses of the phase-2 antigen complex 1,2,.. of Salmonella spp. J. Bacteriol. 177:3863–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei L. N., Joys T. M. 1985. Covalent structure of three phase-1 flagellar filament proteins of Salmonella. J. Mol. Biol. 186:791–803 [DOI] [PubMed] [Google Scholar]
- 37. Wei L. N., Joys T. M. 1986. The nucleotide sequence of the H-1r gene of Salmonella rubislaw. Nucleic Acids Res. 14:8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.