Abstract
Protein deacetylase Sirt1 has been implicated in the regulation of hepatic gluconeogenesis; however, the mechanisms are not fully understood. To further elucidate how Sirt1 regulates gluconeogenesis, we took a loss-of-function approach by deleting the coding DNA sequence for the catalytic domain of the Sirt1 gene in the liver of a wild-type mouse (LKOSirt1) or a genetic diabetic mouse in which hepatic insulin receptor substrates 1 and 2 are deleted (DKOIrs1/2). Whereas LKOSirt1 mice exhibited normal levels of fasting and fed blood glucose, inactivation of Sirt1 in DKOIrs1/2 mice (TKOIrs1/2:Sirt1) reduced blood glucose levels and moderately improved systemic glucose tolerance. Pyruvate tolerance was also significantly improved in TKOIrs1/2:Sirt1 mice, suggesting that Sirt1 promotes hepatic gluconeogenesis in this diabetic mouse model. To understand why inactivation of hepatic Sirt1 does not alter blood glucose levels in the wild-type background, we searched for a potential cause and found that expression of small heterodimer partner (SHP, encoded by the Nr0b2 gene), an orphan nuclear receptor, which has been shown to suppress the activity of forkhead transcription factor FoxO1, was decreased in the liver of LKOSirt1 mice. Furthermore, our luciferase reporter assays and chromatin immunoprecipitation analysis revealed that the Nr0b2 gene is a target of FoxO1, which is also regulated by Sirt1. After the gene is upregulated, Nr0b2 can feed back and repress FoxO1- and Sirt1-activated G6pc and Pdk4 gene expression. Thus, our results suggest that Sirt1 can both positively and negatively regulate hepatic gluconeogenesis through FoxO1 and Nr0b2 and keep this physiological process in control.
Keywords: diabetes, insulin receptor substrate, small heterodimer partner, pyruvate dehydrogenase kinase-4
in diabetes, hyperglycemia is partly attributed to dysregulated hepatic glucose production, particularly elevated gluconeogenesis (10, 41). Hepatic gluconeogenesis is regulated by several key enzymes, such as phosphoenoylpyruvate carboxykinase-1 (Pck1), glucose-6-phosphatase (G-6-Pase, the catalytic subunit is encoded by the G6pc gene), and pyruvate dehydrogenase kinase-4 (Pdk-4). Pdk-4 enhances hepatic gluconeogenesis through inhibition of pyruvate dehydrogenase and therefore conservation of pyruvate as gluconeogenic substrates (18, 43). The genes that encode these enzymes are transcriptionally regulated by a number of key transcription factors and coregulators, including FoxO1 (forkhead box O1), HNF-4α (hepatocyte nuclear factor 4α), PGC-1α (peroxisome proliferator-activated receptor-γ coactivator-1α), C/EBPα (CCAAT/enhancer-binding protein-α), CRTC2 (CREB-regulated transcription coactivator 2), STAT3 (signal transducer and activator of transcription 3), and Sirt1 (sirtuin 1) (1, 11, 15, 19, 25, 26, 29–31, 35, 38, 39, 42, 44, 49–52, 54). The critical role of FoxO1 in hepatic gluconeogenesis has been demonstrated using liver-specific Foxo1 knockout mouse models (12, 26). Inactivation of hepatic FoxO1 normalizes blood glucose levels and significantly improves glucose tolerance and insulin sensitivity in diabetic mice with hepatic or systemic insulin resistance (12, 26). Additionally, transcriptional activity of FoxO1 is subject to negative regulation by an orphan nuclear receptor called SHP (small heterodimer partner), which is encoded by the Nr0b2 gene (47, 48). PGC-1α that is regulated by FoxO1 and Sirt1 at transcriptional and posttranslational levels, respectively, also strongly promotes expression of Pck1 and G6pc genes (9, 35, 39). CRTC2 has been shown to play a critical role in short-term gluconeogenesis in response to glucagon (25). Intriguingly, Sirt1 has been shown to confer both positive and negative effects on hepatic gluconeogenesis through differential modulation of the above mentioned factors (2, 13, 15, 28, 30, 39, 40). For example, on the one hand, Sirt1 activates FoxO1 and PGC-1α for the promotion of hepatic gluconeogenesis; on the other hand, Sirt1 suppresses the activity of CRTC2 and HNF-4α to downregulate gluconeogenic gene expression (25, 51).
Previously, several studies were conducted to directly assess the role of Sirt1 in gluconeogenesis in vivo; however, the results have not been quite consistent (6, 13, 25, 36, 37, 40). Acute knockdown of Sirt1 in mouse liver leads to a moderate decrease in blood glucose levels under fed and fasting conditions in addition to moderately improved glucose and pyruvate tolerance (40). In contrast, liver-specific Sirt1 knockout mice maintain normal blood glucose levels (6, 36, 37). Whereas Sirt1 knockdown in liver and adipose tissues by using specific antisense oligonucleotides reduces hepatic glucose production in a rat model of type 2 diabetes (13), adenovirus-mediated overexpression of Sirt1 in liver also lowers blood glucose levels (25). Therefore, further investigation is needed to clarify the role of Sirt1 in gluconeogenesis under physiological and pathological conditions.
We (12) have recently developed a diabetic mouse model that is deficient in insulin receptor substrates (IRS)-1 and -2 specifically in hepatocytes (DKOIrs1/2). DKOIrs1/2 mice manifest hyperglycemia and severe insulin resistance. Strikingly, inactivation of hepatic FoxO1 largely reverses the diabetic phenotype (12). Since FoxO1 is regulated by Sirt1 through deacetylation (15, 21, 28), we hypothesized that inactivation of Sirt1 in DKOIrs1/2 mouse liver might produce similar outcomes to FoxO1 inactivation. However, our results showed that the effect from Sirt1 inactivation was quite different from what we observed in the Foxo1 gene deletion in DKOIrs1/2 mice. Moreover, we have uncovered a novel feedback pathway in the regulation of hepatic gluconeogenesis.
MATERIALS AND METHODS
Animals, blood chemistry, and metabolic analysis.
Irs1, Irs2, and Sirt1 floxed mice were generated as previously described (7, 12, 24). Transgenic mice that carry a Cre coding sequence plus the albumin gene promoter were purchased from the Jackson Laboratory. Irs1 and Irs2 liver-specific double knockout mice (DKOIrs1/2) were generated as described previously (12). To generate Irs1, Irs2, and Sirt1 liver-specific triple knockout mice (TKOIrs1/2:Sirt1), Irs1 and Irs2 double floxed mice were crossed with liver-specific Sirt1 knockout mice (LKOSirt1), and the resultant triple heterozygotes were intercrossed to obtain TKOIrs1/2:Sirt1 mice. All procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Institutional Animal Use and Care Committee of Indiana University School of Medicine. Blood glucose levels were measured using a glucose meter (Contour from Bayer) under ad libitum (fed) or overnight 16-h fasting conditions. Serum insulin was measured using a commercial assay kit (ALPCO). Glucose and insulin tolerance tests, fasting, and refeeding were performed as previously described (12). Pyruvate tolerance tests were essentially similar to glucose tolerance tests except 2 g/kg body wt pyruvate solution used for the injection.
Cell culture and DNA transfection.
Mouse H2.35 cell line was obtained from the American Type Culture Collection (ATTC), and they were maintained in DMEM containing 100 U/ml penicillin and 100 μg/ml streptomycin, 1 g/l glucose, 200 nM dexamethasone, and 4% fetal bovine serum (FBS). Human Hep G2 cells (ATCC) was cultured in DMEM containing 100 U/ml penicillin and 100 μg/ml streptomycin, 4.5 g/l glucose, and 10% FBS. Human HEK 293A cell line was purchased from Invitrogen (Carlsbad, CA), and they were maintained in DMEM containing 100 U/ml penicillin and 100 μg/ml streptomycin, 4.5 g/l glucose, and 10% FBS. DNA transfections were performed using the TurboFect reagent from Fermentas (Glen Burnie, MD). DNA constructs that were used for transfections were cloned into pcDNA3 (Invitrogen) by PCR using the following primers. GFP forward ATGGTGAGCAAGGGCGA, reverse TTACTTGTACAGCTCGTCCATG; mouse Sirt1 forward GCGGACGAGGTGGCGCTCG, reverse TTATGATTTGTCTGATGGATAGTTTACATCTG; mouse Foxo1 forward GCCGAGGCGCCCCAG, reverse TTAGCCTGACACCCAGCTGTGTG; mouse Nr0b2 forward ATGAGCTCCGGCCAGTC, reverse TCACCTCAGCAAAAGCATGTC.
Protein analysis.
Liver tissue or cell culture was homogenized in the lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 1 mM EGTA, 10 mM sodium pyrophosphate, 100 mM sodium fluoride and freshly added 100 μM sodium vanadate, 1 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Protein extracts were resolved on an SDS-PAGE gel and transferred to nitrocellulose membrane. Proteins were probed using the following antibodies: Irs1, Irs2 (Millipore), FoxO1 (Cell Signaling Technology), FLAG (Sigma), Sirt1, PGC-1α, Gck, Nr0b2, β-actin and actinin (Santa Cruz Biotechnology), and Pck1 (Abcam). Protein signals were detected by incubation with horseradish peroxidase-conjugated secondary antibodies, followed by ECL detection reagent (Pierce).
Real-time RT-PCR.
Mouse liver RNA isolation was performed as previously described (12). Real-time RT-PCR (RT-PCR) was performed in two steps: first, cDNA was synthesized using a cDNA synthesis kit (Applied Biosystems); second, cDNA was analyzed by real-time PCR using SYBR Green Master Mix (Promega). Primer sequences for the specific genes are as follows: mouse Ppia forward 5′-CACCGTGTTCTTCGACATCA-3′, reverse 5′-CAGTGCTCAGAGCTCGAAAGT-3′; mouse Irs1 forward 5′-GCCAGAGGATCGTCAATAGC-3′, reverse 5′-AGACGTGAGGTCCTGGTTGT-3′; mouse Irs2 forward 5′-GGTCCAGGCACTGGAGCTTTG-3′, reverse 5′-GGGGCTGGTAGCGCTTCACT-3′; mouse Sirt1 forward 5′-CCCTCAAGCCATGTTTGATA-3′, reverse 5′-ACACAGAGACGGCTGGAACT-3′; mouse Pgc-1α forward 5′-TGAAGTGGTGTAGCGACCAA-3′, reverse 5′-CGCTAGCAAGTTTGCCTCAT-3′; mouse Pck1 forward 5′-ATCATCTTTGGTGGCCGTAG-3′, reverse 5′-TGATGATCTTGCCCTTGTGT-3′; mouse G6pc forward 5′-TCGGAGACTGGTTCAACCTC-3′, reverse 5′-TCACAGGTGACAGGGAACTG-3′; mouse Gck forward 5′-AAGGACAGGGACCTGGGTTCCA-3′, reverse 5′-TCACTGGCTGACTTGGCTTGCA-3′; mouse Pdk4 forward GATTGACATCCTGCCTGACC, reverse CATGGAACTCCACCAAATCC; mouse Nr0b2 forward ACGATCCTCTTCAACCCAGA, reverse AGGGCTCCAAGACTTCACAC; human PPIA forward AGGTCCCAAAGACAGCAGAA, reverse GAAGTCACCACCCTGACACA; human G6PC forward AAGCCGACCTACAGATTTCG, reverse GAGGAAAATGAGCAGCAAGG; human PDK4 forward TGCCTTTGAGTGTTCAAGGA, reverse TGTGAATTGGTTGGTCTGGA.
Primary hepatocyte preparation and adenoviral transduction.
Mouse primary hepatocytes were isolated and cultured as previously described (46). Briefly, primary hepatocytes were isolated from C57BL/6J mice by use of collagenase perfusion under anesthesia. The viability of hepatocytes was assessed by the trypan blue exclusion method. Cells with viability >95% were used for the experiments. The adenoviruses carrying GFP, Sirt1, and FoxO1 coding sequences (with a FLAG tag) were generated using pAdEasy system (Agilent). The adenoviruses carrying GFP (GCATCAAGGTGAACTTCAAGA), Sirt1 (GCACCGATCCTCGAACAATTC), and Foxo1 (GAGCGTGCCCTACTTCAAGGA) shRNA sequences were generated using BLOCK-iT (Invitrogen). Generally, we used 100 multiplicity of infection (MOI) for overexpression and 600 MOI for shRNA knockdown experiments.
Luciferase reporter assay.
Human NR0B2 gene promoter (−606 bp) was cloned by PCR using the following primers: forward ATTCTGTGAGTTCTTCTCAGCA, reverse CAACAACCTTGACTCCAGAAG. Human PDK4 gene promoter (−1322 bp) was cloned using the following primers: forward GTATGACAGGGTAATGTGTCTCA, reverse GTCCCAAACAGGAGGAGTCA. Firefly luciferase reporter system (pGL4.10luc2 and pGL4.74hRluc/TK) was purchased from Promega (Madison, WI). DNA constructs were transfected into mouse H2.35 hepatocytes or HEK 293 cells, and luciferase activity was analyzed using Dual-Luciferase Assay System from Promega.
ChIP assay.
Chromatin immunoprecipitaion (ChIP) assays were performed as previously described (16). Briefly, mouse primary hepatocytes were grown to 90% confluence before they were treated with 1% formaldehyde at 37°C for 15 min. The cross-link reaction was stopped by adding glycine to a final concentration of 125 mM. Chromatin was sonicated to an average size of 150–300 bp. Immunoprecipitation was performed using M2-FLAG antibody (Sigma) following manufacturer's manual. ChIP DNA was analyzed by real-time PCR using the following primers: Nr0b2 proximal forward primer 5′-CATGGAAATGGGCATCAATAG, Nr0b2 proximal reverse primer 5′-TGCCCTTTATCGGATGACTC; Nr0b2 upstream forward primer 5′-GTGCCTGTTAGCCACCAGTT, Nr0b2 upstream reverse primer 5′-GGTGTGTCGGACCTCAAAGT; the internal control Ppia gene promoter forward primer 5′-cagacccacattcctgaggt, reverse primer 5′-aagtcggtgctgtggaagac.
Statistical analysis.
Data are presented as means ± SE. Significance between two groups was assessed using a two-tailed unpaired Student's t-test, and P < 0.05 was considered significant.
RESULTS
Inactivation of hepatic Sirt1 in vivo.
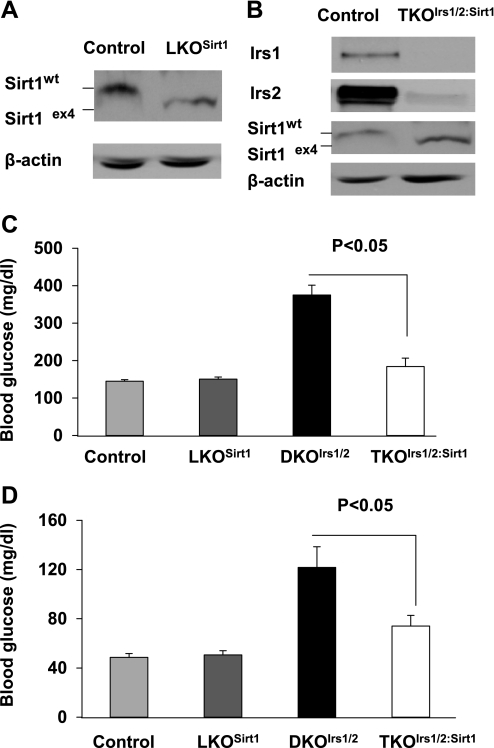
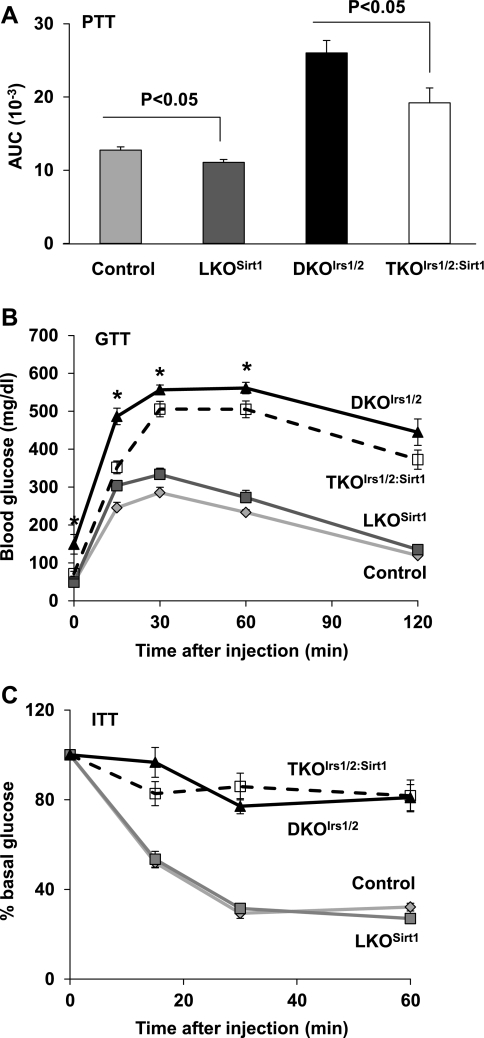
To examine the role of hepatic Sirt1 in glucose homeostasis in vivo, we set out to inactivate Sirt1 in livers of wild-type (LKOSirt1) and Irs1/2-deficient diabetic DKOIrs1/2 mice (TKOIrs1/2:Sirt1). RT-PCR results indicated that Sirt1 mRNA was reduced by 98% in the liver of LKOSirt1 mice, and Irs1, Irs2, and Sirt1 mRNA levels were decreased >95% in the liver of TKOIrs1/2:Sirt1 mice relative to controls (Supplementary Fig. S1, A–D; supplementary materials are found with the online version of this paper at the Jounal website). Western blot analysis also showed that Irs1 and Irs2 proteins were abolished and that Sirt1 mutant protein [exon 4 deletion leads to a 51-amino acid shorter inactive mutant (7)] was smaller in size than the wild-type protein (Fig. 1, A and B). To assess glucose homeostasis, we monitored blood glucose levels in the control and knockout mice. Consistent with previous reports (6, 37), LKOSirt1 mice had normal fed and fasting blood glucose levels (Fig. 1, C and D). As previously reported (12), DKOIrs1/2 mice exhibited hyperglycemia under ad libitum and overnight fasting conditions (Fig. 1, C and D); however, inactivation of hepatic Sirt1 lowered fed and fasting blood glucose levels by 51 and 39% in TKOIrs1/2:Sirt1 mice relative to controls, respectively (Fig. 1, C and D). To examine what caused blood glucose lowering in TKOIrs1/2:Sirt1 mice, we performed the following several tests. Pyruvate tolerance tests showed that inactivation of Sirt1 improved tolerance to exogenous pyruvate injection by 13 and 26% in LKOSirt1 and TKOIrs1/2:Sirt1 mice relative to controls, respectively (Fig. 2A), suggesting that hepatic gluconeogenesis may be decreased in the absence of Sirt1. Glucose tolerance tests showed that LKOSirt1 mice had slight glucose intolerance compared with control mice, whereas TKOIrs1/2:Sirt1 mice had moderate improvement in glucose tolerance relative to DKOIrs1/2 mice (Fig. 2B). However, insulin tolerance tests (ITT) revealed no significant difference between LKOSirt1 and control mice, and DKOIrs1/2 and TKOIrs1/2:Sirt1 mice as well (Fig. 2C). In line with the ITT results, blood insulin levels were not significantly different between LKOSirt1 and control mice and remained high in TKOIrs1/2:Sirt1 mice relative to DKOIrs1/2 mice (Supplementary Fig. S2). The persistent systemic insulin resistance might partly explain the glucose intolerance in the TKOIrs1/2:Sirt1 mice.
Fig. 1.
Sirt1, Irs1, and Irs2 (A and B) protein levels were analyzed in control, LKOSirt1, and TKOIrs1/2:Sirt1 mice by immunoblot analysis. Blood glucose levels were measured in ad libitum-fed 6-wk-old (C) and overnight fasted 2-mo-old (D) control loxp, LKOSirt1, DKOIrs1/2, and TKOIrs1/2:Sirt1 mice (n = 8–10), respectively. Values are presented as means ± SE.
Fig. 2.
A: pyruvate tolerance tests (PTT) were performed on 4-mo-old control and knockout mice (n = 6–8), and data are presented as areas under curve (AUC, artificial units). B: glucose tolerance tests (GTT) were performed on 2-mo-old control and knockout mice (n = 6–9). C: insulin tolerance tests (ITT) were performed on 2-mo-old control and knockout mice (n = 6–8). Values are presented as means ± SE. *P < 0.05 indicates a significant difference between DKOIrs1/2 and TKOIrs1/2:Sirt1 mice.
Hepatic deficiency in Sirt1 and gluconeogenic gene expression.
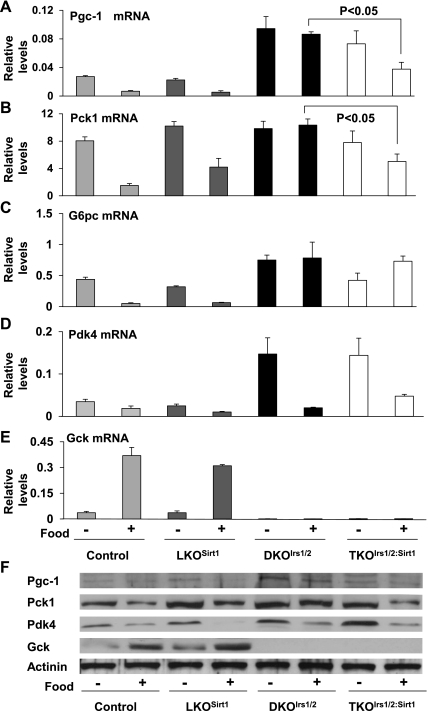
To elucidate the molecular events in glucose metabolism in livers of LKOSirt1 and TKOIrs1/2:Sirt1 mice, we analyzed expression of several genes involved in gluconeogenesis and glycolysis in livers from overnight fasted and 4-h refed mice. Hepatic deficiency of Sirt1 alone did not cause any significant changes in expression of gluconeogenic genes of Pgc1α, Pck1, G6pc, and Pdk4 and the glycolytic gene Gck (Fig. 3, A–E). Consistent with the previous report (12), hepatic deficiency of Irs1 and Irs2 led to elevated expression of Pgc1α, G6pc, and Pck1 but diminished Gck gene expression; however, inactivation of Sirt1 in the DKOIrs1/2 mice resulted in 57 and 51% decreases in Pgc1α and Pck1 gene expression after refeeding, respectively (Fig. 3, A and B). Interestingly, Pdk4 gene expression still responded to food cue even in the livers of DKOIrs1/2 and TKOIrs1/2:Sirt1 mice (Fig. 3D), suggesting that pathways independent of Irs1/2 and Sirt1 might be involved in feeding-mediated Pdk4 gene regulation. Sirt1 inactivation did not improve Gck gene expression under severe deficiency of insulin signaling in TKOIrs1/2:Sirt1 livers (Fig. 3E). Protein levels for Pgc-1a, Pck1, Pdk-4, and Gck were generally consistent with their corresponding mRNA levels described above, and it was notable that Pdk-4 protein diminished much fast in the liver of that LKOSirt1 than that in the control mice in response to feeding (Fig. 3F). Together, these results suggest that hepatic Sirt1 may be involved in dysregulated hepatic gluconeogenesis in the diabetic DKOIrs1/2 mice through modulation of some of the gluconeogenic genes.
Fig. 3.
Hepatic mRNA levels for Pgc1α (A), Pck1 (B), G6pc (C), Pdk4 (D), and Gck (E) genes were analyzed in control loxp, LKOSirt1, DKOIrs1/2, and TKOIrs1/2:Sirt1 mice by real-time RT-PCR after being fasted overnight for 16 h with free access to water or refed ad libitum for another 4 h immediately after fasting. Values are presented as means ± SE; n = 3. Protein levels for Pgc-1α, Pck1, Pdk-4, Gck, and the internal control actinin (F) were also analyzed in the above fasted and refed liver lysates.
Nr0b2 gene is downregulated in the Sirt1-deficient liver.
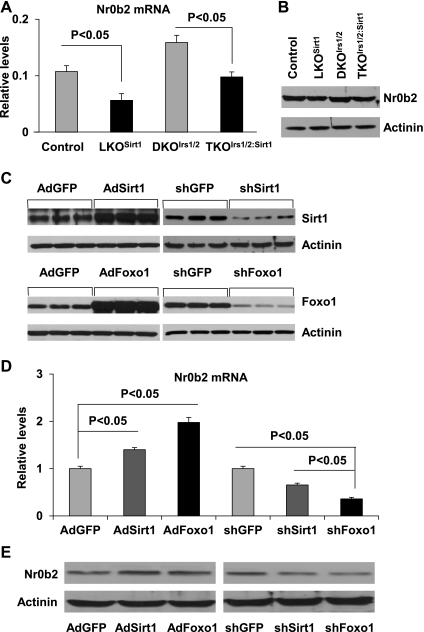
Since inactivation of Sirt1 has less dramatic impact on gluconeogenesis than FoxO1 does as previously reported (12, 26), we thought that there might be a negative feedback mechanism in the Sirt1-FoxO1 pathway. It has been reported that SHP/Nr0b2 can inhibit the transcriptional activity of FoxO1 on the G6pc gene promoter (47). To test whether Sirt1 regulates Nr0b2, we first analyzed the Nr0b2 gene expression in the liver of Sirt1-deficient mice. Indeed, mRNA levels of the Nr0b2 gene were decreased by 47% in LKOSirt1 livers relative to controls (Fig. 4A). Compared with controls, DKOIrs1/2 livers had a 48% increase in the Nr0b2 gene expression, and this was normalized in TKOIrs1/2:Sirt1 livers (Fig. 4A). Consistently, Nr0b2 protein levels were also decreased in the liver of LKOSirt1 and TKOIrs1/2:Sirt1 mice relative to the controls (Fig. 4B). To further verify this observation, we overexpressed or knocked down Sirt1 and FoxO1 in mouse primary hepatocytes (Fig. 4C). Overexpression of Sirt1 and FoxO1 upregulated the Nr0b2 gene expression by 40 and 98% at mRNA levels, respectively, and knockdown of Sirt1 and Foxo1 suppressed the Nr0b2 gene expression by 35 and 64%, respectively (Fig. 4D). The regulation of Nr0b2 gene by Sirt1 and FoxO1 was also confirmed by immunoblot analysis (Fig. 4E). These results suggest that the Nr0b2 gene might be a target gene of Sirt1 and FoxO1.
Fig. 4.
Nr0b2 mRNA (A) and protein levels (B) were analyzed in the liver of control loxp, LKOSirt1, DKOIrs1/2, and TKOIrs1/2:Sirt1 mice by real-time RT-PCR (Values are presented as means ± SE; n = 3) and immunoblotting, respectively. Immunoblot analysis of Sirt1, FoxO1, and Nr0b2 proteins (C and E) and real-time RT-PCR analysis of Nr0b2 mRNA (D) in mouse primary hepatocytes infected with adenoviruses carrying control GFP-, Sirt1-, and Foxo1-overexpressing or shRNA constructs. Values are presented as means ± SE; n = 3.
FoxO1 interacts with the Nr0b2 gene promoter.
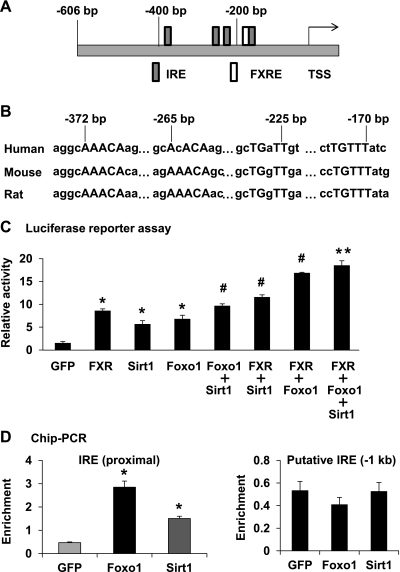
To further investigate the Nr0b2 gene regulation, we first performed an in silico analysis of mammalian Nr0b2 gene promoters and found that there are four conserved putative FoxO1 binding sites (insulin-responsive element, IRE) nearby the previously characterized farnesoid X receptor response element (FXRE) in the proximal promoters of human, mouse, and rat Nr0b2 genes (Fig. 5, A and B). Luciferase reporter assays confirmed that the proximal promoter of human NR0B2 gene could be activated by FXR, FoxO1, and Sirt1 (Fig. 5C). Interestingly, Sirt1 and FXR had additive effects on top of the FoxO1 induction of the NR0B2 gene promoter (Fig. 5C). To examine whether Sirt1 and FoxO1 are associated with the Nr0b2 gene promoter in the chromatin context, we performed ChIP analysis in mouse primary hepatocytes. Indeed, the DNA fragments flanking the conserved IREs (from −300 to −400 bp in mouse Nr0b2 gene promoter) were enriched six- and threefold by FoxO1 and Sirt1 immunoprecipitation, respectively, whereas no specific enrichment was observed in an upstream putative IRE (near −1 kb) (Fig. 5D). These results further suggest that FoxO1 and Sirt1 may regulate the Nr0b2 gene through an interaction with its promoter.
Fig. 5.
A schematic diagram (A) of the proximal promoter of the human NR0B2 gene with locations of putative insulin responsive elements (IRE) and Farnesoid X receptor response element (FXRE) relative to the transcription start site (TSS). DNA sequence alignments (B) among human, mouse, and rat Nr0b2 gene promoters in the four putative IREs with consensus sequences in uppercase. Luciferase reporter assays (C) using human NR0B2 gene promoter were performed in mouse H2.35 hepatocytes by transfection of the reporter constructs together with a single or combined constructs of GFP, FXR, Sirt1, Foxo1. *P < 0.05 for FXR, Sirt1, and Foxo1 vs. GFP overexpression; #P < 0.05 for cotransfection of two factors vs a single factor; **P < 0.05 for the triple transfection vs. Sirt1 cotransfection with Foxo1 or FXR. ChIP PCR analysis of Nr0b2 promoter association (D) was performed in mouse primary hepatocytes infected with GFP, Foxo1, or Sirt1 adenoviruses. DNA enrichment relative to the promoter of the internal control Ppia gene was quantified using real-time PCR. Values are presented as means ± SEM, n = 3. *P < 0.05 indicates a significant difference between enrichments by GFP and Sirt1or Foxo1.
Nr0b2 inhibits Sirt1 and FoxO1activated gluconeogenesis.
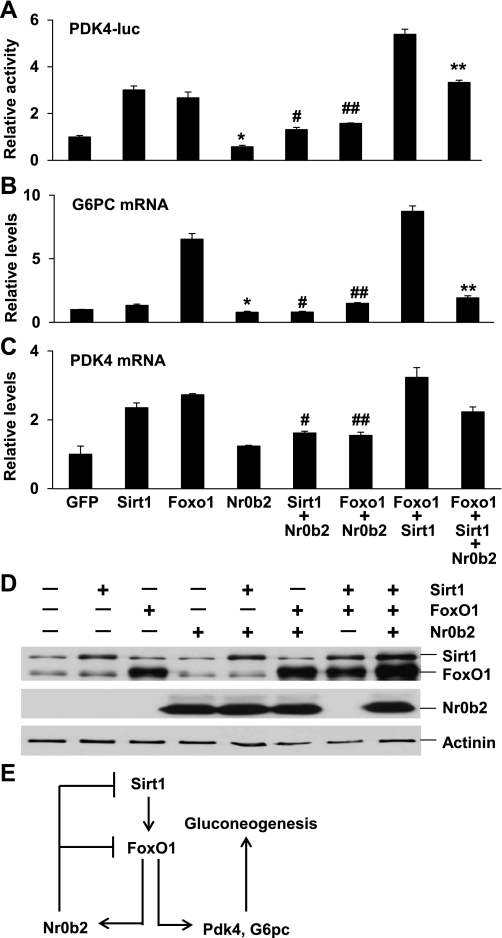
Previously, Nr0b2 has been shown to mediate bile acid inhibition of FoxO1 activity in the regulation of G6pc gene expression (47). To test whether Nr0b2 also represses other gluconeogenic genes, we generated a human PDK4 promoter reporter construct. As previously reported (22), FoxO1 activated the human PDK-4 promoter 2.7-fold relative to GFP control (Fig. 6A). Notably, Sirt1 also induced the promoter activity threefold. In contrast, Nr0b2 suppressed the basal or FoxO1- and Sirt1-activated promoter activity by 42, 56, and 41%, respectively. FoxO1 and Sirt1 also exhibited an additive effect on the activation of the PDK4 promoter; however, this was repressed 38% by Nr0b2. To verify the luciferase reporter results, we analyzed endogenous G6PC and PDK4 gene expression in Hep G2 cells after transfection of relevant DNA plasmids (Fig. 6D). As expected, cotransfection of Nr0b2 with Sirt1, FoxO1, or both led to 40, 77, and 78% reduction of G6PC mRNAs, respectively (Fig. 6B). Consistent with the PDK4 promoter assay results, Sirt1 and FoxO1 individually or collectively induced expression of the PDK4 gene, and their activation was again suppressed 31, 43, and 31% by Nr0b2, respectively (Fig. 6C). These data demonstrate that Nr0b2 can negatively regulate FoxO1- and Sirt1-mediated gluconeogenic gene expression.
Fig. 6.
Luciferase activity (A) driven by the human PDK4 gene promoter was analyzed in HEK293 cells after transfection with indicated DNA plasmids. mRNA levels of G6PC (B) and PDK4 (C) genes were analyzed by real-time RT-PCR in HepG2 cells after transfection with indicated DNA plasmids. Values are presented as means ± SEM. *P < 0.05 for Nr0b2 vs. GFP overexpression; #P < 0.05 for Sirt1+Nr0b2 vs. Sirt1 overexpression; ##P < 0.05 for Foxo1+Nr0b2 vs. Foxo1 overexpression; **P < 0.05 for Foxo1+Sirt1 vs. Foxo1+Sirt1+Nr0b2 overexpression. Overexpression of Sirt1, Foxo1, and Nr0b2 (D) in HepG2 cells was analyzed by immunoblotting with Sirt1 and FoxO1 total protein antibodies and FLAG antibodies (for Nr0b2). A diagram (E) depicting a feedback regulation of hepatic gluconeogenesis through the Sirt1-FoxO1-Nr0b2 pathway. In addition to activation of gluconeogenic genes including Pdk4 and G6pc, FoxO1 also upregulates the Nr0b2 gene, which feeds back onto Sirt1 and FoxO1 to modulate their activity.
DISCUSSION
As an NAD+-dependent deacetylase, Sirt1 has been suggested to play critical roles in nutrient and energy homeostasis (17, 45, 53). However, the role of Sirt1 in glucose homeostasis is not totally clear (2, 3, 13–15, 25, 27, 28, 30, 39, 40, 51). Recently, it has been reported that Sirt1 activators such as SRT1720 can improve type 2 diabetes by increasing systemic insulin sensitivity and mitochondrial capacity and by lowering hepatic glucose production in the Zucker fa/fa obese rat model (27); however, the specificity of SRT1720 is still a matter of debate (8, 32). Systemic overexpression of Sirt1 in mice has been shown to have protective effects against diabetes and insulin resistance-induced inflammation (2, 34). The increase in circulated adiponectin levels is thought to be part of the anti-diabetes mechanism, whereas induction of antioxidant proteins including MnSOD (manganese superoxide dismutase) and NRF1 (nuclear respiratory factor 1) and downregulation of NF-κB (nuclear factor of κ light polypeptide gene enhancer in B-cells) may contribute to lower inflammation in the Sirt1 transgenic mice (2, 34). In the present study, we have used liver-specific knockout mouse models to address hepatic functions of Sirt1 in glucose homeostasis. Although inactivation of Sirt1 alone in the liver does not cause any significant changes in expression of gluconeogenic genes and blood glucose levels in LKOSirt1 mice, pyruvate tolerance is slightly improved, which is consistent with the previous reports by downregulation of Sirt1 using specific shRNA or antisense oligonucleotides (13, 40). Remarkably, Sirt1 inactivation in the DKOIrs1/2 liver significantly improves hyperglycemia in those mice. Whereas glucose tolerance is only moderately improved, pyruvate tolerance is significantly improved, possibly due to attenuated gluconeogenesis. This phenotype is consistent with gene expression profiles, because expression of gluconeogenic genes is decreased in TKOIrs1/2:Sirt1 mice compared with DKOIrs1/2 mice, although glucokinase gene expression is still suppressed. With regard to glucose tolerance, because inactivation of hepatic Sirt1 cannot improve systemic insulin resistance and impaired hepatic glucose utilization, TKOIrs1/2:Sirt1 mice remain largely glucose intolerant.
Although Sirt1 regulates the activities of several key factors involved in hepatic gluconeogenesis, including FoxO1, PGC-1α, STAT3, CRTC2, and HNF-4α (15, 25, 30, 39, 40, 51), the phenotypes of LKOSirt1 and TKOIrs1/2:Sirt1 mice are surprisingly moderate. In contrast, deletion of FoxO1 in the liver of DKOIrs1/2 mice largely normalizes insulin-regulated gene expression and systemic glucose (12). By identifying the Nr0b2 gene as a target gene of Sirt1/FoxO1, now we understand better about the difference in the regulation of hepatic gluconeogenesis by Sirt1 and FoxO1 (Fig. 6E). In addition to regulation through the Sirt1-FXR pathway (20), the Sirt1-FoxO1 pathway also plays an important role in the regulation of the Nr0b2 gene expression. As an orphan nuclear receptor, Nr0b2 has been shown to inhibit numerous nuclear receptors and transcription factors, including HNF-4α, CREB, and FoxO1 (4, 5, 23, 33, 47). Inactivation of hepatic Sirt1 not only attenuates the functions of PGC-1α and FoxO1 due to their hyperacetylation but may also dampen Nr0b2-mediated negative feedback on transcriptional activities of HNF-4α, CREB, and FoxO1 due to decreased gene expression of Nr0b2. This may partly explain why LKOSirt1 mice can still maintain normal glycemia, but we cannot rule out that adaptation might have developed due to chronic Sirt1 inactivation, because acute knockdown of Sirt1 in mouse liver lowers both fasting and fed glucose levels (40). In the case of DKOIrs1/2 mice, expression of gluconeogenic genes is highly elevated, and inactivation of Sirt1 might have a greater impact on gluconeogenesis in TKOIrs1/2:Sirt1 mice than in LKOSirt1 because both mRNA and protein levels of PGC-1α and Pck1 are decreased in TKOIrs1/2:Sirt1 mice. Since Nr0b2 mRNA and protein levels are higher in TKOIrs1/2:Sirt1 than those in LKOSirt1 liver, feedback inhibition on gluconeogenesis by Nr0b2 might be greater as well. Consistently, Sirt1 knockdown in the liver by specific antisense oligonucleotides lowers hepatic glucose production only in diabetic but not in normal rats (13). However, in the case of FoxO1 knockout, because Nr0b2 can no longer impact FoxO1-mediated gluconeogenesis, hepatic gluconeogenesis is remarkably decreased (12, 47). In addition, unlike downregulation of gluconeogenic genes, inactivation of Sirt1 in the DKOIrs1/2 liver does not alter Gck gene expression as FoxO1 does (12), although knockdown of Sirt1 can upregulate Gck gene expression in mouse liver (40), suggesting that insulin plays a predominant role in the regulation of Gck gene, partly through FoxO1.
In summary, hepatic Sirt1 normally modulates hepatic glucose production during fasting through balanced regulation of gluconeogenic genes. One of the mechanisms that confer feedback regulation of hepatic gluconeogenesis is through Sirt1- and FoxO1-controlled Nr0b2 gene expression. This feedback mechanism ensures gluconeogenesis under control.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R00-DK-077505 to X. C. Dong.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Fred Alt for providing the Sirt1 floxed mice, Dr. Anna Depaoli-Roach for helping with the import of the animal colony, and Dr. Robert Harris for providing the PDK4 antibody.
REFERENCES
- 1. Altomonte J, Richter A, Harbaran S, Suriawinata J, Nakae J, Thung SN, Meseck M, Accili D, Dong H. Inhibition of Foxo1 function is associated with improved fasting glycemia in diabetic mice. Am J Physiol Endocrinol Metab 285: E718–E728, 2003 [DOI] [PubMed] [Google Scholar]
- 2. Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab 8: 333–341, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caton PW, Nayuni N, Kieswich J, Khan N, Yaqoob M, Corder R. Metformin suppresses hepatic gluconeogenesis through induction of SIRT1 and GCN5. J Endocrinol 205: 97–106, 2010 [DOI] [PubMed] [Google Scholar]
- 4. Chanda D, Park JH, Choi HS. Molecular basis of endocrine regulation by orphan nuclear receptor small heterodimer partner. Endocr J 55: 253–268, 2008 [DOI] [PubMed] [Google Scholar]
- 5. Chanda D, Xie YB, Choi HS. Transcriptional corepressor SHP recruits SIRT1 histone deacetylase to inhibit LRH-1 transactivation. Nucleic Acids Res 38: 4607–4619 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev 22: 1753–1757, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA 100: 10794–10799, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Perni RB, Riera TV, Szczepankiewicz B, Vlasuk GP, Stein RL. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem 285: 32695–32703, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes 52: 642–649, 2003 [DOI] [PubMed] [Google Scholar]
- 10. DeFronzo RA. Current issues in the treatment of type 2 diabetes. Overview of newer agents: where treatment is going. Am J Med 123: S38–S48, 2010 [DOI] [PubMed] [Google Scholar]
- 11. Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449: 366–369, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Dong XC, Copps KD, Guo S, Li Y, Kollipara R, Depinho RA, White MF. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab 8: 65–76, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, Murray SF, Bhanot S, Monia BP, Horvath TL, Gao Q, Samuel VT, Shulman GI. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci USA 106: 11288–11293, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 8: 347–358, 2008 [DOI] [PubMed] [Google Scholar]
- 15. Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem 280: 20589–20595, 2005 [DOI] [PubMed] [Google Scholar]
- 16. Ghoshal K, Datta J, Majumder S, Bai S, Dong X, Parthun M, Jacob ST. Inhibitors of histone deacetylase and DNA methyltransferase synergistically activate the methylated metallothionein I promoter by activating the transcription factor MTF-1 and forming an open chromatin structure. Mol Cell Biol 22: 8302–8319, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5: 253–295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harris RA, Bowker-Kinley MM, Huang B, Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv Enzyme Regul 42: 249–259, 2002 [DOI] [PubMed] [Google Scholar]
- 19. Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, Hashimoto N, Kido Y, Mori T, Sakaue H, Teshigawara K, Jin S, Iguchi H, Hiramatsu R, LeRoith D, Takeda K, Akira S, Kasuga M. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med 10: 168–174, 2004 [DOI] [PubMed] [Google Scholar]
- 20. Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab 10: 392–404, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab 2: 153–163, 2005 [DOI] [PubMed] [Google Scholar]
- 22. Kwon HS, Huang B, Unterman TG, Harris RA. Protein kinase B-alpha inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes 53: 899–910, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Lee JM, Seo WY, Song KH, Chanda D, Kim YD, Kim DK, Lee MW, Ryu D, Kim YH, Noh JR, Lee CH, Chiang JY, Koo SH, Choi HS. AMPK-dependent repression of hepatic gluconeogenesis via disruption of CREB/CRTC2 complex by orphan nuclear receptor SHP. J Biol Chem 285: 32182–32191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin X, Taguchi A, Park S, Kushner JA, Li F, Li Y, White MF. Dysregulation of insulin receptor substrate 2 in beta cells and brain causes obesity and diabetes. J Clin Invest 114: 908–916, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, III, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456: 269–273, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab 6: 208–216, 2007 [DOI] [PubMed] [Google Scholar]
- 27. Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450: 712–716, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell 116: 551–563, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Nakae J, Kitamura T, Silver DL, Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest 108: 1359–1367, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol 11: 492–500, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Onuma H, Vander Kooi BT, Boustead JN, Oeser JK, O'Brien RM. Correlation between FOXO1a (FKHR) and FOXO3a (FKHRL1) binding and the inhibition of basal glucose-6-phosphatase catalytic subunit gene transcription by insulin. Mol Endocrinol 20: 2831–2847, 2006 [DOI] [PubMed] [Google Scholar]
- 32. Pacholec M, Chrunyk BA, Cunningham D, Flynn D, Griffith DA, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem 285: 8340–8351, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park MJ, Kong HJ, Kim HY, Kim HH, Kim JH, Cheong JH. Transcriptional repression of the gluconeogenic gene PEPCK by the orphan nuclear receptor SHP through inhibitory interaction with C/EBPalpha. Biochem J 402: 567–574, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA 105: 9793–9798, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423: 550–555, 2003 [DOI] [PubMed] [Google Scholar]
- 36. Purushotham A, Schug TT, Li X. SIRT1 performs a balancing act on the tight-rope toward longevity. Aging (Albany NY) 1: 669–673, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 9: 327–338, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiao L, Macdougald OA, Shao J. CCAAT/enhancer-binding protein alpha mediates induction of hepatic phosphoenolpyruvate carboxykinase by p38 mitogen-activated protein kinase. J Biol Chem 281: 24390–24397, 2006 [DOI] [PubMed] [Google Scholar]
- 39. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434: 113–118, 2005 [DOI] [PubMed] [Google Scholar]
- 40. Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA 104: 12861–12866, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Savage DB, Petersen KF, Shulman GI. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 87: 507–520, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schilling MM, Oeser JK, Boustead JN, Flemming BP, O'Brien RM. Gluconeogenesis: re-evaluating the FOXO1-PGC-1alpha connection. Nature 443: E10–E11, 2006 [DOI] [PubMed] [Google Scholar]
- 43. Sugden MC, Holness MJ. Therapeutic potential of the mammalian pyruvate dehydrogenase kinases in the prevention of hyperglycaemia. Curr Drug Targets Immune Endocr Metabol Disord 2: 151–165, 2002 [PubMed] [Google Scholar]
- 44. Wang JC, Stromstedt PE, Sugiyama T, Granner DK. The phosphoenolpyruvate carboxykinase gene glucocorticoid response unit: identification of the functional domains of accessory factors HNF3 beta (hepatic nuclear factor-3 beta) and HNF4 and the necessity of proper alignment of their cognate binding sites. Mol Endocrinol 13: 604–618, 1999 [DOI] [PubMed] [Google Scholar]
- 45. Westphal CH, Dipp MA, Guarente L. A therapeutic role for sirtuins in diseases of aging? Trends Biochem Sci 32: 555–560, 2007 [DOI] [PubMed] [Google Scholar]
- 46. Xiong Y, Collins QF, An J, Lupo E, Jr, Liu HY, Liu D, Robidoux J, Liu Z, Cao W. p38 Mitogen-activated protein kinase plays an inhibitory role in hepatic lipogenesis. J Biol Chem 282: 4975–4982, 2007 [DOI] [PubMed] [Google Scholar]
- 47. Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem 279: 23158–23165, 2004 [DOI] [PubMed] [Google Scholar]
- 48. Yamagata K, Yoshimochi K, Daitoku H, Hirota K, Fukamizu A. Bile acid represses the peroxisome proliferator-activated receptor-gamma coactivator-1 promoter activity in a small heterodimer partner-dependent manner. Int J Mol Med 19: 751–756, 2007 [PubMed] [Google Scholar]
- 49. Yamamoto T, Shimano H, Nakagawa Y, Ide T, Yahagi N, Matsuzaka T, Nakakuki M, Takahashi A, Suzuki H, Sone H, Toyoshima H, Sato R, Yamada N. SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem 279: 12027–12035, 2004 [DOI] [PubMed] [Google Scholar]
- 50. Yang J, Kalhan SC, Hanson RW. What is the metabolic role of phosphoenolpyruvate carboxykinase? J Biol Chem 284: 27025–27029, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang J, Kong X, Martins-Santos ME, Aleman G, Chaco E, Liu GE, Wu SY, Samols D, Hakimi P, Chiang CM, Hanson RW. Activation of SIRT1 by resveratrol represses transcription of the gene for the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) by deacetylating hepatic nuclear factor 4alpha. J Biol Chem 284: 27042–27053, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413: 131–138, 2001 [DOI] [PubMed] [Google Scholar]
- 53. Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann NY Acad Sci 1173, Suppl 1: E10–E19, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem 281: 10105–10117, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.