Abstract
Purpose
The purpose of this study was to evaluate baseline RRM2 protein and gene expression in tumors of patients receiving 3-AP.
Experimental Design
Tumor blocks from patients enrolled in phase I and II clinical studies using 3-AP, were evaluated for RRM2 gene and protein expression by quantitative real time polymerase chain reaction (Q-RTPCR) and automated quantitative analysis, (AQUA).
Results
Esophageal and gastric cancers overexpressed RRM2 protein when compared to prostate cancer (Z-score, 0.68 ± 0.94 SD, vs 0.41 ± 0.84SD, respectively,; p=0.04). Esophageal and gastric cancers also overexpressed RRM2 mRNA when compared to prostate cancer (relative gene expression 2.56 ± 1.49 SD, vs 0.29 ± 0.20 SD, respectively,; p=0.02). Protein and gene expression were moderately associated (Spearman's rank correlation =0.30; p=0.12).
Conclusion
RRM2 gene and protein expression varies by tumor type and may predict response to 3-AP.
Keywords: Triapine®, 3-aminopyridine-2-carboxaldehyde thiosemicarbazone, ribonucleotide reductase, automated quantitative immunohistochemistry (AQUA), quantitative real time PCR
Introduction
Ribonucleotide reductase (RR) is the enzyme that catalyzes the conversion of ribonucleoside 5′-diphosphates into 2′-deoxyribonucleotides (1). RR consists of one regulatory subunit, RRM1, and one catalytic subunit, RRM2. Together, RRM1 and RRM2 form the catalytically active RR enzyme. RRM1 has binding sites for ribonucleotides, and its expression in proliferating cells remains consistent throughout the cell cycle. RRM2 contains a tyrosyl-free radical that is stabilized by a non-heme iron center, which is essential for ribonucleotide reduction and conversion of nucleotides to deoxynucleotides (2). This conversion is a rate-limiting step in the production of 2′-deoxyribonucleoside 5′-triphosphates that are necessary for DNA replication, making RRM2 essential in DNA synthesis.
Overexpression of RR2M is associated with increased cell proliferation (3) and malignant potential in certain cancers and inhibition of RRM2 reduces cellular proliferation in vitro and in vivo (4,5). RRM2 interacts with a variety of oncogenes which promotes tumor progression (6,7), enhances the invasiveness of cancer cells (8), reduces radiosensitivity in human solid tumors (9) and increases the drug-resistant properties of cancer cells to various chemotherapeutic reagents, including hydroxyurea and gemcitabine (10-13). Therefore, inhibition of RRM2 is a potential therapeutic target for new anticancer agents. Triapine© (3-aminopyridine-2-carboxaldehyde thiosemicarbazone, 3-AP, Vion Pharmaceuticals, Inc., New Haven, CT), is a novel small molecule inhibitor of RRM2 that is being evaluated in Phase I and II clinical trials. In this study, we hypothesized that baseline tumor RRM2 expression varies by tumor and RRM2 expression may be used to identify tumors sensitive to 3-AP.
Materials and Methods
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were obtained from 40 of the 43 patients with locally advanced, unresectable or metastatic solid tumors who participated in three clinical trials at the University of Wisconsin: [a phase I combination of 3-AP and doxorubicin (14), a phase I combination of 3-AP and irinotecan (15) and a phase II single agent study of 3-AP in pancreatic cancer (16)]. There were thirteen tissue blocks from patients with pancreatic cancer (four from primary sites and nine from metastatic sites). Other tissue types included,:one primary bladder cancer; one primary cervical cancer; four cholangiocarcinoma (one primary, three metastatic); two primary colon cancers; three primary esophageal cancers, one primary gastric cancer, one metastatic lymphoma, three primary melanomas, two primary mesotheliomas, one primary non-small cell lung cancer (NSCLC), two primary prostate cancers, one primary sarcoma and one primary small cell lung cancer (SCLC). All samples were available for automated quantitative immunohistochemistry (AQUA) analysis. Seven individuals with pancreas cancer, two with breast cancer and one with lymphoma did not have sufficient material for RNA analysis. The Health Sciences Institutional Review Board of the University of Wisconsin approved these trials prior to their implementation, and all patients gave informed written consent.
Laser Capture Microdissection
Sections were prepared from each FFPE tissue blocks, and hematoxylin and eosin (H&E) staining was performed. H&E stained slides were reviewed by a pathologist to determine the location of tumor tissue on each slide. Laser capture microdissection (LCM) with the SL μCut Laser Microdissection System (Molecular Machines & Industries, Glattbrug, Switzerland) was utilized to ensure the isolation of only tumor cells. Briefly, the sections were melted at 60°C for 30 min and deparaffinized in xylene. Sections were then rehydrated in graded ethanols, rinsed with DEPC-treated water, stained with toluidine blue, rinsed in DEPC-treated water, dehydrated in graded ethanols followed by isopropanol and placed in a desiccator until dry.
RRM2 Gene Expression Analysis by Quantitative Reverse Transcriptase Polymerase Chain Reaction (Q-RTPCR)
After LCM, RNA extraction was performed using the Paradise Whole Transcript RT Reagent System (Arcturus Bioscience, Sunnyvale, CA) which processes FFPE tissue scrapes. Caps were placed in a microcentrifuge tube-containing proteinase K and incubated at 37°C for 16-20 hours. After centrifugation, the caps were removed and the RNA was isolated and treated with DNase following the manufacture's instructions. The total RNA was resuspended and then treated with DNase. Total RNA was stored at -80°C until analyzed. The RNA was quantified via NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE), and the total RNA extracted from FFPE tumor tissue was reverse transcribed using random primers by standard methods.
TaqMan assays were designed for the genes listed in Table 1 using Integrated DNA Technologies (Coralville, IA), keeping the amplicon length < 100bp. Quantitative real-time PCR was performed using the Bio-Rad iCycler IQ system (Hercules, CA). Due to the limited sample supply, the target gene (RRM2) and the endogenous reference gene (YWHAZ) were amplified in a single well. Each well contained 5 pmol/μL of the probes, 5 pmol/μL of the primers, and 12.5 μL of iQ Multiplex Powermix (Bio-Rad) in a 25 μL final reaction mixture. The Multiplex Powermix was heat-activated for 3 min at 95°C. Each of the 50 PCR cycles consisted of 15 sec of denaturation at 95°C and hybridization of primers and probes for 45 sec at 60°C.
Table 1. Sequences of the primers and probes used in quantitative PCR studies.
| Gene | Oligonucleotide | Sequence | PCR product size (bp) | GenBank Accession Number |
|---|---|---|---|---|
| YWHAZ | Forward primer | 5′-CCAATGCTTCACAAGCAGAGAGCA-3′ | 99 | NM_003406 |
| Reverse primer | 5′-CTTTCTTGTCATCACCAGCGGCAA-3′ | |||
| Probe | 5′-fAGGAGATTACTACCGTTACTTGGCTGAGGq-3′ | |||
| RRM2 | Forward primer | 5′-TTTAGTGAGCTTAGCACAGCGGGA-3′ | 89 | NM_001034 |
| Reverse primer | 5′-AAATCTGCGTTGAAGCAGTGAGGC-3′ | |||
| Probe | 5′-rACAGTCCTTTAACCAGCACAGCCAGTq-3′ |
f denotes the fluorescein FAM, r denotes the fluorescein TET, q denotes Black Hole Quencher 1
Expression levels in the patient samples were determined by the standard curve method using standard cDNA solutions which were serially diluted 5-fold from HepG2 carcinoma cell line. The standard curve and samples were run in triplicate, and nontemplate controls were included in each run. The data was analyzed with the standard curve line equations generated by iQ5 software (Bio-Rad). The starting mass value for the gene of interest and reference gene were calculated by substituting the threshold cycle (Ct) values generated by the iQ5 software into the standard curve formula. The Ct (threshold cycle) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The data was expressed as target gene (RRM2)/endogenous gene (YWHAZ). The correlation coefficient (r) for each standard curve exceeded 0.99, and the coefficient of variation for the Ct values was less than 3.5% for all replicates.
Validation Summary
Linearity was determined with five standard curves, and the mean coefficient of determination (r2) for RRM2 was 0.990 (range 0.982-0.998) and for YWHAZ was 0.996 (range 0.994-0.998). Triplicate determinations of each standard were run on a single plate on five separate occasions over a two-week period. The intra-day variability in the standard Ct readings for RRM2 averaged 0.65% (range 0.09-2.84%), while for YWHAZ the standard Ct readings averaged 0.61% (range 0.06-1.96%). The mean CV in the Ct values for RRM2 over this time ranged from 0.32% (range 0.09-0.68%) for the 400 ng standard to 1.47% (range 0.66-2.84%) for the 0.128 ng standard. For YWHAZ, the mean CV in the Ct values over the standard curve ranged from 0.54% (range 0.31-1.03%) for the 400 ng standard to 1.11% (0.33-1.96%) for the 0.128 ng standard. Triplicate determinations of Ct readings in five patient samples on a single plate had a mean CV for RRM2 of 1.18% (range 0.25-2.21%) over the mass range of 0.19-114.43 ng (all samples were diluted). In the same five patient samples, the mean CV for YWHAZ was 0.70% (range 0.03-1.14%) over the mass range of 0.13-116.12 ng (all samples were diluted). The variability did not change with concentration.
M2 Protein Analysis by Automated Quantitative Analysis (AQUA)
RRM2 expression was determined using the AQUA system (HistoRx, New Haven, CT) as previously described (17-19). Initially, target compartments were localized using a fluorescently tagged rabbit anti-cytokeratin antibody (or the anti-S100 antibody for melanoma cells). 4,6-Diamidino-2-phenylindole (DAPI) was added to visualize nuclei. RRM2 was visualized with an Alexa Fluor 488 labeled tyramide which, like diaminobenzidine, is activated by horseradish peroxidase and results in the deposition of numerous covalently associated Alexa Fluor 488 dyes immediately adjacent to the horseradish peroxidase-conjugated secondary antibody (Figure 1). Using this approach, classical compartments are defined on the basis of molecular co-localization. The cytokeratin compartment is equivalent to all epithelial cells in the tissue section. The S-100 compartment is equivalent to melanoma cells in tissue section. DAPI is the area defined as the cell nucleus. A matched set of H&E sections were used for locating tumor. RRM2 was tagged and measured within the subcellular compartments by the PLACE algorithm as previously described. Data is represented as a Z-score, which was calculated by subtracting the mean AQUA score from the individual AQUA score and dividing by the standard deviation (18,19). Therefore a negative Z-score indicates the protein expression was less than the average and a positive Z-score means it is greater than the average.
Figure 1.
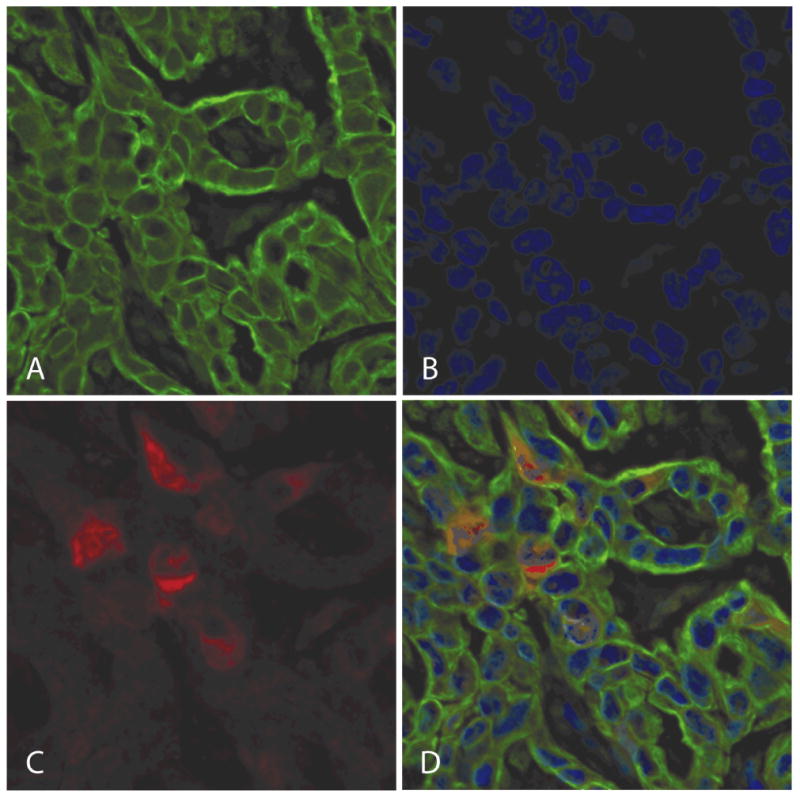
Protein expression of ribonucleotide reductase M2 (RRM2) was determined using an automated in situ quantitative measurement of protein analysis, automated quantitative immunohistochemistry on the basis of immunofluorescence.
a. Target compartments were localized using a fluorescently tagged Alexa Fluor 555, rabbit anti-cytokeratin antibody (green). b. 4,6-Diamidino-2-phenylindole was added to visualize nuclei (blue). c. RRM2 was visualized with Alexa Fluor 488-tyramide, human anti-RRM2 (red).
d. A three color overlay shows localization of RRM2 to the cytoplasm.
Statistical Methods
RRM2 gene expression and protein expression were summarized in terms of number of observations, means and standard deviations. The data were presented in graphical format using boxplots. The comparisons between groups were performed using a non-parametric Wilcoxon Rank Sum test or the Kruskal-Wallis test. Exact p-values were computed for all comparisons. All statistical tests were two-sided, and P < .05 was used to indicate statistical significance. Due to the exploratory nature of this study, no adjustments for multiple comparisons were made. Non-parametric Spearman's rank correlation analysis was used to examine the association between protein and gene expression levels. Statistical analysis was performed using SAS® (SAS Institute Inc., Cary, North Carolina) version 9.1 software.
Results
RRM2 Expression by Tumor Type
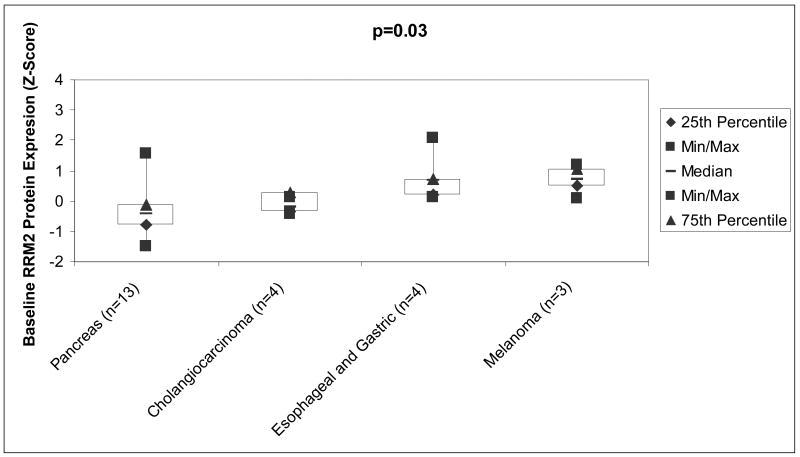
AQUA demonstrated that RRM2 protein localized to the cytoplasm (Figure 1), where it is produced. Table 2 lists the number and types of samples that were analyzed. Comparisons were made for tumor types where at least three tissue blocks were available. The median Z-scores were as follows: pancreatic cancers (n=13) -0.41± 0.84, for cholangiocarcinoma (n=4) -0.21 ± 0.23, for esophageal and gastric cancers (n=4) 0.68 ± 0.95, and for melanoma (n=3) it 0.74 ± 0.57. The p-value for the comparison across all four tumor types is 0.03 (Kruskal-Wallis test), suggesting that at least one tumor type is different than the others (Figure 2). Pairwise comparisons between the groups with the Wilcoxan Rank Sum test demonstrate cholangiocarcinoma and esophageal cancer are significantly different (p=0.03), pancreatic cancer and esophageal/gastric cancer are significantly different (p=0.04), and that pancreatic cancer and melanoma show a trend towards difference (p=0.06).
Table 2. Ribonucleotide Reductase M2 Protein and Gene Expression by Tumor Type.
| Tumor Type (n=Protein/mRNA) | RRM2 Protein Expression (Z-Score) Mean ± SD | RRM2 Gene Expression Relative to YWHZ Mean ± SD |
|---|---|---|
| Unknown Primary (2) | -7.8±1.0 | 0.03±0.01 |
| Prostate (2) | -0.77±0.1 | 0.35±0.14 |
| Sarcoma (1) | -0.57 | 3.3 |
| Cervical (1) | -0.56 | 0.35 |
| Lymphoma (1/0) | -0.52 | |
| Breast (2/0) | -0.42±0.41 | |
| Pancreas (13/6) | -0.41±0.84 | 0.29±0.20 |
| Cholangiocarcinoma (4) | -0.21±0.23 | 0.54±0.52 |
| Mesothelioma (2) | 0.26±0.23 | 0.16±0.14 |
| Colon (2) | 0.29±0.46 | 0.31±0.12 |
| Bladder (1) | 0.57 | 2.17 |
| Esophageal/Gastric (4) | 0.68±0.95 | 2.56±1.49 |
| Melanoma (3) | 0.74±0.57 | 0.79±0.22 |
| NSCLC (1) | 0.78 | 2.43 |
| Small Cell Lung Cancer (1) | 3.8 | 0.03 |
RRM2: Ribonucleotide Reductase M2
NSCLC: non small cell lung cancer
SD: standard deviation
Figure 2.
Boxplot of RRM2 Protein Expression Evaluated by automated quantitative immunohistochemistry. The median Z score for pancreas cancers (n=13) was -0.41± 0.84, cholangiocarcinoma (n=4) was -0.21 ± 0.23, esophageal and gastric (n=4) was 0.68 ± 0.95, and melanoma (n=3) was 0.74 ± 0.57. The p-values for the comparisons across all four tumor types is 0.03 (Kruskal-Wallis test), suggesting that at least one tumor type is different than the others. Abbreviations: Min, minimum; Max, maximum; n, number; RRM2, ribonucleotide reductase M2.
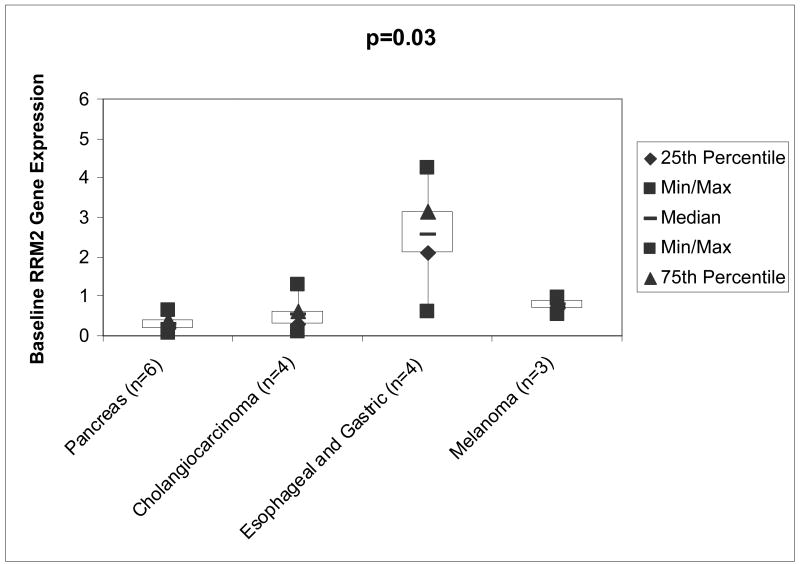
The RRM2 gene expression relative to the housekeeping gene YWHAZ for pancreatic cancers (n=6) was 0.29± 0.20, for cholangiocarcinoma (n=4) was 0.54 ± 0.52, for esophageal/gastric cancers (n=4) 2.56 ± 1.49, and for melanoma (n=3) it was 0.79 ± 0.22. The p-value for the comparison across all 4 tumor types is 0.03 (Wilcoxon Rank Sum test), suggesting that at least one tumor type is different than the others. See Figure 3. Pairwise comparisons between the groups with the Kruskal-Wallis test demonstrate that pancreas cancer and esophageal/gastric cancer (p=0.02), and prostate cancer and melanoma (p=0.05) are significantly different. Cholangiocarcinoma and esophageal cancers show a trend towards difference (p=0.06).
Figure 3.
Boxplot of RRM2 Gene Expression Evaluated by RT- PCR. The relative ribonucleotide reductase M2 gene expression for pancreas cancers (n=6) was 0.29± 0.20, cholangiocarcinoma (n=4) was 0.54 ± 0.52, esophageal and gastric (n=4) was 2.56 ± 1.49, and melanoma (n=3) was 0.79 ± 0.22. The p-values for the comparisons across all 4 tumor types is 0.03 (Kruskal-Wallis test), suggesting that at least one tumor type is different than the others. Abbreviations: RRM2, ribonucleotide reductase M2; n, number; Min, minimum; Max, maximum.
Spearman's rank correlation analysis was performed to examine the association between RRM2 protein expression as evaluated by AQUA and gene expression evaluated by Q-RTPCR in baseline tumor specimens. This analysis demonstrated a moderate correlation (Spearman's rank correlation =0.30, p=0.12) between the cancers, and two tumor types (SCLC and sarcoma) had highly discordant results (Table 2).
Patient Response
Two patients (5%) achieved a partial response by Response Evaluation Criteria in Solid Tumors (RECIST) (20). One patient with NSCLC, who received 3-AP at 85 mg/m2 on days 1-3 of a 21 day cycle and irinotecan 150 mg/m2 on day 1 of a 21 day cycle had a Z-score of 0.78 and a relative gene expression value of 2.43. Another patient with bladder cancer, who received 3-AP at 25 mg/m2 Days 1-4 of a 21 day cycle and doxorubicin at 60 mg/m2, on day 1 of a 21 day cycle had an unconfirmed PR and a Z-score of 0.57 and relative gene expression value of 2.17. While response rates were too low to perform a statistical analysis comparing responders to non-responders, both patients achieving a partial response had relatively high expression levels compared to other patients without a response (Table 2).
Discussion
In this study, we evaluated differences in RRM2 mRNA and protein expression in baseline tumor samples. Our findings demonstrated that RRM2 levels are higher in esophageal/gastric cancer and melanomas when compared to pancreatic cancer. Additionally, esophageal cancers have more expression when compared to cholangiocarcinomas. These differences in RRM2 were significant despite small sample sizes and suggest that relative RRM2 overexpression may play a role in tumor development of esophageal and gastric cancers and melanoma, although confirmatory studies are required.
As RRM2 is the intracellular target of 3-AP, one potential application would be to evaluate RRM2 expression to predict 3-AP -sensitive tumors. Gandera and colleagues recently reported preliminary findings suggesting inhibition of RRM2 gene expression was associated with clinical response to the antisense agent GTI-2040 targeting RRM2 (21). Two phase II trials of 3-AP have recently been reported (22,23) with disappointing results. In a trial of single agent 3-AP in advanced renal cell cancer, Knox and colleagues demonstrated a 7% (1/19) partial response rate (22). Mackenzie and colleagues studied 3-AP in combination with gemcitabine for advanced pancreatic cancer with no responses observed (23). While RRM2 expression was not evaluated, the clinical results are consistent with our own phase II trial of 3-AP in advanced pancreas cancer where no responses were observed (16). Since RRM2 gene expression and protein expression in pancreas cancer was found to be significantly less than in other solid tumors, low RRM2 expression may be a potential mechanism of 3-AP resistance.
While a statistical analysis could not be performed due to a limited sample size, both patients with a partial response had high RRM2 mRNA and protein levels. Since both patients achieving a partial response received chemotherapy in addition to 3-AP, the observed responses cannot be attributed solely to 3-AP relatively high baseline RRM2 expression. The patient with NSCLC received concurrent 3-AP and irinotecan, and the patient with bladder cancer received 3-AP and doxorubicin. However, given the single agent response rate of second-line irinotecan in metastatic NSCLC is less than 5% (24) and the single agent response rate for doxorubicin in bladder cancer is 17% (25), our data suggests that increased baseline expression of RRM2 and 3-AP may contribute to the activity of the regimen.
In this analysis, protein expression and mRNA expression were moderately correlated. This association can likely be explained by the SCLC and sarcomas that had highly discordant results. The data suggests that for the majority of tumors, transcription is an important mechanism controlling RRM2 protein expression. In some tumors, however, transcription and expression do not appear to be linked, and unknown mechanisms may control protein expression. This analysis also raises an important question regarding the preferred method for expression analysis, which cannot be answered in the current study. Further studies comparing protein expression to mRNA expression to 3-AP response and mechanistic studies to determine the relationship between mRNA expression and protein expression for RRM2 are necessary.
In conclusion, both RRM2 gene and protein expression vary by tumor type in baseline tumor samples. Given the poor phase II activity of 3-AP in renal cell and pancreatic cancer, one potential application of our findings would be evaluating baseline RRM2 to predict tumors sensitive to 3-AP.
Acknowledgments
Supported by: U01CA062491 “Early Clinical Trials of Anti-Cancer Agents with Phase I Emphasis” NCI; CTEP Translational Research Initiative Funding 24XS090, and 1ULRR025011 Clinical and Translational Science Award of the National Center for Research Resources, NIH; and NIH grant T32 CA009614 Physician Scientist Training in Cancer Medicine (Dr. Attia).
References
- 1.Thelander M, Gräslund A, Thelander L. Subunit M2 of mammalian ribonucleotide reductase. J Biol Chem. 1985;260:2737–2741. [PubMed] [Google Scholar]
- 2.Larsson A, Sjöberg BM. Identification of the stable free radical tyrosine residue in ribonucleotide reductase. EMBO J. 1986;5:2037–2040. doi: 10.1002/j.1460-2075.1986.tb04461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elford HL, Freese M, Passamani E, Morris HP. Ribonucleotide reductase and cell proliferation. J Biol Chem. 1970;245:5228–5233. [PubMed] [Google Scholar]
- 4.Heidel JD, Liu JY, Yen Y, Zhou B, Heale BS, Rossi JJ, Bartlett DW, Davis ME. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res. 2007 Apr 1;13(7):2207–15. doi: 10.1158/1078-0432.CCR-06-2218. [DOI] [PubMed] [Google Scholar]
- 5.Avolio TM, Lee Y, Feng N, Xiong K, Jin H, Wang M, Vassilakos A, Wright J, Young A. RNA interference targeting the R2 subunit of ribonucleotide reductase inhibits growth of tumor cells in vitro and in vivo. Anticancer Drugs. 2007 Apr;18(4):377–88. doi: 10.1097/CAD.0b013e328013c04f. [DOI] [PubMed] [Google Scholar]
- 6.Fan H, Villegas C, Huang A, Wright JA. The mammalian ribonucleotide reductase R2 component cooperates with a variety of oncogenes in mechanisms of cellular transformation. Cancer Res. 1998;58:1650–1653. Bibliographic Links. [PubMed] [Google Scholar]
- 7.Fan H, Villegas C, Wright JA. Ribonucleotide reductase R2 component is a novel malignancy determinant that cooperates with activated oncogenes to determine transformation and malignant potential. Proc Natl Acad Sci U S A. 1996;93:14036–14040. doi: 10.1073/pnas.93.24.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu J, et al. Overexpression of transfected human ribonucleotide reductase M2 subunit in human cancer cells enhances their invasive potential. Clin Exp Metastasis. 1998;16:43–49. doi: 10.1023/a:1006559901771. [DOI] [PubMed] [Google Scholar]
- 9.Kuo ML, Hwang HS, Sosnay PR, Kunugi KA, Kinsella TJ. Overexpression of the R2 subunit of ribonucleotide reductase in human nasopharyngeal cancer cells reduces radiosensitivity. Cancer J. 2003;9:277–285. doi: 10.1097/00130404-200307000-00010. [DOI] [PubMed] [Google Scholar]
- 10.Huang A, Fan H, Taylor WR, Wright JA. Ribonucleotide reductase R2 gene expression and changes in drug sensitivity and genome stability. Cancer Res. 1997;57:4876–4881. [PubMed] [Google Scholar]
- 11.Zhou BS, Hsu NY, Pan BC, Doroshow JH, Yen Y. Overexpression of ribonucleotide reductase in transfected human KB cells increases their resistance to hydroxyurea: M2 but not M1 is sufficient to increase resistance to hydroxyurea in transfected cells. Cancer Res. 1995;55:1328–1333. [PubMed] [Google Scholar]
- 12.Goan YG, Zhou B, Hu E, Mi S, Yen Y. Overexpression of ribonucleotide reductase as a mechanism of resistance to 2,2-difluorodeoxycytidine in the human KB cancer cell line. Cancer Res. 1999;59:4204–4207. [PubMed] [Google Scholar]
- 13.Jung CP, Motwani MV, Schwartz GK. Flavopiridol increases sensitization to gemcitabine in human gastrointestinal cancer cell lines and correlates with down-regulation of ribonucleotide reductase M2 subunit. Clin Cancer Res. 2001;7:2527–2536. [PubMed] [Google Scholar]
- 14.Schelman WR, Holen K, Mulkerin D, Kolesar J, Thomas J, Kruse M, Oliver K, Marnocha R, Eickhoff J, Wilding G. Journal of Clinical Oncology; 2006 ASCO Annual Meeting Proceedings Part I; 2006. p. 12011. [Google Scholar]
- 15.Chang JE, Morgan Meadows S, Traynor A, Kolesar J, Marnocha R, Lee F, Eickoff J, Beth E, Binger K, Wilding G. Journal of Clinical Oncology; 2006 ASCO Annual Meeting Proceedings Part I; 2006. p. 13168. [Google Scholar]
- 16.Attia S, Kolesar J, Mahoney M, Pitot H, Laheru D, Heun J, Huang W, Antholine W, Erlichman C, Holen K. A Phase 2 Consortium (P2C) trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) for advanced adenocarcinoma of the pancreas. Investigational New Drugs. 2008 Feb 16; doi: 10.1007/s10637-008-9123-6. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 18.Rubin MA, Zerkowski MP, Camp RL, et al. Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol. 2004;164(3):831–40. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren M, Twohig M, Pier T, Eickhoff J, Lin CY, Jarrard D, Huang W. Protein Expression of Matriptase and Its Cognate Inhibitor HAI-1 in Human Prostate Cancer: a tissue microarray and automated quantitative analysis. Applied Immunohistochemistry and Molecular Morphology 2008. doi: 10.1097/PAI.0b013e31817c3334. Accepted for publication. [DOI] [PubMed] [Google Scholar]
- 20.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. Journal of the National Cancer Institute. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 21.Juhasz A, Vassilakos A, Chew HK, Gandara D, Yen Y. Analysis of ribonucleotide reductase M2 mRNA levels in patient samples after GTI-2040 antisense drug treatment. Oncol Rep. 2006 May;15(5):1299–304. [PubMed] [Google Scholar]
- 22.Knox JJ, Hotte SJ, Kollmannsberger C, Winquist E, Fisher B, Eisenhauer EA. Phase II study of Triapine(R) in patients with metastatic renal cell carcinoma: a trial of the National Cancer Institute of Canada Clinical Trials Group (NCIC IND.161) Invest New Drugs. 2007 Mar 28; doi: 10.1007/s10637-007-9044-9. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie MJ, Saltman D, Hirte H, Low J, Johnson C, Pond G, Moore MJ. A Phase II study of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) and gemcitabine in advanced pancreatic carcinoma. A trial of the Princess Margaret Hospital Phase II consortium. Invest New Drugs. 2007 Jun 22; doi: 10.1007/s10637-007-9066-3. [DOI] [PubMed] [Google Scholar]
- 24.Georgoulias V, Kouroussis C, Agelidou A, Boukovinas I, Palamidas P, Stavrinidis E, Polyzos A, Syrigos K, Veslemes M, Toubis M, Ardavanis A, Tselepatiotis E, Lung Cancer Committee of the Hellenic Oncology Research Group Irinotecan plus gemcitabine vs irinotecan for the second-line treatment of patients with advanced non-small-cell lung cancer pretreated with docetaxel and cisplatin: a multicentre, randomised, phase II study. Br J Cancer. 2004 Aug 2;91(3):482–8. doi: 10.1038/sj.bjc.6602010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosl GJ, Fair WR, Herr HW, Bajorin DF, Dalbagni G, Sarkis AS, Reuter VE, Cordon-Cardo C, Sheinfeld J, Scher HI. Bladder cancer: advances in biology and treatment. Crit Rev Oncol Hematol. 1994 Feb;16(1):33–70. doi: 10.1016/1040-8428(94)90041-8. [DOI] [PubMed] [Google Scholar]