The acetylase inhibitor spermidine and the sirtuin-1 activator resveratrol disrupt the antagonistic network of acetylases and deacetylases that regulate autophagy.
Abstract
Autophagy protects organelles, cells, and organisms against several stress conditions. Induction of autophagy by resveratrol requires the nicotinamide adenine dinucleotide–dependent deacetylase sirtuin 1 (SIRT1). In this paper, we show that the acetylase inhibitor spermidine stimulates autophagy independent of SIRT1 in human and yeast cells as well as in nematodes. Although resveratrol and spermidine ignite autophagy through distinct mechanisms, these compounds stimulate convergent pathways that culminate in concordant modifications of the acetylproteome. Both agents favor convergent deacetylation and acetylation reactions in the cytosol and in the nucleus, respectively. Both resveratrol and spermidine were able to induce autophagy in cytoplasts (enucleated cells). Moreover, a cytoplasm-restricted mutant of SIRT1 could stimulate autophagy, suggesting that cytoplasmic deacetylation reactions dictate the autophagic cascade. At doses at which neither resveratrol nor spermidine stimulated autophagy alone, these agents synergistically induced autophagy. Altogether, these data underscore the importance of an autophagy regulatory network of antagonistic deacetylases and acetylases that can be pharmacologically manipulated.
Introduction
Macroautophagy (which we refer to as autophagy) is a cellular self-cannibalistic pathway in which parts of the cytosol or cytoplasmic organelles are enwrapped in double-membraned vesicles, autophagosomes, which then fuse with lysosomes (Klionsky, 2007). Autophagy plays a major role in the maintenance of cellular homeostasis, allows for the mobilization of energy reserves when external resources are limited, and is essential for the removal of damaged organelles and potentially toxic protein aggregates (Levine and Kroemer, 2008). At the organismal level, autophagy can mediate cytoprotection (for instance neuroprotection and cardioprotection in the context of ischemic preconditioning; Moreau et al., 2010) and delay the pathogenic manifestations of aging (Levine and Kroemer, 2009). Given the potential health and longevity-promoting effects of autophagy, pharmacological agents that stimulate autophagy at a low level of toxicity are urgently needed.
Rapamycin and the so-called rapalogs are the most effective clinically used inducers of autophagy yet have severe immunosuppressive effects (Hartford and Ratain, 2007). Thus, alternative, nontoxic autophagy inducers (such as rilmenidine or carbamazepine) are being characterized for their pharmacological profile in suitable preclinical models (Hidvegi et al., 2010; Rose et al., 2010). Nontoxic compounds, such as resveratrol and spermidine, are also being evaluated for their potential to induce autophagy in vivo (Eisenberg et al., 2009; Morselli et al., 2010). Resveratrol is a natural polyphenol found in grapes, red wine, berries, knotweed, peanuts, and other plants. The interest in this molecule rose because it was suggested to mediate the cardioprotective effects of red wine (Baur and Sinclair, 2006). Resveratrol is also a potent inducer of autophagy (Scarlatti et al., 2008a,b), and this effect is mediated through the activation of sirtuin 1 (SIRT1), a NAD+-dependent deacetylase (Morselli et al., 2010). Resveratrol has been suggested to directly activate SIRT1 (Baur and Sinclair, 2006; Lagouge et al., 2006), although indirect effects may actually be preponderant (Beher et al., 2009; Pacholec et al., 2010). Spermidine is polyamine found in citrus fruit and soybean, which has recently been shown to increase the lifespan of yeast, nematodes, and flies in an autophagy-dependent fashion (Eisenberg et al., 2009).
The transfection-enforced expression of SIRT1 is sufficient to stimulate autophagy in human cells (Lee et al., 2008). Starvation-induced autophagy (but not autophagy induced by rapamycin) requires SIRT1, both in vitro (in mammalian cells; Lee et al., 2008) and in vivo (in Caenorhabditis elegans; Morselli et al., 2010). Activated SIRT1 induces autophagy via its capacity to deacetylate acetyl lysine residues in other proteins (Lee et al., 2008). Conversely, knockdown of the acetyltransferase EP300 (Lee and Finkel, 2009), as well as inhibition of histone acetylases, potently induces autophagy (Eisenberg et al., 2009), indicating that protein deacetylation may play a general role in the initiation of the autophagic cascade. EP300 acetylates several autophagy-relevant proteins, including autophagy-related 5 (ATG5), ATG7, ATG12, and microtubule-associated protein 1 light chain 3 β (LC3; Lee and Finkel, 2009), whereas SIRT1 deacetylates ATG5, ATG7, LC3 (Lee et al., 2008), and the transcription factor forkhead box O3, which can stimulate the expression of proautophagic genes (Kume et al., 2010). As a result, protein (de)acetylation reactions influenced by sirtuins and other enzymes control autophagy at multiple levels, including the modification of autophagy core proteins and/or of transcriptional factors that control the expression of autophagic genes.
Driven by these premises and incognita, we comparatively assessed the mechanisms of autophagy induction mediated by two distinct compounds that modulate protein acetylation, namely resveratrol and spermidine. We found that both agents induce autophagy through initially distinct yet convergent pathways that culminate in the acetylation and deacetylation of hundreds of proteins, with opposed patterns in distinct subcellular compartments. Based on this characterization, we demonstrated that these agents can stimulate autophagy in a synergistic fashion, both in vitro, in cultured human cells, and, in vivo, in mice.
Results
Sirtuin-dependent versus -independent autophagy induced by resveratrol and spermidine
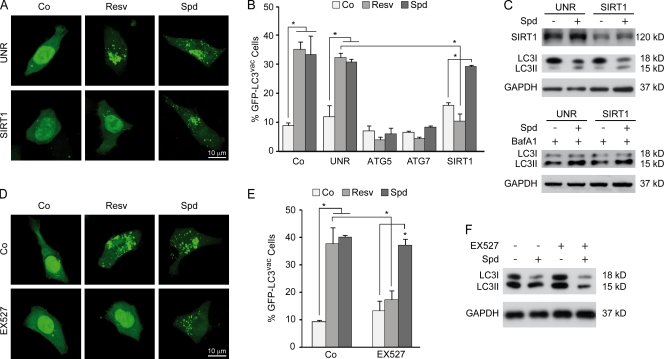
Spermidine and resveratrol were comparable in their autophagy stimulatory potency and induced hallmarks of autophagy with similar kinetics in human colon cancer HCT 116 cells. These signs included the redistribution of a GFP-LC3 chimera, which is usually diffuse, to cytoplasmic puncta and the lipidation of endogenous LC3, increasing its electrophoretic mobility (Fig. 1 and Fig. S1 A). In these conditions, neither spermidine nor resveratrol impaired oxidative phosphorylation (Fig. S1 B), ruling out that resveratrol might induce autophagy via mitochondriotoxicity (Dörrie et al., 2001). Knockdown of SIRT1 with a specific siRNA suppressed the proautophagic activity of resveratrol (Fig. 1, A and B) yet failed to affect spermidine-induced autophagy (Fig. 1 C). Similarly, the SIRT1 inhibitor EX527 (Peck et al., 2010) abolished autophagy induction by resveratrol but not by spermidine (Fig. 1, D–F). These results indicate that resveratrol and spermidine trigger autophagy through distinct mechanisms.
Figure 1.
SIRT1 activity is required for resveratrol-induced autophagy but not for spermidine-mediated autophagy induction in mammalian cultured cells. (A–C) Human colon carcinoma HCT 116 cells were left untransfected (Co, control) or transfected with an irrelevant siRNA (UNR, unrelated) or siRNAs specific for ATG5, ATG7, or SIRT1 and then retransfected with a GFP-LC3–encoding plasmid, cultured in complete medium for 24 h, and left untreated or treated for 4 h with 100-µM resveratrol (Resv) or spermidine (Spd). The same experiment was performed in the presence of bafilomycin A1 (BafA1), which inhibits the fusion between lysosomes and autophagosomes, to evaluate the autophagic flux. (A) Representative images. (B) Quantitative data. (C) Representative immunoblots of HCT 116 cells transfected either with an unrelated siRNA or with a SIRT1-specific siRNA showing LC3 lipidation after treatment with 100-µM spermidine in the presence or absence of bafilomycin A1. (D–F) HCT 116 cells were left transfected with a GFP-LC3 plasmid, cultured in complete medium for 24 h, and treated with either vehicle (Co), 100-µM resveratrol, or 100-µM spermidine in the presence or absence of the SIRT1 inhibitor EX527 for 4 h. (D) Representative images. (E) Quantitative data. (B and E) Bars depict the percentages of cells showing accumulation of GFP-LC3 in puncta (GFP-LC3vac; means ± SEM; n = 3; *, P < 0.05). (F) Representative immunoblots showing LC3 lipidation in HCT 116 cells treated with 100-µM spermidine in the presence or absence of EX527. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Phylogenetic conservation of sirtuin-independent autophagy induction by spermidine
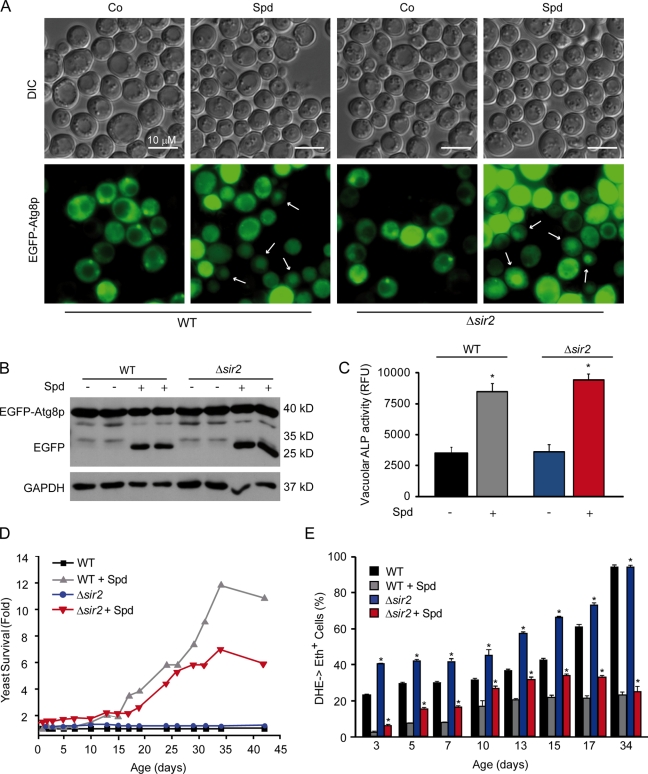
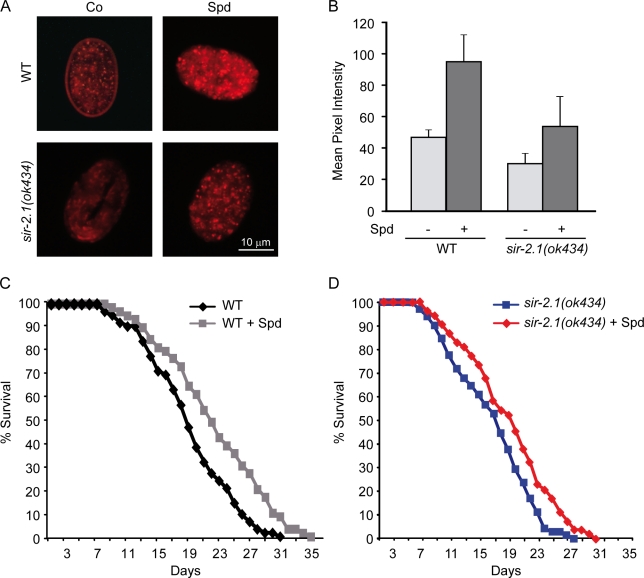
We next investigated whether the orthologues of sirt1 in Saccharomyces cerevisiae and C. elegans (sir2 and sir-2.1, respectively) are required for the proautophagic activity of spermidine. In yeast, spermidine caused the redistribution of a GFP-Atg8p chimera from a diffuse to a vacuolar localization (Fig. 2 A), the autophagy-dependent proteolytic liberation of GFP from GFP-Atg8p (Fig. 2 B; Suzuki et al., 2004), as well as an autophagy-related increase in vacuolar AP (Fig. 2 C; Noda et al., 1995). These effects were similar in wild-type (WT) and Δsir2 yeast strains (Fig. 2, A–C). Moreover, spermidine significantly improved the survival of aging WT yeast cultures, a beneficial effect that was attenuated, yet remained significant, in aging Δsir2 yeast cultures (Fig. 2 D). Accordingly, spermidine reduced the aging-associated overproduction of reactive oxygen species (measured by assessing the conversion of nonfluorescent dihydroethidine into fluorescent ethidium) both in WT and Δsir2 cells (Fig. 2 E). In C. elegans embryos, spermidine induced the autophagy-related expression and cytoplasmic aggregation of DsRed::LGG-1 (Fig. 3, A and B; Eisenberg et al., 2009). This effect was significant in both WT and sir-2.1 mutant nematodes, although the sir-2.1 mutation attenuated autophagy induction by spermidine (Fig. 3, C and D). Consistently, spermidine prolonged the lifespan of WT and sir-2.1–deficient worms by 18 and 13%, respectively. Collectively, these results indicate that spermidine can stimulate autophagy and extend the lifespan of yeast cells and nematodes that lack SIRT1 orthologues.
Figure 2.
The lifespan-extending and autophagy-inducing effects of spermidine in yeast are not mediated by Sir2. (A–E) EGFP-Atg8p was ectopically expressed in wild-type (WT) or Δsir2 S. cerevisiae undergoing chronological aging on small synthetic 2% glucose media with or without (Co, control) supplementation of 4-mM spermidine (Spd). (A) Representative images. EGFP-Atg8p localization (bottom) was visualized by fluorescence microscopy. Yeast cells undergoing autophagy (in which EGFP-Atg8p exhibits a prominent vacuolar localization) are indicated by arrows. Yeast morphology was monitored by differential interference contrast (DIC; top). (B) Representative immunoblots against EGFP. Free EGFP indicates the vacuolar degradation of EGFP-Atg8p fusion, thereby representing the autophagic flux. Notice that both WT and Δsir2 yeast cells show similar free EGFP levels after spermidine-mediated autophagy induction. (C) Relative alkaline phosphatase (ALP) activity indicative of autophagy. n = 3. (D) Survival data. n = 4. (E) Quantification of reactive oxygen species. Bars indicate the percentages of cells exhibiting the reactive oxygen species–mediated conversion of dihydroethidine (DHE) into ethidium (Eth; n = 4). Data represent means ± SEM; *, P < 0.001 as compared with untreated cells of the same genotype. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. RFU, relative fluorescence unit.
Figure 3.
The life-extending and autophagy-inducing effects of spermidine in C. elegans are not mediated by Sir2. (A) Fluorescence microscopy of C. elegans transgenic embryos expressing a full-length plgg-1DsRed::LGG-1 fusion protein indicative of autophagic activity. Two representative pictures of wild-type (WT) and sir-2.1 embryos untreated (Co, control) or treated with 0.2-mM spermidine (Spd) supplementation of food are shown. (B) Quantification of autophagic activity through the measurement of DsRed::LGG-1 pixel intensity from images of WT animals shown in A. Data represent means ± SEM (n = 3) with ≥25 images processed for each trial. (C) Survival of WT C. elegans during aging with and without (control) supplementation of food (UV-killed E. coli) with 0.2-mM spermidine (n = 110; P < 0.005). (D) Survival of sir-2.1 C. elegans (ok434 phenotype) during aging with and without (control) supplementation of food (UV-killed E. coli) with 0.2-mM spermidine (n = 110; P < 0.01). P-values were calculated using the log-rank test as described in Materials and methods.
Resveratrol and spermidine induce autophagy through convergent pathways
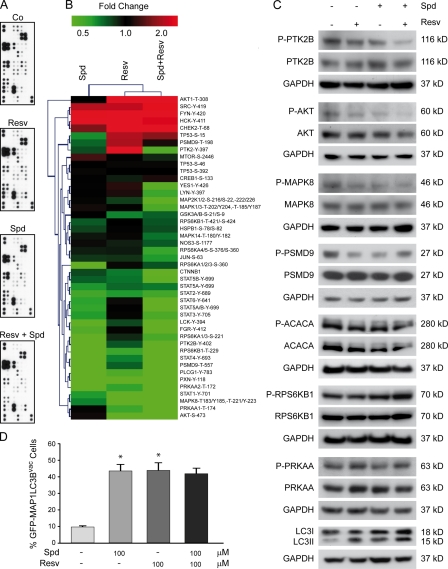
To investigate the signal transduction pathway stimulated by resveratrol and spermidine, the phosphorylation status of multiple cellular proteins was analyzed in human colon cancer HCT 116 cells by means of an antibody array. Surprisingly, spermidine and resveratrol, alone or in combination, elicited similar changes in the phosphorylation status of multiple kinases and their substrates (Fig. 4, A–C). For example, both spermidine and resveratrol mediated the dephosphorylation of the protein tyrosine kinase 2 β (also known as PYK2) and the cyclin-dependent kinase inhibitor 1B (better known as p27Kip1). However, neither of the two agents had major effects on the phosphorylation levels of the regulatory subunit of AMP-dependent kinase and its substrate acetyl–coenzyme A (CoA) carboxylase, which was in line with the hypothesis that the energy metabolism of the cells was normal. Moreover, spermidine and resveratrol did not affect the phosphorylation of mechanistic target of rapamycin (mTOR) nor that of its substrate ribosomal protein S6 kinase (also known as p70S6K; Fig. 4, A–C), which suggests that resveratrol and spermidine induce autophagy through AMP-dependent kinase/mTOR-independent convergent pathways. Accordingly, the administration of an optimal dose of resveratrol and spermidine (100 µM for both agents) did not result in higher levels of autophagy than that of either agent alone (Fig. 4 D). This kind of epistatic analysis confirms the suspected convergence of the proautophagic pathways elicited by both agents.
Figure 4.
Convergent alterations in the phosphoproteome status after resveratrol and/or spermidine treatment. (A–C) Human colon carcinoma HCT 116 cells were treated for 2 h with vehicle (Co, control), 100-µM resveratrol (Resv), and 100-µM spermidine (Spd), alone or in combination (Resv + Spd). (A) Representative phosphoprotein arrays are shown. (B) Clustering analysis for the effects on protein kinase phosphorylation. (C) Representative immunoblots of selected kinases whose phosphorylation status was unaffected (PRKAA1, RPS6KB1, and acetyl-CoA carboxylase a [ACACA]) or affected by resveratrol or/and spermidine treatment (PTKB, AKT1, MAPK8, and CDKN1B), validating phosphoprotein array data. (D) Human colorectal carcinoma HCT 116 cells were transfected with a GFP-LC3–encoding plasmid, cultured in complete medium for 24 h, and then treated with either vehicle or the indicated dose of resveratrol or spermidine, alone or in combination, for 2 h. Quantitative data. Bars depict the percentages (means ± SD; n = 3; *, P < 0.05) of cells showing the accumulation of GFP-LC3 in puncta (GFP-LC3vac). GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Convergent action of resveratrol and spermidine on the acetylproteome
Next, we comparatively explored the effects of resveratrol and spermidine on the acetylation patterns of cytosolic, mitochondrial, and nuclear proteins. To that purpose, we performed stable isotope labeling with amino acids in cell culture (SILAC) and then purified the proteins/peptides containing acetylated lysine residues and identified them by quantitative mass spectrometry (MS). Resveratrol or spermidine induced changes in the acetylation of 560 lysine-containing motifs corresponding to 375 different proteins (Table S1). Surprisingly, 170 proteins whose acetylation status was modified in response to resveratrol or spermidine treatment are part of the recently elucidated human autophagy protein network (Behrends et al., 2010). Many of the (de)acetylated proteins identified in our study are central to the network because 89 among them interact with at least 10 proteins in the network (Table S2).
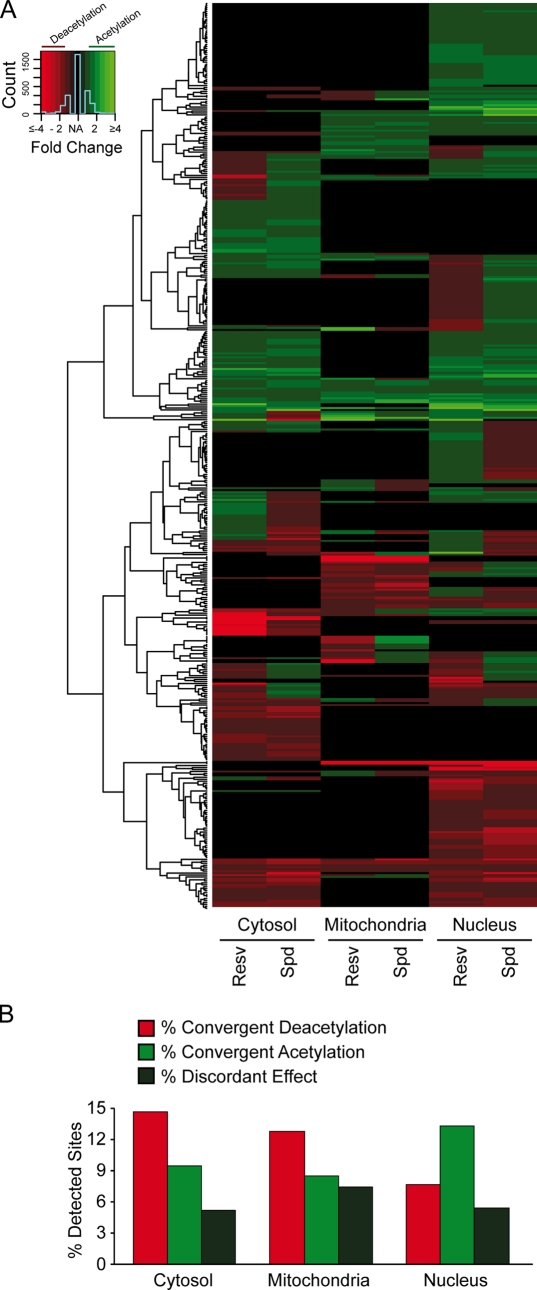
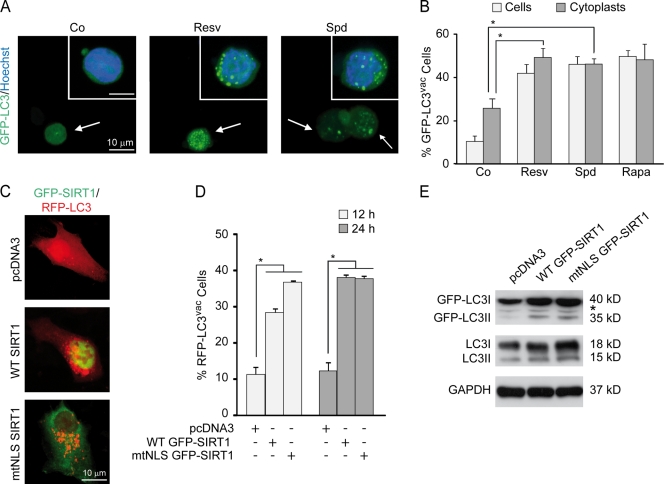
Both resveratrol and spermidine tended to induce the (de)acetylation of similar proteins, including that of autophagy-relevant substrates, such as ATG5 and LC3 (Fig. 5, A and B; and Fig. S2 E). Interestingly, no fundamental differences were found in the consensus (de)acetylation sites that were modified in response to resveratrol or spermidine (Fig. 6 and Fig. 7). In the cytosol, resveratrol and spermidine induced convergent deacetylation more frequently than convergent acetylation, whereas in the nucleus, acetylation was dominantly triggered by both agents (Fig. 5 B, P < 0.001, χ2 test). Moreover, when we analyzed the distinct biological processes associated with the observed (de)acetylated proteins after gene ontology (GO) term enrichment (Ashburner et al., 2000), deacetylated proteins often fell in the category of metabolism (which includes autophagy; Fig. S3). Therefore, we investigated whether short-term autophagy induction by spermidine and/or resveratrol is a transcription-dependent or -independent event using cytoplasts (enucleated cells). Cytoplasts were still able to accumulate GFP-LC3 puncta in response to spermidine or resveratrol treatment (Fig. 8, A and B), indicating that nuclei (and by extension transcription) are not required for short-term autophagy stimulation by these two agents. Next, we enforced overexpression of transgenic WT SIRT1 (which although preponderantly localizes to the nucleus, has been reported to efficiently shuttle to the cytoplasm; Tanno et al., 2007) or that of a mutant SIRT1 protein with a mutation in the nuclear localization signal (which is, therefore, virtually restricted to the cytoplasm; Fig. 8 C). Both constructs induced RFP-LC3 punctuation and LC3 lipidation with similar potency and similar kinetics (Fig. 8, C–E), suggesting that autophagy can be efficiently regulated by cytoplasmic (de)acetylation reactions.
Figure 5.
Convergent acetylproteome modification after resveratrol or spermidine treatment. (A–C) Colon carcinoma HCT 116 cells were cultured for 2 wk in three different SILAC media containing different arginine and lysine isotopes. Cells were treated with 100-µM resveratrol (Resv) or spermidine (Spd) for 2 h, fractioned into cytoplasmic, nuclear, and mitochondrial extracts, processed for acetyl lysine peptide enrichment, and analyzed by MS. (A) Hierarchical clustering of drug-specific organellar distributions of all acetylated sites quantified in at least one fraction. Fold changes are calculated relative to untreated cells. Only sites regulated >1.5-fold were included in statistical analyses. (B) Graphical representation of peptides whose acetylation status was affected in a convergent or divergent way. n = 560.
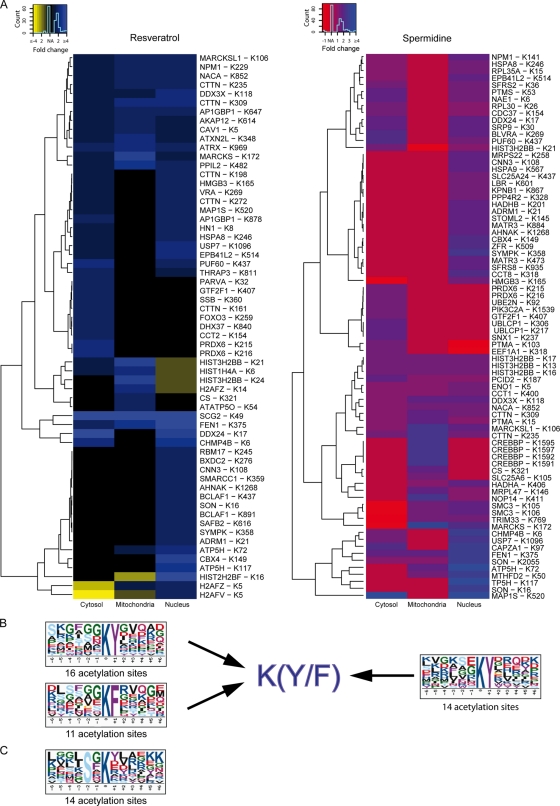
Figure 6.
Significant motifs among sites undergoing acetylation. (A) Hierarchical clustering of the organellar distributions of sites whose acetylation was increased by >1.5-fold in response to resveratrol or spermidine, at least in one organellar fraction. (B) Consensus acetylation motifs identified upon resveratrol (left) or spermidine (right) treatment are depicted using the MotifX algorithm (Schwartz and Gygi, 2005). Among sites that were hyperacetylated in response to both agents, the K(F/Y) motif is significantly enriched when tested against the whole proteome (P < 0.00001). (C) When testing against the largest acetylation site dataset from Choudhary et al. (2009) (acetylation background dataset [ABD]), the SxK motif is significant (P < 0.0001) for sites whose acetylation increased upon spermidine treatment. No general consensus motifs were found for sites whose acetylation increased in response to both agents.
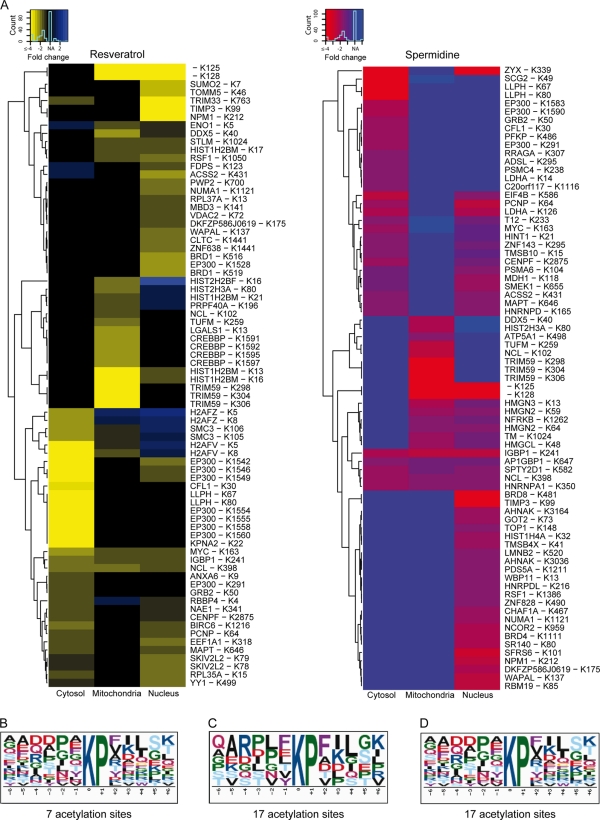
Figure 7.
Significant motifs among sites undergoing deacetylation. (A) Hierarchical clustering of the organellar distributions of sites whose deacetylation was increased by >1.5-fold in response to resveratrol or spermidine, at least in one organellar fraction. (B) Among sites that were hypoacetylated in response to both agents, the KP motif is significantly enriched when tested against ABD (P < 0.001). (C and D) When tested against the ABD (C) or the whole proteome (D), the KP motif is significant for sites undergoing hypoacetylation upon spermidine treatment (P < 0.0001). No general consensus motifs were found for sites whose acetylation decreased in response to both agents.
Figure 8.
Autophagy can be efficiently regulated by cytoplasmic (de)acetylation reactions. (A and B) HCT 116 cells were transfected with a GFP-LC3–encoding plasmid, cultured in complete medium for 24 h, and enucleated to obtain cytoplasts. The cytoplasts were treated with either vehicle (Co, control), 100-µM resveratrol (Resv), or 100-µM spermidine (Spd) for 4 h. 1-µM rapamycin (Rapa) was used as a positive control. (A) Representative images of cytoplasts indicative of autophagic activity. Arrows indicate GFP-LC3–transfected cytoplasts, whereas insets show nonenucleated transfected cells. (B) Quantitative data. Bars show the percentages of cells or cytoplasts showing the accumulation of GFP-LC3 in puncta (GFP-LC3vac). (C–E) HCT 116 cells were cotransfected with a plasmid for the expression of RFP-LC3 together with an empty vector (pcDNA3) or plasmids encoding wild-type (WT) SIRT1 or a SIRT1 variant with a mutation in the nuclear localization signal fused to GFP (WT GFP-SIRT1 and mtNLS GFP-SIRT1, respectively) for 12 or 24 h, fixed, and analyzed by fluorescence microscopy. (C) Representative images indicative of 24-h autophagic activity. (D) Quantitative data. Bars depict the percentages of cells exhibiting the accumulation of RFP-LC3 in puncta (RFP-LC3vac). (B and D) Means ± SEM; n = 3; *, P < 0.05. (E) Representative immunoblots for 24-h endogenous LC3 lipidation. The asterisk stands for a nonspecific band. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Synergistic induction of autophagy by low doses of resveratrol and spermidine
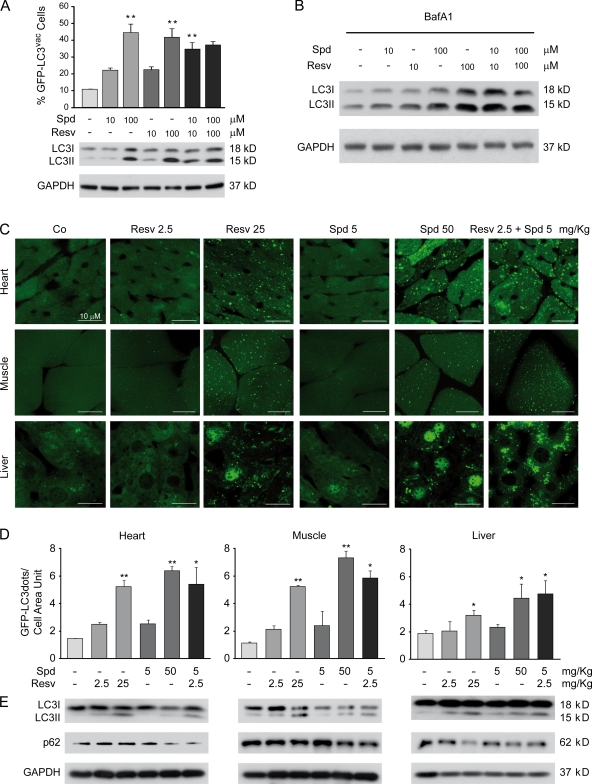
Resveratrol (but not spermidine) induces autophagy through the activation of the deacetylase SIRT1 (Morselli et al., 2010), whereas spermidine is thought to act as an inhibitor of acetylases (Eisenberg et al., 2009). We reasoned that low doses of resveratrol and spermidine might synergistically induce autophagy by affecting the equilibrium state of (de)acetylation. To assess this possibility, we treated HCT 116 cells with different concentrations of resveratrol or spermidine, alone or in combination, and analyzed the effects of the different pharmacological combinations in terms of autophagy induction. As expected, both spermidine and resveratrol used at high doses (100 µM) induced GFP-LC3 punctuation and LC3 lipidation (Fig. 9 A) in cultured cells. Interestingly, although none of the two agents at low doses (10 µM) was able to significantly up-regulate autophagic flux, the combination of spermidine and resveratrol at low doses (10 µM) was as efficient in enhancing GFP-LC3 puncta formation, LC3 lipidation, and an increase in autophagic flux as were high doses of spermidine or resveratrol (Fig. 9, A and B).
Figure 9.
Low doses of resveratrol and spermidine synergistically induce autophagy in vitro and in vivo. (A) Human colorectal carcinoma HCT 116 cells were transfected with a GFP-LC3–encoding plasmid, cultured in complete medium for 24 h, and then treated with either vehicle (Co, control) or the indicated dose of resveratrol (Resv) or spermidine (Spd), alone or in combination, for 2 h. (top) Quantitative data. Bars depict the percentages (means ± SD; n = 3; **, P < 0.05) of cells showing the accumulation of GFP-LC3 in puncta (GFP-LC3vac). (bottom) Representative immunoblots showing endogenous LC3 lipidation. (B) Representative immunoblots showing endogenous LC3 lipidation in the presence of bafilomycin A1 (BafA1). (C–E) Transgenic C57BL/6 mice expressing a GFP-LC3 fusion protein were injected with resveratrol and spermidine at the indicated concentrations. 3 h later, mice were killed, and tissues were processed for immunofluorescence microscopy determinations of GFP-LC3vac. (C) Representative images. (D) Quantitative results. Bars represent the percentages (means ± SEM; n = 3; *, P < 0.05; and **, P < 0.01 as compared with the same tissue from untreated animals) of cells exhibiting GFP-LC3vac. (E) Representative immunoblots showing endogenous LC3 lipidation and p62 protein content in WT C57BL/6 mouse tissues. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
To try to extend these results to a physiological setting, we intraperitoneally injected optimal doses of resveratrol (25 mg/kg) or spermidine (50 mg/kg) into mice expressing a GFP-LC3 transgene to induce autophagy in an array of organs. One tenth of this optimal dose (2.5 mg/kg resveratrol or 5 mg/kg spermidine) had no major proautophagic effect in vivo when either compound was injected alone. However, the combination of low doses of both agents was highly efficient in triggering autophagy in vivo (Fig. 9, C and D). Similar results were obtained when these agents were injected into WT mice, as shown by means of LC3 lipidation and p62 degradation (Fig. 9 E). In conclusion, low doses of spermidine and resveratrol can induce autophagy in a synergistic fashion.
Discussion
Resveratrol can induce autophagy only in the presence of SIRT1 (Morselli et al., 2010), whereas SIRT1 (or its orthologues in yeast and nematodes) is dispensable for spermidine-stimulated autophagy. Thus, these agents clearly ignite distinct pathways across a large phylogenetic distance. In spite of the difference in the primary targets of resveratrol and spermidine, both agents activated convergent pathways in that thus far they both stimulated mTOR-independent autophagy and elicited rather similar changes in the phosphoproteome and, more importantly, in the acetylproteome. Both agents provoked multiple changes (increases or decreases) in the lysine acetylation of hundreds of proteins, and the convergent changes induced by both agents largely outnumbered discordant modifications. When combined between each other, high doses of spermidine and resveratrol did not induce higher levels of autophagy than each of the two agents alone, which is in line with the idea that the terminal pathways stimulated by these compounds overlap. Spermidine and resveratrol modulated the acetylation of >100 proteins that are part of the central network of autophagic regulators/executioners (Behrends et al., 2010). This suggests that both agents stimulate autophagy through a multipronged mechanism that involves a large number of (de)acetylation reactions.
Although resveratrol can (directly or indirectly) activate SIRT1, a deacetylase (Baur and Sinclair, 2006; Lagouge et al., 2006; Beher et al., 2009; Pacholec et al., 2010), spermidine has been shown to inhibit acetylases (Erwin et al., 1984; Eisenberg et al., 2009). Based on this consideration, it appears paradoxical that neither of these two agents was able to provoke a general deacetylation state and that both of them actually stimulated a similar shift in the acetylation pattern, in which hundreds of proteins were deacetylated (more in the cytosol than in the nucleus), whereas several others were acetylated (more in the nucleus than in the cytosol). Cells harbor multiple deacetylases and acetylases (Hassig and Schreiber, 1997; Katan-Khaykovich and Struhl, 2002; Nakamura et al., 2010), and it appears plausible, yet remains to be proven, that inhibition of one (or a few) acetylase will activate compensatory reactions by other acetylases and/or impact the action of deacetylases so that the global cellular level of protein acetylation remains near to constant. As a significant trend, however, we observed that both resveratrol and spermidine stimulated the deacetylation of cytosolic proteins, such as ATG5 and LC3, and the acetylation of nuclear proteins, including multiple histones.
It has been recently reported that lifespan extension by spermidine treatment (during conditions of chronological aging) is linked to deacetylation of nuclear histones and to an increase in the transcription of different autophagy-related genes (Eisenberg et al., 2009). Interestingly, autophagy was rapidly induced by both spermidine and resveratrol in cytoplasts prepared from proliferating human cells, and an extranuclear variant of SIRT1 was as efficient in inducing autophagy as the predominantly nuclear WT SIRT1. Collectively, these data suggest that protein deacetylation first stimulates autophagy predominantly through a cytosolic mechanism. These results not only illustrate the differences between quiescent and proliferating cells in terms of autophagy modulation but also suggest that after a fast and nuclear-independent autophagic response transcriptional reprogramming is required to maintain an increased basal autophagic activity (Kroemer et al., 2010), thus contributing to the previously reported lifespan extension.
Although we have few mechanistic cues to understand the discrepancy in cytosolic versus nuclear (de)acetylation reactions induced by resveratrol and spermidine, it is tempting to explain the synergistic proautophagic action of both compounds by the network properties of acetylases and deacetylases. One tenth of the dose of spermidine or resveratrol, which optimally stimulates autophagy, has no major proautophagic effects, meaning that dose–response curves are rather steep (most likely caused by compensatory reactions that tend to maintain the homeostasis of the acetylproteome). However, the partial yet simultaneous activation of the deacetylase activity of SIRT1 by resveratrol and the concomitant inhibition of acetylases by spermidine can unbalance the acetylproteome, thereby synergistically stimulating autophagy.
Resveratrol is a natural polyphenol contained in red wine and vegetables, whereas spermidine is a polyamine found in other healthy food, such as citrus fruit and soybean. When analyzed as individual compounds, neither polyphenols nor polyamines consumed with the normal diet may reach concentrations high enough to mediate pharmacological effects. Nonetheless, it is tempting to speculate that combinations of these agents—and perhaps that of other proautophagic dietary components—may affect the autophagic rheostat, as based on their distinct yet convergent mode of action.
Materials and methods
Chemical, cell line, and culture conditions
Unless otherwise specified, chemicals were purchased from Sigma-Aldrich, culture media and supplements for cell culture were obtained from Invitrogen, and plasticware was purchased from Corning. Human colon carcinoma HCT 116 cells (gift from B. Volgelstein, Howard Hughes Medical Institute, Johns Hopkins University, Baltimore, MD; Bunz et al., 1999) were cultured in McCoy’s 5A medium containing 10% fetal bovine serum, 100 mg/liter sodium pyruvate, 10-mM Hepes buffer, 100 U/ml penicillin G sodium, and 100 µg/ml streptomycin sulfate (5% CO2 at 37°C). Cells were seeded in 6- and 12-well plates or in 10- and 15-cm dishes and grown for 24 h before treatment with 10-µM EX527 (Tocris Bioscience), 10- or 100-µM resveratrol, 10- or 100-µM spermidine, and 1-µM rapamycin or 1-nM bafilomycin A1 (Tocris Bioscience) for the time indicated in each experimental figure legend.
Plasmids, transfection, and RNA interference in human cell cultures
Cells were cultured in 12-well plates and transfected at 50% confluence with siRNAs targeting human SIRT1 (Ford et al., 2005), ATG5, or ATG7 (Thermo Fisher Scientific) or with an unrelated control siRNA by means of a transfection reagent (Oligofectamine; Invitrogen) following the manufacturer’s instructions. After 24 h, cells were transfected with a plasmid coding for a GFP-LC3 fusion (Kabeya et al., 2000). Transient plasmid transfections were performed with the Attractene reagent (QIAGEN) as suggested by the manufacturer, and unless otherwise indicated, cells were analyzed 24 h after transfection. Cells were transfected with a plasmid coding for RFP fused to LC3 (RFP-LC3; obtained from Invitrogen) in the presence of an empty vector (pcDNA3) or of different constructs for the overexpression of GFP-tagged WT SIRT1 or a SIRT1 variant mutated in the nuclear localization signal, which mostly localizes in the nucleus (Tanno et al., 2007). For fluorescence microscopy determinations, cells cultured on coverslips were fixed in paraformaldehyde (4% wt/vol) for 15 min at RT, washed three times in PBS, and mounted with mounting medium (Vectashield; Vector Laboratories).
Fluorescence microscopy
Confocal fluorescent images were captured using a confocal fluorescence microscope (TCS SP2; Leica). For experiments with HCT 116 cells, an Apochromat 63× 1.3 NA immersion objective was used, whereas for the analysis of GFP-LC3 mice tissue sections, an Apochromat 40× 1.15 NA immersion objective was used. All the acquisitions were made at RT with fixed cells/tissue slides. Images were acquired with a camera (DFC 350 FX 1.8.0; Leica) using LAS AF software (Leica) and processed with Photoshop (CS2; Adobe) software. Specifically, picture processing involved cropping of representative areas and linear adjustments of contrast and brightness and was performed using Photoshop (with equal adjustment parameters for all pictures); no explicit γ correction was used. Nonconfocal microscopy of yeast strains carrying the EGFP-tagged Atg8 protein was performed with a microscope (Axioskop; Carl Zeiss, Inc.) using a Plan Neofluar objective lens (Carl Zeiss, Inc.) with a 63× magnification and 1.25 NA in oil at RT. Images were taken with a camera (SPOT 9.0 Monochrome 6; Diagnostic Instruments, Inc.), acquired using the Metamorph software (6.2r4; Universal Imaging Corp.), and processed with IrfanView (version 3.97) and Photoshop (CS2) software. Specifically, picture processing involved coloring and cropping of representative areas and was performed with IrfanView. In addition, linear adjustments of contrast and brightness were applied with Photoshop (using equal adjustment parameters for all pictures); no explicit γ correction was used. Nonconfocal microscopy of C. elegans was performed with a microscope (AxioImager Z2; Carl Zeiss, Inc.) using a Plan Neofluar 40× objective with a 0.75 NA and a 63× Plan Neofluar objective with an NA of 1.25 in oil at RT. Images were taken with a camera (AxioCam MRc5; Carl Zeiss, Inc.) with Axiovision software (Carl Zeiss, Inc.) without further processing. The different fluorophores used in this work were GFP and RFP for HCT 116 cells, EGFP for yeast experiments, DsRed for C. elegans analyses, and GFP for mice tissue sections. Nuclei were counterstained by Hoechst (Invitrogen). Immersion oil (Immersol; Carl Zeiss, Inc.) was used for all microscopy analyses.
Immunoblotting, immunoprecipitation, and phosphokinase array
For immunoblotting, cells were collected, washed with cold PBS, and lysed as previously described (Criollo et al., 2007). 25 µg of protein was then separated on 4–12% Bis-Tris acrylamide (Invitrogen) or 12% Tris-glycine SDS-PAGE precast gels (Bio-Rad) and electrotransferred to Immobilon membranes (Millipore) according to standard procedures. Unspecific binding sites were saturated by incubating membranes for 1 h in 0.05% Tween 20 (vol/vol in TBS) supplemented with 5% nonfat powdered milk (wt/vol in TBS) followed by overnight incubation with primary antibodies specific for acetylated lysine, phosphoacetyl-CoA carboxylase α (Ser79), acetyl-CoA carboxylase α, phospho-AKT1 (Thr308), AKT1, LC3, phosphomitogen-activated protein kinase 8 (Tyr183/Thr185), phospho-PRKAA1 (Thr172), PRKAA1, cyclin-dependent kinase inhibitor 1B, phosphoprotein tyrosine kinase 2 β (Tyr402), phosphoribosomal protein S6 kinase (Thr421/Ser424), ribosomal protein S6 kinase, stress-induced phosphoprotein 1 (Cell Signaling Technology), ATG7 (Sigma-Aldrich), phosphocyclin-dependent kinase inhibitor 1B (Abcam), TP53 (DO7), p62, SIRT1 (Santa Cruz Biotechnology), and peptidylprolyl isomerase A (cyclophilin A; Enzo Life Sciences, Inc.). Revelation was performed with appropriate HRP-labeled secondary antibodies (SouthernBiotech) plus a chemoluminescent substrate (SuperSignal West Pico; Thermo Fisher Scientific). An antiglyceraldehyde-3 phosphate dehydrogenase antibody (Millipore) was used to control the equal loading of lanes. For immunoprecipitation, extracts from 8 × 106 cells were lysed, and 500 µg of protein was precleaned for 1 h with 15 µl protein G–Sepharose 4 Fast Flow (GE Healthcare) followed by incubation for 2 h in the presence of an antiacetylated lysine (Cell Signaling Technology) antibody. Subsequent immunoblotting was performed by means of TrueBlot-HRP (eBioscience) secondary antibodies. For phosphokinase array analyses, the Proteome Profiler kit (R&D Systems) was used according to the manufacturer’s instructions.
Yeast aging and autophagy measurements
For chronological aging experiments, WT BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) S. cerevisiae or the respective sir2 deletion mutant (Δsir2) from the European Saccharomyces Cerevisiae Archive for Functional Analysis were inoculated from fresh overnight cultures to an absorbance of 0.1 (∼106 cells/ml) and grown at 28°C on Synthetic Complete 2% glucose medium. Aliquots were taken to perform survival plating at the indicated time points (Fig. 2 D, line graph) as previously described (Herker et al., 2004). Representative aging experiments are shown with at least three independent samples. Spermidine was added to stationary cultures at day 1 of the aging experiments (Eisenberg et al., 2009). Dihydroethidium staining was performed as previously described (Büttner et al., 2007), and the superoxide-driven conversion to ethidium was quantified either on a fluorescence plate reader (GeniosPro; TECAN) or on a cytofluorometer (FACSAria; BD) followed by first-line statistical analysis by means of the FACSDiva software (BD). Autophagy was monitored by vacuolar localization of Atg8p using fluorescence microscopy or by immunoblotting of cells ectopically expressing an EGFP-Atg8 chimera (Eisenberg et al., 2009) with anti-GFP (Sigma-Aldrich) and antiglyceraldehyde-3 phosphate dehydrogenase antibodies. For biochemical quantifications of the autophagic flux, AP activity was assayed according to published methods (Noda et al., 1995). In brief, WT or Δsir2 BY4741 cells were transformed and selected for stable insertion of a pTN9 HindIII fragment encoding for the cytosolic Pho8ΔN60 protein. AP activity was then assessed in 1.5 µg of crude protein extracts by measuring the conversion of α-naphtyl phosphate to naphthalene using a GeniosPro fluorescence plate reader with excitation and emission wavelengths at 340 nm and 485 nm, respectively (Noda et al., 1995). To correct for intrinsic AP activity, WT or Δsir2 yeast cells lacking the pTN9 HindIII fragment were simultaneously assayed, and these values were used for background subtraction, giving the vacuolar (autophagic) AP activity.
C. elegans strains, genetics, and pharmacology
We followed standard procedures for C. elegans strain maintenance. Nematode-rearing temperature was kept at 20°C. The following strains were used in this study: N2, WT Bristol isolate, and VC199, sir-2.1(ok434)IV. The VC199 strain was provided by the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation, which is part of the International C. elegans Gene Knockout Consortium and the Caenorhabditis Genetics Center and is funded by the National Institutes of Health National Center for Research Resources. The construction of the plgg-1DsRed::LGG-1 reporter plasmid has been described previously (Samara et al., 2008). Spermidine was dissolved in sterilized water to a stock solution concentration of 100 mM. Escherichia coli (OP50) bacteria on seeded nematode growth medium (NGM) plates were killed by UV irradiation for 10 min (0.5 J) using a UV cross-linker (Bio-Link BLX-E365; Vilber Lourmat). A range of spermidine concentrations was prepared by dilutions in 100 µl of sterilized water and applied to the top of the agar medium (7-ml NGM plates). Plates were then gently swirled to allow the drug to spread to the entire NGM surface. Identical drug-free water solutions were used for the control plates. Plates were then allowed to dry overnight. The procedure was repeated each time worms were transferred to fresh plates (every 2–3 d during the first 2 wk and every week thereafter). Worms were incubated at 20°C.
C. elegans autophagy measurements
Images from transgenic embryos expressing a DsRed::LGG-1 fusion protein were acquired using a 540 ± 15–nm band-pass excitation filter and a 575-nm long-pass emission filter. Experiments were performed at 20°C, with photography exposure time kept identical for each embryo. Emission intensity was measured on grayscale images with a pixel depth of 8 bits (256 shades of gray). We calculated the mean and maximum pixel intensity for each embryo in these images using the ImageJ software (National Institutes of Health). For each transgenic line, we processed ≥25 images over at least three independent trials.
C. elegans lifespan analysis
Lifespan assays were performed at 20°C. Synchronous animal populations were generated by hypochlorite treatment of gravid adults to obtain tightly synchronized embryos that were allowed to develop into adulthood under appropriate conditions. Progeny were grown at 20°C through the L4 larval stage and then transferred to fresh plates in groups of 10–20 worms per plate for a total of 100–150 individuals per experiment. The day of egg harvest was set as t = 0. Animals were transferred to fresh plates every 2–4 d and were examined every day for touch-provoked movement and pharyngeal pumping until death. Worms that died (because of internally hatched eggs, extruded gonads, or desiccation upon crawling on the plate edge) were censored and incorporated as such into the dataset. Each survival assay was repeated at least three times.
SILAC cell culture, sample processing, and analysis
HCT 116 cells were cultured for 2 wk in three different SILAC media (Invitrogen) containing either (1) light isotopes ofl-arginine and l-lysine (Arg0/Lys0), (2) l-arginine–13C6 HCl (Euroisotop) and l-lysine 2HCl 4,4,5,5-D4 (Arg6/Lys4; Euroisotop), or (3) l-arginine–13C615N4 HCl and l-lysine 13C615N4 HCl (Arg10/Lys8; Invitrogen) and complemented as previously described (Blagoev and Mann, 2006). Cells were treated for 2 h with 100 µM spermidine (Arg10/Lys8) or 100 µM resveratrol (Arg6/Lys4) and then lysed and subdivided in nuclear, mitochondrial, and cytosolic fractions as previously described (Gurbuxani et al., 2003). The cytosolic fraction was precipitated using ice-cold (−20°C) acetone (4 vol of the sample extract), vortexed, and placed for 2 h at −20°C. The resulting solution was centrifuged for 10 min at 16,000 g, and the supernatant was removed. The pellet was subsequently washed twice with ice-cold 4:1 acetone/water. The final pellet was dried using a concentrator (SpeedVac; Thermo Fisher Scientific) for 10–15 min. All fractions were dissolved in denaturant 6:2-M urea/thiourea (both obtained from Merck), and Benzonase (Merck) was added to the nuclear fraction. All steps were performed at RT to avoid carbamoylation of amines. Reduction of cysteines was performed with 5-mM DTT (Sigma-Aldrich) for 30 min followed by alkylation with 11-mM iodoacetamide for 20 min in the dark. The proteins were digested with 1:100 protease LysC (Wako Chemicals USA, Inc.) for 3.5 h, diluted four times with 50-mM ammonium bicarbonate, and then digested with 1:100 trypsin (Promega) overnight. The nuclear fraction mixture was centrifuged at 10,000 g for 10 min, and the supernatant was filtered through a 0.45-µm MillexHV filter (Millipore). From each fraction, ∼100 µg of digested protein was collected for isoelectric focusing, a step required for subsequent normalization. The acetyl lysine peptide enrichment was performed as previously described (Choudhary et al., 2009). After peptide enrichment and isoelectric focusing, samples were subjected to MS analysis, and data were processed as described in the next paragraph.
Preparation of cytoplasts
Trypsinized cells previously transfected with GFP-LC3 cDNA were incubated in 3 ml of complete medium supplemented with 7.5 mg/ml cytochalasin B for 45 min at 37°C. This cell suspension was layered onto a discontinuous Ficoll density gradient (3 ml of 55%, 1 ml of 90%, and 3 ml of 100% Ficoll; GE Healthcare) in complete medium containing 7.5 mg/ml cytochalasin B. Gradients were prepared in ultracentrifuge tubes and preequilibrated at 37°C in a CO2 incubator overnight. Gradients containing cell suspensions were centrifugated in a prewarmed rotor (SW41; Beckman Coulter) at 100,000 g for 20 min at 32°C. The cytoplast-enriched fraction was collected from the interface between 55 and 90% Ficoll layers, washed in complete RPMI 1640 medium, and incubated for 4 h at 37°C before resveratrol, spermidine, or rapamycin treatment.
Mouse experiments and tissue processing
C57BL/6 mice obtained from Charles River Laboratory were bred and maintained according to both the Federation for Laboratory Animal Science Associations and the Animal Experimental Ethics Committee guidelines. They were housed in a temperature-controlled environment with 12-h light/dark cycles and received food and water ad libitum. Mice were injected intraperitoneally with 5 or 50 mg/kg spermidine and with 2.5 or 25 mg/kg 3 h before anesthetization and killing. Mice tissues were immediately frozen in liquid nitrogen after extraction and homogenized in a 20-mM Tris buffer, pH 7.4, containing 150-mM NaCl, 1% Triton X-100, 10-mM EDTA, and protease inhibitor cocktail (Complete; Roche). Tissue extracts were then centrifuged at 12,000 g at 4°C, and supernatants were collected. Protein concentration in the supernatants was evaluated by the bicinchoninic acid technique (BCA protein assay kit; Thermo Fisher Scientific).
Quantitative analysis of GFP-LC3 dots in mice tissue sections
To avoid postmortem autophagy induction, dead mice were immediately perfused with 4% paraformaldehyde (wt/vol in PBS, pH 7.4). Tissues were then harvested and further fixed with the same solution for ≥4 h followed by treatment with 15% sucrose (wt/vol in PBS) for 4 h and with 30% sucrose (wt/vol in PBS) overnight. Tissue samples were embedded in Tissue-Tek OCT compound (Sakura) and stored at −70°C. 5-µm-thick tissue sections were generated with a cryostat (CM3050 S; Leica), air dried for 1 h, washed in PBS for 5 min, dried at RT for 30 min, and mounted with Vectashield antifading medium. In each organ, the number of GFP-LC3 dots was counted in five independent visual fields from at least five mice using a confocal fluorescence microscope (TCS SP2).
SILAC sample processing and analysis
Upon digestion, peptides were concentrated and desalted on SepPak C18 (Waters) purification cartridges and eluted using a highly organic buffer (80% acetonitrile and 0.5% acetic acid). Eluates were lyophilized in a vacuum centrifuge, dissolved in immunoprecipitation buffer (50-mM MOPS, pH 7.2, 10-mM sodium phosphate, and 50-mM sodium chloride), and mixed with anti–acetyl lysine antibodies conjugated to agarose beads. The mixture was left for 12 h at 4°C on a rotation wheel. The flow-through (containing nonbound peptides) was removed, and beads were washed four times with the immunoprecipitation buffer and twice with deionized water. Bead-bound peptides were eluted with 0.1% trifluoroacetic acid. The same enrichment procedure was performed again on the flow-through containing the unbound peptides from the first enrichment. Six eluates and three separately collected digested protein samples were desalted using SepPak C18 cartridges, and 5% of each acetyl lysine–enriched eluate was used for MS analysis. The rest of the samples were subsequently separated into 12 fractions by isoelectric focusing using the 3100 OFFGEL Fractionator (Agilent Technologies) as previously described (Hubner et al., 2008). Gel strips and amfolyte buffer were purchased from GE Healthcare. Focusing was performed for 20 kV/h at a maximum current of 50 µA and maximum power of 200 mW. 10 µl of 10% trifluoroacetic acid was added to each fraction and STAGE (stop and go extraction) tipped as previously described (Rappsilber et al., 2007) before MS analysis, which was performed on either an LTQ Orbitrap XL spectrometer (Thermo Fisher Scientific) connected to a 1200 Series Nanoflow HPLC system (Agilent Technologies) or an LTQ Orbitrap Velos spectrometer (Thermo Fisher Scientific; Olsen et al., 2009) connected to an 1100 Series Nanoflow HPLC system (Agilent Technologies) using a nanoelectrospray ion source (Thermo Fisher Scientific). Peptides were separated by reversed phase chromatography using an in-house–made fused silica emitter (75-µm internal diameter) packed with 3-µm reversed phase material (Reprosil-Pur C18-AQ; Dr. Maisch GmbH). Peptides were loaded in 98% solvent A (0.5% acetic acid) followed by a 100-min linear gradient to 50% solvent B (80% acetonitrile and 0.5% acetic acid). Survey full-scan MS spectra (mass to charge [m/z] range of 300–200, with a resolution of 60,000 at an m/z of 400) were acquired followed by fragmentation of the 10 (in the case of using the LTQ Orbitrap XL) or the 20 (in the case of using the LTQ Orbitrap Velos) most intense multiply charged ions. Ions selected for MS/MS were placed on a dynamic exclusion list for 45 s. Real-time internal lock mass recalibration was used during data acquisition (Olsen et al., 2005). For unfractionated acetyl lysine–enriched eluates, an additional MS analysis was performed on the LTQ Orbitrap Velos using higher energy collision dissociation fragmentation (normalized collision energy of 40) of the 10 most intense ions from each MS spectrum creating MS/MS spectra at a resolution of 7,500. All samples used for protein normalization were analyzed on a mass spectrometer (LTQ FT ULTRA; Thermo Finnigan) in which the five most intense ions from each precursor scan were selected for fragmentation in the LTQ. In this case, no real-time lock mass recalibration was used. Reverse-phase chromatography settings were the same as described for the analysis performed on the Orbitrap spectrometers.
SILAC data processing
Raw files were processed with MaxQuant v.1.0.13.13 (Cox and Mann, 2008) into centroided data and submitted to database searching with Mascot v.2.2 (Matrix-Science). Preprocessing by MaxQuant was performed to determine charge states, miscleavages, and SILAC states and to filter the MS/MS spectra, keeping the six most intense peaks within a 100-D bin. Cysteine carbamidomethyl was chosen as a fixed modification, whereas N-terminal acetylation and methionine oxidation were chosen as variable modifications. Furthermore, acetylation of light, medium, and heavy isotope lysines (Lys0/Lys4/Lys8) was chosen as a variable modification. Processed MS/MS spectra were searched against a concatenated target decoy database of forward and reversed sequences from the International Protein Index database (152,616 sequences; FASTA file created 5/6/2008). For the search, trypsin/P + DP was chosen for the in silico protein digestion allowing four miscleavages. The mass tolerance for the MS spectra acquired in the Orbitrap was set to 7 ppm, whereas the MS/MS tolerance was set to 0.6 D for the collision-induced dissociation MS/MS spectra from the LTQ and to 0.04 D for the higher energy collision dissociation MS/MS spectra. Upon peptide search, protein and peptide identification was performed given an estimated maximal false discovery rate of 1% at both the protein and peptide level. For false discovery rate calculation, posterior error probabilities were calculated based on peptides of at least six amino acids having a Mascot score of ≥10. For protein quantification, only unmodified peptides, peptides modified by N-terminal acetylation, and methionine oxidation were calculated. If a counterpart to a given lysine-acetylated peptide was identified, this counterpeptide was also excluded by protein quantitation. According to the protein group assignment performed by MaxQuant, both razor and unique peptides are used for protein quantification. A minimum of two ratio counts was required for protein quantification. For quantification of lysine-acetylated sites, the least modified peptides were used. The ratios for the sites were normalized by the corresponding protein ratios to account for eventual changes in protein abundance. In case a protein ratio was not determined, normalization was based on a logarithm transformation algorithm as previously described (Cox and Mann, 2008).
Cell respiration and mitochondrial substrate oxidations
Cell respiration and mitochondrial substrate oxidation were polarographically measured at 37°C in 250 µl of a buffer containing 0.3-M mannitol, 10-mM KCl, 5-mM MgCl2, 1 mg/ml BSA, and 10-mM KH2PO4, pH 7.4 (Rustin et al., 1994). Respiration was measured on intact cells (final concentration of ∼106/ml), which were subsequently permeabilized by 0.01% digitonin to study mitochondrial substrate oxidation. 10-mM malate plus10-mM glutamate oxidation was measured in the presence of 200-µM ADP. 10-mM succinate oxidation by digitonin-permeabilized cells was measured in the presence of 2-µM rotenone and 200-µM ADP. Sequential addition of 2-µM oligomycin, a specific inhibitor of the mitochondrial ATPase, and 2-µM carbonyl cyanide m-chlorophenyl hydrazone, a potent mitochondrial uncoupler, allowed for the determination of the respiratory control value associated with succinate oxidation.
Functional analysis of proteins regulated by deacetylation or acetylation
To decipher the functional context of the proteins associated with the drug-specific regulation of proteins by deacetylation and acetylation, GO term (Ashburner et al., 2000) enrichment was performed using the Cytoscape (Shannon et al., 2003) plugin BiNGO (Biological Networks Gene Ontology tool; Maere et al., 2005) and PANTHER (Protein Analysis Through Evolutionary Relationships) classification system. For the enrichment analysis, proteins regulated by ≥1.5-fold were included, and p-values were calculated by Fisher’s exact test after the Benjamini–Hochberg adjustment for multiple testing (Benjamini and Hochberg, 1995). A significance level of 0.05 (corresponding to the maximal false discovery rate) and a minimum of five proteins in at least one of the subsets of each given significant GO term were set as thresholds.
Statistical analysis
Statistical analyses were performed using the Prism software package (GraphPad Software, Inc.), the Office 2003 Excel software package (Microsoft), and the statistical environment R (R Development Core Team). In cell culture experiments, values were compared using unpaired Student’s t tests. For multiple comparisons, we used the one-factor analysis of variance corrected by posthoc Bonferroni test. C. elegans survival curves were created using the product-limit method of Kaplan and Meier. The log-rank (Mantel–Cox) test was used to evaluate differences between survival and determine p-values.
Online supplemental material
Fig. S1 describes the kinetics of autophagy induction after 100-µM resveratrol and spermidine treatment and shows measurements of cell respiration and mitochondrial substrate oxidation in the absence (control) or presence of 100-µM resveratrol or spermidine. Fig. S2 shows the measurement of SILAC fraction purity and analysis of the acetylation status of autophagy essential proteins after spermidine and/or resveratrol treatment. Fig. S3 schematically shows the biological processes associated with the differentially acetylated proteins in response to resveratrol and spermidine treatment by means of GO enrichment. Table S1 shows all the detected acetyl lysine–containing motifs in the different subcellular compartments after spermidine or resveratrol treatment and their relative enrichment in treated samples as compared with the corresponding control. Table S2 shows the list of proteins belonging to the human autophagy network interacting with the proteins detected in SILAC analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008167/DC1.
Acknowledgments
The VC199 strain was provided by the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation, which is part of the International C. elegans Gene Knockout Consortium and the Caenorhabditis Genetics Center and is funded by the National Institutes of Health’s National Center for Research Resources. The authors thank Dr. Toren Finkel (National Heart Lung and Blood Institute, National Institutes of Health, Bethesda, MD) for supplying invaluable reagents.
G. Kroemer is supported by the Ligue Nationale contre le Cancer (Equipes labelisée), Agence Nationale pour la Recherche, the European Commission (Active p53, Apo-Sys, ChemoRes, and ApopTrain), Fondation pour la Recherche Médicale, Institut National du Cancer, and Cancéropôle Ile-de-France. N. Tavernarakis is supported by grants from the European Union Marie Curie actions and the European Research Council, and T. Eisenberg and F. Madeo are grateful to the European Commission for the grant Apo-Sys. M.V. Bennetzen is supported by the Danish Ministry of Science, Technology, and Innovation (the EliteResearch initiative). C. López-Otín is supported by grants from Ministerio de Ciencia e Innovación–Spain, the European Union (Microenvimet), and Fundación Marcelino Botín.
Footnotes
Abbreviations used in this paper:
- ABD
- acetylation background dataset
- CoA
- coenzyme A
- GO
- gene ontology
- MS
- mass spectrometry
- mTOR
- mechanistic target of rapamycin
- NGM
- nematode growth medium
- SILAC
- stable isotope labeling with amino acids in cell culture
- WT
- wild type
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur J.A., Sinclair D.A. 2006. Therapeutic potential of resveratrol: the in vivo evidence. Nat. Rev. Drug Discov. 5:493–506 10.1038/nrd2060 [DOI] [PubMed] [Google Scholar]
- Beher D., Wu J., Cumine S., Kim K.W., Lu S.C., Atangan L., Wang M. 2009. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 74:619–624 10.1111/j.1747-0285.2009.00901.x [DOI] [PubMed] [Google Scholar]
- Behrends C., Sowa M.E., Gygi S.P., Harper J.W. 2010. Network organization of the human autophagy system. Nature. 466:68–76 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 57:289–300 [Google Scholar]
- Blagoev B., Mann M. 2006. Quantitative proteomics to study mitogen-activated protein kinases. Methods. 40:243–250 10.1016/j.ymeth.2006.08.001 [DOI] [PubMed] [Google Scholar]
- Bunz F., Hwang P.M., Torrance C., Waldman T., Zhang Y., Dillehay L., Williams J., Lengauer C., Kinzler K.W., Vogelstein B. 1999. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 104:263–269 10.1172/JCI6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., Ruckenstuhl C., Sigrist C., Wissing S., Kollroser M., Fröhlich K.U., et al. 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell. 25:233–246 10.1016/j.molcel.2006.12.021 [DOI] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 325:834–840 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Criollo A., Maiuri M.C., Tasdemir E., Vitale I., Fiebig A.A., Andrews D., Molgó J., Díaz J., Lavandero S., Harper F., et al. 2007. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 14:1029–1039 [DOI] [PubMed] [Google Scholar]
- Dörrie J., Gerauer H., Wachter Y., Zunino S.J. 2001. Resveratrol induces extensive apoptosis by depolarizing mitochondrial membranes and activating caspase-9 in acute lymphoblastic leukemia cells. Cancer Res. 61:4731–4739 [PubMed] [Google Scholar]
- Eisenberg T., Knauer H., Schauer A., Büttner S., Ruckenstuhl C., Carmona-Gutierrez D., Ring J., Schroeder S., Magnes C., Antonacci L., et al. 2009. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11:1305–1314 10.1038/ncb1975 [DOI] [PubMed] [Google Scholar]
- Erwin B.G., Persson L., Pegg A.E. 1984. Differential inhibition of histone and polyamine acetylases by multisubstrate analogues. Biochemistry. 23:4250–4255 10.1021/bi00313a036 [DOI] [PubMed] [Google Scholar]
- Ford J., Jiang M., Milner J. 2005. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 65:10457–10463 10.1158/0008-5472.CAN-05-1923 [DOI] [PubMed] [Google Scholar]
- Gurbuxani S., Schmitt E., Cande C., Parcellier A., Hammann A., Daugas E., Kouranti I., Spahr C., Pance A., Kroemer G., Garrido C. 2003. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene. 22:6669–6678 10.1038/sj.onc.1206794 [DOI] [PubMed] [Google Scholar]
- Hartford C.M., Ratain M.J. 2007. Rapamycin: something old, something new, sometimes borrowed and now renewed. Clin. Pharmacol. Ther. 82:381–388 10.1038/sj.clpt.6100317 [DOI] [PubMed] [Google Scholar]
- Hassig C.A., Schreiber S.L. 1997. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1:300–308 10.1016/S1367-5931(97)80066-X [DOI] [PubMed] [Google Scholar]
- Herker E., Jungwirth H., Lehmann K.A., Maldener C., Fröhlich K.U., Wissing S., Büttner S., Fehr M., Sigrist S., Madeo F. 2004. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164:501–507 10.1083/jcb.200310014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidvegi T., Ewing M., Hale P., Dippold C., Beckett C., Kemp C., Maurice N., Mukherjee A., Goldbach C., Watkins S., et al. 2010. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 329:229–232 10.1126/science.1190354 [DOI] [PubMed] [Google Scholar]
- Hubner N.C., Ren S., Mann M. 2008. Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics. 8:4862–4872 10.1002/pmic.200800351 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y., Struhl K. 2002. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 16:743–752 10.1101/gad.967302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., Levine B. 2010. Autophagy and the integrated stress response. Mol. Cell. 40:280–293 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S., Uzu T., Horiike K., Chin-Kanasaki M., Isshiki K., Araki S., Sugimoto T., Haneda M., Kashiwagi A., Koya D. 2010. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J. Clin. Invest. 120:1043–1055 10.1172/JCI41376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 127:1109–1122 10.1016/j.cell.2006.11.013 [DOI] [PubMed] [Google Scholar]
- Lee I.H., Finkel T. 2009. Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 284:6322–6328 10.1074/jbc.M807135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.H., Cao L., Mostoslavsky R., Lombard D.B., Liu J., Bruns N.E., Tsokos M., Alt F.W., Finkel T. 2008. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA. 105:3374–3379 10.1073/pnas.0712145105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2009. Autophagy in aging, disease and death: the true identity of a cell death impostor. Cell Death Differ. 16:1–2 10.1038/cdd.2008.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 21:3448–3449 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- Moreau K., Luo S., Rubinsztein D.C. 2010. Cytoprotective roles for autophagy. Curr. Opin. Cell Biol. 22:206–211 10.1016/j.ceb.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E., Maiuri M.C., Markaki M., Megalou E., Pasparaki A., Palikaras K., Criollo A., Galluzzi L., Malik S.A., Vitale I., et al. 2010. Caloric restriction and resveratrol prolong longevity via the sirtuin-1 dependent induction of autophagy. Cell Death Dis. 1:e10 10.1038/cddis.2009.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Kawakami K., Kametani F., Nakamoto H., Goto S. 2010. Biological significance of protein modifications in aging and calorie restriction. Ann. NY Acad. Sci. 1197:33–39 10.1111/j.1749-6632.2009.05374.x [DOI] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. 1995. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210:126–132 10.1006/bbrc.1995.1636 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., de Godoy L.M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. 2005. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 4:2010–2021 10.1074/mcp.T500030-MCP200 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Schwartz J.C., Griep-Raming J., Nielsen M.L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., et al. 2009. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics. 8:2759–2769 10.1074/mcp.M900375-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholec M., Bleasdale J.E., Chrunyk B., Cunningham D., Flynn D., Garofalo R.S., Griffith D., Griffor M., Loulakis P., Pabst B., et al. 2010. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285:8340–8351 10.1074/jbc.M109.088682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck B., Chen C.Y., Ho K.K., Di Fruscia P., Myatt S.S., Coombes R.C., Fuchter M.J., Hsiao C.D., Lam E.W. 2010. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol. Cancer Ther. 9:844–855 10.1158/1535-7163.MCT-09-0971 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Rose C., Menzies F.M., Renna M., Acevedo-Arozena A., Corrochano S., Sadiq O., Brown S.D., Rubinsztein D.C. 2010. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum. Mol. Genet. 19:2144–2153 10.1093/hmg/ddq093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin P., Chretien D., Bourgeron T., Gérard B., Rötig A., Saudubray J.M., Munnich A. 1994. Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta. 228:35–51 10.1016/0009-8981(94)90055-8 [DOI] [PubMed] [Google Scholar]
- Samara C., Syntichaki P., Tavernarakis N. 2008. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 15:105–112 10.1038/sj.cdd.4402231 [DOI] [PubMed] [Google Scholar]
- Scarlatti F., Maffei R., Beau I., Codogno P., Ghidoni R. 2008a. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 15:1318–1329 10.1038/cdd.2008.51 [DOI] [PubMed] [Google Scholar]
- Scarlatti F., Maffei R., Beau I., Ghidoni R., Codogno P. 2008b. Non-canonical autophagy: an exception or an underestimated form of autophagy? Autophagy. 4:1083–1085 [DOI] [PubMed] [Google Scholar]
- Schwartz D., Gygi S.P. 2005. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23:1391–1398 10.1038/nbt1146 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Noda T., Ohsumi Y. 2004. Interrelationships among Atg proteins during autophagy in Saccharomyces cerevisiae. Yeast. 21:1057–1065 10.1002/yea.1152 [DOI] [PubMed] [Google Scholar]
- Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. 2007. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 282:6823–6832 10.1074/jbc.M609554200 [DOI] [PubMed] [Google Scholar]