Growth factor signaling pathways regulate a broad spectrum of cellular processes ranging from proliferation to differentiation and tissue homeostasis. Activation of a signaling pathway ultimately leads to transcriptional changes in specific target genes. Although the molecular identities of many signaling pathway components have been revealed over the last years, there is still very little knowledge of how these components induce changes in the chromatin structure of target genes, a requirement for activation or repression of these genes. A new link is provided by an interesting paper in PNAS that demonstrates that in the context of Wnt signaling the histone methyltransferase SETD8 (PR-SET7, KMT5a, SET8) is recruited to enhancer regions of Wnt-regulated genes. SETD8 establishes H4K20 monomethylation (H4K20me1) at these regulatory regions, which is crucial for full activation of these target genes (1).
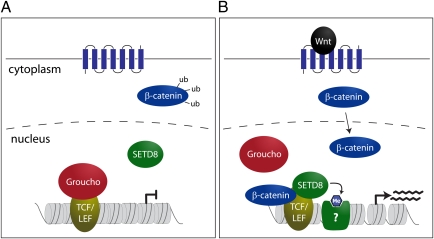
The canonical Wnt signaling pathway plays important roles for cellular differentiation during embryogenesis and tissue homeostasis in adults. Dysregulation of this pathway is often found in cancer and other diseases. The Wnt-dependent activation of target genes is mediated through the action of T cell-specific transcription factor (TCF) and lymphoid enhancer-binding factor (LEF), which constitutively bind to TCF binding elements (TBEs). In the absence of Wnt signaling, TCF/LEF cannot induce transcriptional activation due to interaction with the corepressor Groucho. Under conditions of active Wnt signaling, β-catenin is stabilized and enters the nucleus. It will then displace Groucho from TCF/LEF and recruit other proteins, leading to transcriptional activation of target genes (Fig. 1). Activation of Wnt target genes requires complex changes in the underlying chromatin structure. In this context, β-catenin was previously implicated in the recruitment of chromatin remodelers and activating histone modifications to the c-myc gene (2).
Fig. 1.
Wnt signaling stimulates SETD8-mediated H4K20me1 at TCF/LEF binding sites (TBEs). (A) In the absence of Wnt ligand, cellular β-catenin is destabilized and cannot enter the nucleus. Wnt target genes are constitutively bound by TCF/LEF transcription factors; however, transcription is blocked by binding of the repressor protein Groucho. (B) Under active Wnt signaling, β-catenin can enter the nucleus and displace Groucho from TCF/LEF. This allows for complex formation with the histone methyltransferase SETD8, which induces H4K20me1 at TBEs. Increased H4K20me1 is a prerequisite for full transcriptional activity of the Wnt target gene, possibly due to recruitment of currently unknown binding proteins.
To understand which histone modifications are implicated in the activation of Wnt target genes, Li et al. (1) probe 11 different histone methylation marks at the TBE of the AXIN2 gene in HEK293 cells. After stimulation of these cells with Wnt-conditioned medium, of all tested modifications, only H4K20me1 was significantly increased. This is surprising, as H4K20me1 is not a well-established activation mark. On the one hand, there are correlative studies that link H4K20me1 with transcriptional activity (3, 4). On the other hand, several publications demonstrate strong links of H4K20me1 with transcriptional repression and chromatin compaction (5–7). H4K20me1 is a very dynamic modification and could possibly play different roles, depending on the cell-cycle stage. Very low levels can be detected in G1 and S phase, followed by a massive increase in the G2/M transition (8). Previous analyses considered changes only in bulk H4K20me1 levels. Cell cycle-dependent changes at single loci have not been analyzed yet.
In metazoans, H4K20me1 is controlled by the histone methyltransferase SETD8. Using a variety of knockdown and overexpression approaches, Li et al. (1) demonstrate that SETD8 is responsible for H4K20me1 at human TBEs. Knockdown of SETD8 reduces H4K20me1 at these regulatory regions, and, consequently, several of the tested Wnt target genes show reduced activation. Expression of catalytically inactive SETD8 protein cannot rescue these defects, and, therefore, H4K20me1 must directly play important roles in activating transcription. This finding is difficult to explain, as currently there is only limited knowledge about the mechanisms by which H4K20me1 can affect chromatin structure. In general, histone modifications are thought to mediate effects on chromatin as interaction platforms for specific binding proteins, which then mediate changes in the underlying chromatin structure. Three binding proteins for H4K20me1 have been identified so far. N-CAPD3 and N-CAPG2 are subunits of the condensin II complex. Binding of these proteins to H4K20me1 in G2/M phase is important for mitotic chromosome condensation (6). L3MBTL1 is a three malignant brain tumor (MBT) domain protein and recognizes H4K20me1 in the context of additional modifications (7). L3MBTL1 was shown to negatively regulate gene expression by inducing a compact chromatin structure (7, 9). It is not clear whether any of these binding proteins is recruited to TBEs upon Wnt signaling; however, due to their roles in gene repression and chromatin condensation, this seems rather unlikely. Therefore, mechanistic insight into the activating roles of H4K20me1 can be revealed only upon identification of other binding proteins.
Li et al. (1) then go on to test whether SETD8-mediated H4K20me1 regulates Wnt signaling in a more physiological setting and decide to analyze zebrafish development. Morpholino knockdown of the SETD8 homolog Setd8a results in reduced H4K20me1 at the Wnt8 target gene tbx6. Consistent with a Wnt-dependent recruitment mechanism for Setd8a, H4K20me1 is reduced upon knockdown of Wnt8. Setd8a morphant embryos show developmental defects, such as a shorter trunk and tail region. These phenotypes mimic aspects of Wnt8 morphants; however, due to additional functions of Setd8a outside of Wnt signaling, the phenotypes are not expected to fully match. Notably, disruption of Setd8 in the mouse results in a much stronger phenotype. Setd8 mouse mutant embryos die at the four-cell stage due to accumulation of DNA damage and mitotic defects (10). Also, in Drosophila, which is a phylogenetically even more distant organism, Setd8 has important functions in ensuring chromosome segregation and genome integrity (11, 12). Why is the developmental phenotype of zebrafish Setd8a morphants comparably mild? One possibility is that, in a knockdown situation, residual Setd8a activity could prevent appearance of stronger phenotypes. Another explanation is that Setd8b, a second SETD8 homolog in zebrafish, could have partially redundant functions (13). Future experiments are needed to distinguish these possibilities.
In their PNAS paper, Li et al. (1) provide several lines of evidence that SETD8-mediated H4K20me1 at TBEs is important for full activation of Wnt target genes. But how is SETD8 recruited to TBEs? To address this question, the authors test whether SETD8 can interact with the
The study by Li et al. identifies a new function for H4K20me1 in transcriptional activation.
TCF/LEF transcription factors. In a series of interaction tests, they are able to demonstrate that SETD8 can form complexes with TCF4 and LEF1. These complexes exist only when Wnt signaling is active, and it was therefore plausible to test whether SETD8 is recruited through interaction with β-catenin, as shown before for the Mll complex (2). However, no direct interaction of SETD8 with β-catenin could be demonstrated. Therefore, Li et al. (1) test the intriguing hypothesis that the displacement of Groucho by β-catenin could facilitate SETD8 binding. Groucho was shown before to interact with the high-mobility group (HMG) domain of TCF4 (14). The very same region is now mapped by Li et al. (1) to bind SETD8. Therefore, Groucho might block the SETD8 binding site in TCF4. Immunoprecipitation of TCF4 in cells that coexpress constitutively active β-catenin, SETD8, and Groucho revealed that SETD8 can participate in the TCF4 complex only when β-catenin is present and Groucho is displaced, suggesting a novel mechanism for recruitment of chromatin-modifying activities in Wnt signaling (Fig. 1).
The study by Li et al. (1) identifies a new function for H4K20me1 in transcriptional activation that will stimulate many follow-up experiments. A central question will be the mechanism of transcriptional activation. Is there a specific binding protein, and how does it contribute to opening the chromatin structure? Another question regards the inactivation of Wnt signaling: Is there active removal of H4K20me1 when Wnt target genes should be shut down? Two scenarios are possible. It might be that H4K20me1 is converted to a higher methylation state by the action of Suv4-20h enzymes (15). This could induce a heterochromatin-like repressive state around the TBE. Alternatively, H4K20me1 could be actively removed by histone demethylases (6). An intriguing hypothesis is that such H4K20me1-antagonizing activities could be targeted by Groucho complexes that return to TBEs when β-catenin has left.
Acknowledgments
Work in the G.S. laboratory is supported by grants from the Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung und Forschung, and the Munich Center for Integrated Protein Science.
Footnotes
The author declares no conflict of interest.
See companion article on page 3116.
References
- 1.Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci USA. 2011;108:3116–3123. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talasz H, Lindner HH, Sarg B, Helliger W. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J Biol Chem. 2005;280:38814–38822. doi: 10.1074/jbc.M505563200. [DOI] [PubMed] [Google Scholar]
- 4.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houston SI, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trojer P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 8.Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalakonda N, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda H, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakaguchi A, Steward R. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J Cell Biol. 2007;176:155–162. doi: 10.1083/jcb.200607178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun XJ, et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One. 2008;3:e1499. doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brantjes H, Roose J, van de Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]