Abstract
We describe patterns of DNA sequence diversity in a newly identified sex-linked gene, SlX9/SlY9, in Silene latifolia (Caryophyllaceae). The copies on both sex chromosomes seem to be functional, and each maps close to the respective X- and Y-linked copy of another sex-linked gene pair, SlCypX/SlCypY. The Y-linked copy has low diversity, similar to what has been found for several other Y-linked genes in S. latifolia, and consistent with the theoretical expectations of hitch-hiking processes occurring on a non-recombining chromosome. However, SlX9 has higher diversity than other genes on the S. latifolia X chromosome. We evaluate the hypothesis of introgression from the closely related species S. dioica as an explanation for the high sequence diversity observed.
Keywords: Silene latifolia, sex chromosomes, introgression, SlX9
Introduction
Measurements of neutral nucleotide diversity have been used to infer that Y chromosomes in some species, including neo-Y chromosomes are undergoing hitch-hiking processes that are expected to cause genetic degeneration in these non-recombining genome regions. In contrast to the situation for autosomal and X-linked loci, the non-recombining region of a Y chromosome is expected to experience selective interference effects among linked loci, such as weak selection Hill–Robertson interference (Hill and Robertson, 1966; McVean and Charlesworth, 2000; Comeron, 2008), genetic hitch-hiking because of positive selection (Maynard Smith and Haigh, 1974; Kaplan et al., 1989) or the elimination of strongly deleterious variants (background selection and Muller's ratchet (Muller, 1964; Charlesworth et al., 1993; Gordo et al., 2002)). These effects will reduce diversity values compared with those of genes on the X or autosomes, in addition to the expected reduction in diversity because of the smaller Y effective population size caused by the smaller number of Y chromosomes in the population.
Silene latifolia is a dioecious plant used to study the evolution of young sex chromosomes. Synonymous site diversity values of six X-linked genes studied (SlX1, SlX4, DD44-X, SlssX, SlCyt and SlCyp) vary between 0.07 and 5.1% (Atanassov et al., 2001; Laporte et al., 2005; Bergero et al., 2008; Kaiser et al., 2009); the lowest value (for SlssX) may be due to a recent selective sweep in the genomic region, which did not affect diversity at the nearby DD44-X (Filatov, 2008). In contrast, four Y-linked genes studied (SlY1, SlY4, DD44-Y and SLAP3Y) have silent diversity values between 0 and 0.28% (Atanassov et al., 2001; Matsunaga et al., 2003; Laporte et al., 2005), consistent with the predicted reduced Y chromosome Ne.
Interpretation of within-species polymorphism in S. latifolia, and comparison of X and Y diversity values, are, however, complicated by the possibility of introgression from its closely related sister species Silene dioica, which could increase diversity for some loci. S. latifolia forms natural hybrids with S. dioica (Desfeux et al., 1996; Minder et al., 2007; Minder and Widmer, 2008), although the species differ ecologically (Baker, 1947, 1948), with S. latifolia having white flowers, a generally wider distribution and growing in dry, open habitats, whereas S. dioica has red flowers and grows at the margins of woodlands. The pollinators also differ, with S. latifolia being mainly visited by the dusk-flying moth Hadena bicruris, and S. dioica visited during the day by bumblebees and butterflies (Bopp and Gottsberger, 2004; Minder et al., 2007). There are also some geographic distribution differences, with S. dioica being found mainly in Northern Europe (Baker, 1947, 1948; Karrenberg and Favre, 2008). However, hybrids with pink flowers are common in the wild, and data from three sex-linked genes, DD44, SlX1 and SlX4 (Ironside and Filatov, 2005; Laporte et al., 2005), as well as AFLP markers (Minder et al., 2007; Karrenberg and Favre, 2008; Minder and Widmer, 2008), suggest that introgression of S. dioica sequences into S. latifolia is common in nature.
To understand the early stages of sex chromosome evolution and specifically to test for Y chromosome degeneration and the effects of different chromosomal environments on nucleotide diversity, we have isolated genes from the S. latifolia X and Y chromosomes. In this study, we describe a newly identified sex-linked gene pair in S. latifolia, SlX9/SlY9, and analyze its diversity levels, which suggest Y chromosome degeneration and X chromosome introgression.
Materials and methods
Plant materials
Sex-linkage of SlX9 in S. latifolia was established using the mapping family H2005-1 (Bergero et al., 2007), which is a full-sib cross between F1 offspring whose parents came from different European populations (male E2004-17-1, from the Netherlands, and female E2004-11-1, from Canche, Northern France). Ninety-two plants from this family were used to map its location on the X chromosome. The mother of the mapping family is a heterozygote for two X-linked alleles that produced PCR products of different lengths (bands of approximately 450 and 600 bp, see Supplementary Figure 1), which were used for genetic mapping, as described below. The panel of 38 deletion mutants used to find the location of SlY9 on the Y chromosome is described in Bergero et al. (2008).
To study sequence diversity, we used a sample of 46 S. latifolia males from 24 European populations, covering most of the range of the species. Nineteen S. dioica individuals were also sampled, from Scotland, France and Finland, including 13 males (Supplementary Tables 1 and 2).
PCR amplifications and primers
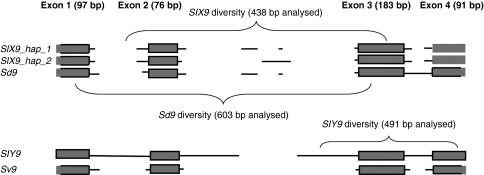
SlX9 was identified from a S. latifolia complementary DNA (cDNA) library derived from male leaf primordia, and shown to be a sex-linked gene (Kaiser et al., 2009). The complete cDNA sequence was obtained, and primers were designed based on this sequence. As described below, it proved very difficult to sequence this gene in its entirety from all our sampled individuals, and only partial genomic sequences were obtained, with different regions sequenced from different species, and from the X and Y copies. To obtain sequences, new primers were designed from the sequences yielded by the initial primers; these are listed in Supplementary Table 3. Figure 1 (below) diagrams the regions amplified for the diversity study, which differ for the X- and Y-linked alleles as follows. For SlX9, the sequences include part of intron 1, exon 2, intron 2, exon3, and part of intron 3 (totalling 255 coding and 178 non-coding sites). For SlY9, they include part of intron 2, exon 3, intron 3 and part of intron 4 (270 coding and 222 non-coding sites).
Figure 1.
Schematic view of the alignment of SlX9, SdX9, SlY9 and Sv9. Sizes of introns and exons are not drawn to scale. Both X-linked intronic variants are shown (SlX9_hap_1 and SlX9_hap_2). Thick lines around exons: sequences available for at least one individual. (The original S. latifolia cDNA clone from which the gene was identified contained the whole open reading frame; sizes of exons for which we do not have complete sequences are drawn based on the assumption that exon sizes are the same as for the cDNA.) Note: sequences used in the diversity studies of SlX9, SdX9 and SlY9 only cover parts of the gene sequences, as indicated.
PCR amplification was generally carried out using Taq JumpStart (Sigma-Aldrich, Poole, UK), and the following conditions: initial denaturation at 95 °C for 5 min, 10 cycles of denaturation at 95 °C (30 s), annealing at 55–58 °C (30 s), extension at 72 °C (1–1.5 min), final extension at 72 °C for 15 min. PCR amplicons were cleaned using ExoSAP-IT (GE Healthcare, Little Chalfont, UK) and sequenced on an ABI 3730 capillary sequencer (Applied Biosystems, Foster City, CA, USA) and sequences edited using Sequencher 4.7 (Gene Codes Corporation, Ann Arbor, MI, USA).
The two X-linked alleles in the mapping family (see above) were cloned from PCR products and sequenced. Primers used to amplify across introns 1, 2 and 3 are listed in Supplementary Table 3. The gene structure of SlX9 was then inferred by comparing the SlX9 genomic sequence with its cDNA sequence, as well as by comparisons with Arabidopsis thaliana and Silene vulgaris gene structures. A BLASTN search was performed at www.ncbi.nlm.nih.gov to identify homologous genes in other organisms and their functions.
Obtaining and mapping the Y-linked homologue
As described in Kaiser et al. (2009), using primers RB18_F and RB18_R (Supplementary Table 3) to amplify DNA from male and female plants from family H2005-1, yielded a male-specific PCR product of approximately 1.2 kb. To sequence the Y-linked homologue of SlX9, the longer, male-specific allele (see Supplementary Figure 1) was cut from the agarose gel, cloned and sequenced. To obtain the 5′ coding sequence of SlY9, which was not present in the region initially sequenced, the cloned PCR products from family H2005-1 were also used to design new, Y-specific primers from this sequence (Supplementary Table 3). These primers were also used for the SlY9 diversity study (see below).
To test whether the Y-linked gene was expressed, we did nested PCR using cDNA derived from male flower tissue. The first round of PCR amplification used the primers TsShort and RB18_R (Supplementary Table 1), in which TsShort matched the sequence to which the cDNA was ligated. The second round used TsShort and the Y-specific primer RB18_Y_E3-R (Supplementary Table 3).
For deletion mapping SlY9 on the Y chromosome, PCR amplifications were scored in the deletion mutants (see above) using the primers RB18_F and RB18R (Supplementary Table 3). The location of SlY9 was inferred by comparing the presence–absence of the Y-linked, larger, fragment to the presence–absence of other Y-linked genes (Zluvova et al., 2005, 2007; Bergero et al., 2008).
Diversity of SlX9 and SlY9 and linkage disequilibrium analysis
To study sequence diversity of SlX9 and SlY9, parts of the genes (listed in Supplementary Table 3) were amplified in 46 S. latifolia male individuals (Supplementary Table 1). The X-linked copies were amplified using the primers RB18_exon_1_F and RB18_exon_4_R, and the Y-linked copies with either RB18-Intron2-male-F-2 and RB18-3′UTR, or RB18-Intron2-male-F-3 and RB18-3′UTR, respectively (Supplementary Table 3). The sequence fragments were assembled and aligned manually using the programme Se-Al v.2.0a11 (Se-Al: sequence alignment editor, http://evolve.zoo.ox.ac.uk/). The sequences have been deposited in GenBank under Accession numbers HM141608–HM141717.
Sequence diversity was analyzed using DnaSP 4.0 software (Rozas and Rozas, 1999), excluding indel polymorphisms. To estimate between-population differentiation, the SlX9 and SlY9 sequences were divided into four broad geographic groups based on their location of origin (‘Northern Europe', ‘North-Eastern Europe', ‘Mediterranean group', and ‘Spain and Portugal', see Supplementary Table 1), KST statistics were computed in DnaSP 4.0. An NJ tree of the X-linked sequences was constructed in MEGA 3.1 (Kumar et al., 2004).
To test for introgression from the sister species S. dioica, we used the principle that introgression will cause variants from one species to be found in the same haplotype more often than expected based on their frequencies in the hybrid population, that is, there will be positive linkage disequilibrium among segregating sites, specifically in regions containing multiple successive variants from S. dioica. The associations are expected to last until they are broken up by recombination events, so that the physical distance over which we observe positive LD can be used as a measure of the time when hybridization occurred and/or the strength of selection against hybrid individuals. We used DnaSP 4.0 to calculate D′, a measure of linkage disequilibrium among segregating sites, standardized relative to its maximum possible value.
Tests of neutrality and recombination estimates
Using population samples of SlX9 and SlY9, several neutrality test were performed in DnaSP 4.0, including Tajima's D, Fu and Li's D* and F*, Fu's F and Fay and Wu's H statistics (Tajima, 1989; Fu and Li, 1993; Fu, 1997; Fay and Wu, 2000). Levels of statistical significance were estimated using coalescent simulations in DnaSP 4.0, conservatively assuming no recombination (Tajima, 1989; Wall, 1999). We also used DnaSP 4.0 to calculate an estimate of the recombination parameter, R=3Ner (for SlX9); the minimum number of recombination events, and Strobeck's S statistic, which gives the probability of sampling the same or smaller number of haplotypes as observed in the population sample, given an estimate of θ (Strobeck, 1987).
Comparisons with outgroup species
Using the primer pair RB18_exon_1_F and RB18_exon_4_R (Supplementary Table 3), the homologue of SlX9/SlY9 was amplified in S. vulgaris, a gynodioecious species that lacks sex chromosome and forms an outgroup to the S. latifolia/S. dioica clade. PCR amplification of the homologue of SlX9 in S. dioica was carried out using different combinations of primers, listed in Supplementary Table 3 (RB18-male-exon1-F and RB18_male_exon3-R; RB18_exon_1_F and RB18-E3-R-beg; RB18-E2-F-beg and RB18_exon_4_R; RB18-male-exon1-F and RB18-exon2-R-Male; RB18-male-exon1-F and RB18_exon_4_R).
We call these homologues Sv9 (for S. vulgaris) and SdX9 (for S. dioica); we infer that SdX9 is X-linked, but we did not obtain the Y-linked homologue for this species (see Results). The HKA test (Hudson et al., 1987), as implemented in DnaSP 4.0, was used to compare SlX9 and SlY9 diversity, using Sv9 as the outgroup sequence.
To test whether SlY9 has an accelerated rate of mutation, all fourfold degenerate sites were extracted from the SlX9/Y9 coding sequence using DnaSP 4.0. The baseml programme of PAML was used to compare the rates of evolution along the three branches of the phylogenetic tree, using the Sv9 outgroup sequence. A model that assumes a single rate of evolution for all branches (‘clock=1' in Table 2, below) was compared with a model that assumes a different rate for the Y-linked branch compared with the X-linked genes (‘clock=2').
The codeml programme of PAML was used to estimate KA/KS ratios on the branches leading to SlX9 and SlY9, respectively, again using S. vulgaris as an outgroup, and allowing each branch of the tree to have its own rate of evolution. The likelihood of obtaining the data under a model in which all three branches of the tree have the same KA/KS (‘model=0' in Table 2, below) was compared with a model (‘model=2') in which there was one KA/KS ratio in the SlY9 branch, and one for the branches leading to SlX9 and Sv9.
Divergence between the X- and Y-linked copies of SlX9-Y9 was estimated using DnaSP 4.0. The exonic sequence of male E2004-15-1 (from Serre de Nogere, Portugal) was compared with the set of X-linked sequences amplified for the diversity study (255 coding sites).
Results
Discovery of the new gene
The segregation results for intron size variants in the mapping family (see Materials and methods) are shown in Supplementary Figure 1, and clearly indicate sex-linkage (Kaiser et al., 2009). The new sex-linked gene was named SlXY9. The original cDNA sequence contains a continuous open reading frame of 444-bp coding sites (148 amino acids), and its sequence identifies it as an X-linked allele, similar to SlX9 in the mapping family. It is probably a housekeeping gene, as BLAST searches showed similarity to the photosystem I subunit of A. thaliana (At1g08380) and to undefined membrane proteins of tobacco, wheat and rice.
To test whether the Y-linked gene was expressed, we used nested PCR with cDNA derived from male flower tissue (see Materials and methods). We retrieved a cDNA sequence that overlaps, and is identical to, part of the genomic SlY9 sequence (whereas there were six differences from all X-linked sequences, including the original cDNA and the X-linked sequences in our diversity study, see below). Therefore, both Y and X copies produce transcripts in male flower buds.
Comparisons between the original cDNA sequence and genomic sequences of SlX9 and SlY9 (including those in our diversity study) show that there are four exons in S. latifolia, and three introns, one more than in the A. thaliana putative homologue (Figure 1). Intron 2 of the Y-linked copy in the mapping family has extra sequence (yielding a distinctive large band in males, in addition to length differences between the maternal and paternal X copies, see Supplementary Figure 1). The complete intron 2 genomic sequence was obtained only for this male, and SlY9 in Figure 1 and the sizes in Supplementary Figure 1 are based on this plant (for the other male plants, in the diversity study, the forward primer was within intron 2, and at most 86 bp of intron 2 sequence was obtained, so the full length of intron 2 in these plants is unknown).
No BlastN or BlastX matches were found for the insertion in SlY9, and no repetitive sequences were detected using the RepeatMasker programme (www.repeatmasker.org). However, the insertion may be a TE of a new type, or changed too much to be recognizable. The other two SlY9 introns are also longer than those of SlX9 or the S. vulgaris homologue (Sv9 in Figure 1). This suggests that the intron sizes have expanded in the Y-linked copy, consistent with previous findings of non-coding sequence accumulation on the S. latifolia Y chromosome (Hobza et al., 2006; Cermak et al., 2008), and longer introns for the Y-linked genes DD44Y and SlX3 (Marais et al., 2008), and the observed expansion of the Drosophila miranda neo-Y and the non-recombining Y-like region of papaya (Liu et al., 2004; Bachtrog et al., 2008).
X and Y haplotypes
To study sequence diversity, we used 46 males from different European locations (Supplementary Table 1), and obtained 46 SlY9 and 40 SlX9 sequences (probably underestimating X diversity, because SlX9 failed to amplify from a few plants, suggesting that sequence differences are present in their X alleles). Our PCR amplifications with X- or Y-specific primers always yielded just one amplicon, identifiable as either SlX9 or SlY9 by the intron length variant that distinguishes Y-linked alleles (see above). The gene is therefore single-copy in the genome, and is sex-linked throughout the species' range. No frame shift mutations or premature stop-codons were found in the coding regions in any of the SlY9 (or SlX9) sequences.
The X-linked sequences are of two distinct sequence types. In 13 SlX9 sequences, intron 2 was ∼485-bp long (yielding a band of 585 bp, including the amplified portions of the flanking exons, which accounts for the ∼600-bp X-linked band observed in the mapping family), whereas in the others it was only ∼380 bp (corresponding to the 480-bp X band amplified in the mapping family). The intron sequences of the two types were highly diverged, and were aligned manually (see Materials and methods and Supplementary Figure 2).
Location of the SlX/Y9 gene on the X and Y chromosomes, and X–Y divergence
SlX9 is closely linked to a previously described X-linked gene, SlCypX (Bergero et al., 2007; Kaiser et al., 2009). Deletion mapping (see Materials and methods) showed that SlY9 is always co-deleted with SlCypY, suggesting that these genes have been physically close because recombination stopped in this sex chromosome region. Both genes should therefore have similar X–Y sequence divergence. This region probably stopped recombining before the S. latifolia and S. dioica split, because synonymous divergence between SlCypX and SlCypY is 6.1%, and SlCypY carries a MITE insertion in intron 2 of both species, which is absent from SlCypX (Bergero et al., 2007). Divergence between SlX9 and SlY9 can be estimated only roughly, because only parts of the sequences could be obtained. As discussed below, SlX9 has high sequence diversity (and all non-synonymous differences between SlX9 and SlY9 were polymorphisms in the SlX9 sequences), inflating the raw X–Y divergence estimates in Table 1. We therefore also computed net divergence, which estimates fixed X–Y differences. The results (Table 1) consistently support divergence in line with that of SlCypXY, whether we include all X sequences, or only the longer or shorter type.
Table 1. Sequence divergence estimates between SlX9 and SlY9 (K values), and diversity estimates (π values) within SlX9 and in the Silene dioica homologue.
| Region compared (footnotes indicate the sequences that were used) | KS or πS (synonymous or silent sites, see the left-hand column)a | KA or πA (non-synonymous sites) |
|---|---|---|
| SlX9 versus SlY9: raw divergence values | ||
| 392 coding sitesb | 0.153 | 0.00069 |
| 255 nucleotides in exons 2 and 3 (85 codons)c | 0.144 | 0.00026 |
| SlX9 versus SlY9: net divergence (excluding SlX9 polymorphisms)d | ||
| 90 silent sites, SlX9 versus SlY9 | 0.063 | — |
| 255 coding sequence sites (as above) | 0.053 | 0 (small negative value) |
| Long X-linked copies only, versus SlY9, 255 coding sites | 0.042 | 0 |
| Short X-linked copies only, versus SlY9, 255 coding sites | 0.062 | 0 |
| Diversity within S. latifoliae | ||
| X-linked copy, synonymous sites (85 codons in exons 2 and 3) | 0.092 | 0.0005 |
| X-linked copy, 240 silent sites | 0.040 | — |
| Long X-linked copies, 559 silent sites | 0.032 | — |
| Short X-linked copies, 412 silent sites | 0.033 | — |
| Y-linked copy, 283 silent sitesf | 0.0016 | 0 |
| X-linked gene diversity within S. dioica | ||
| 514 silent sitesg | 0.018 | — |
| 117 coding sequence sites | 0.066 | 0.0035 |
With Jukes–Cantor correction for saturation.
Comparing the longer (Y) sequence from the male parent of the mapping family with the original complementary DNA clone.
SlX9 from the diversity study and SlY9 from the male parent of the mapping family.
Estimated by subtracting the average of the diversity values.
On the basis of single-nucleotide polymorphisms only.
It is to be noted that the X and Y estimates of π are based on different gene regions (see Figure 1).
From our sample of 18 X-linked sequences; as explained in the text, no Y-linked sequences were amplified from this species.
A caveat is that, if SlX9 and SlY9 stopped recombining independently in S. latifolia and S. dioica after their split, the X-linked copy of each species should be more similar to its Y-linked copy than to the X of the other species, and introgression could then inflate the X–Y divergence. Different degrees of introgression of different regions of the X could then obscure the true times when recombination stopped. However, this is unlikely to affect our conclusions, because SlX9–SlY9 divergence is much higher than divergence between the two species. Silent site divergence between the S. dioica sequence and that of SlX9 is 3.0% (based on 227 sites), similar to estimates from other X-linked genes in these species (the highest previous synonymous divergence estimate is 4.4% for SlCyp-X, see Bergero and Charlesworth, 2009).
Divergence from outgroup species
Several sex-linked genes have been found to have higher Y than X mutation rates (Filatov and Charlesworth, 2002; Filatov, 2005), but SlX9 and SlY9 have similar divergence from the S. vulgaris homologue (Jukes–Cantor corrected divergence estimates for synonymous sites were 21% for both SlX9, based on 255 coding sites, and SlY9, using 261 coding sites). PAML analysis confirms that SlY9 has not evolved significantly faster than the X-linked copy, using fourfold degenerate sites (72 sites) or all 444 alignable coding sites (Table 2).
Table 2. PAML-based tests of rate differences between SlX9 and SlY9, using Sv9 as an outgroup sequence.
| Program | Comparison | Number of sites | Model | χ2 | df | P |
|---|---|---|---|---|---|---|
| Baseml | Fourfold degenerate sites | 72 | Clock=1 | 2.67 | 1 | 0.102 |
| Clock=2 | ||||||
| Baseml | All coding sites | 444 | Clock=1 | 0.32 | 1 | 0.57 |
| Clock=2 | ||||||
| Codeml | KA/KS | 438 | Model=0 | 1.51 | 1 | 0.22 |
| Model=2 |
The dN/dS ratio on the branch leading to SlY9 is 0.077, and does not differ significantly from that on the other branches (Table 2). Together with the lack of frame shift mutations or premature stop-codons in SlY9, as well as its expression as mRNA (see above), these results suggest that the SlY9 gene is still functional.
Sequence diversity within S. latifolia
We sequenced a portion of SlX9 including parts of exons 2 and 3 and introns 1–3. Within S. latifolia, the estimated synonymous diversity (πS) of the X-linked copy is much higher than for other X-linked genes (see above), but this is based on few codons; silent site diversity is also high (Table 1). As described further below, the high diversity results partly from the presence of two SlX9 sequence types (the two X haplotypes with different intron sizes described above, see Figure 1). However, most diversity is within the types, not between them: silent site diversity with JC correction within either set of X-linked sequences was 3.2%, and the net silent site divergence is <1%.
The Y-linked copy, SlY9, has substantially lower diversity (Table 1), and there were only two single-nucleotide polymorphisms, both in introns. There was also one indel polymorphism in intron 3 of SlY9: the sequences from two males from a population in Greece had an insertion of the triplet TCA. Although the numbers of sites analyzed in both sets of sequences are small, an HKA test using all site types finds a significant difference in X–Y diversity estimates, taking into account the different ploidy level (χ2=5.5, P<0.05). The X/Y diversity ratio is 24.5, using the species-wide silent site estimates in Table 1. SlX9 showed significant differentiation between populations (KST estimate 0.109, P<0.001). The estimate for SlY9 was only slightly higher (0.122, P<0.05), but is based on only two segregating sites.
Tests for introgression from S. dioica
Given the genetic evidence mentioned above that there are no duplicate copies of these genes, we tested whether the two different SlX9 sequence types in S. latifolia described in the preceding section, and the high SlX9 diversity, reflect introgression from S. dioica. Very recent introgression seems unlikely, because the short length type was found in populations from the Mediterranean region, in which S. dioica is absent (Prentice et al., 2008). Both intron sizes were found within S. latifolia populations from Sweden and Poland (in which S. dioica is present), and in Greece (where S. dioica is absent), and the short SlX9 sequence was also found in Italy and Spain.
To test this further, we sequenced portions of the gene (Figure 1) from S. dioica sampled from several geographic regions of Europe. All 13 S. dioica males yielded only single sequences, whereas 1 of the 6 females (A2009_2_female_1) contained several heterozygous single-nucleotide polymorphisms, and another female (FS14) was heterozygous for a 22-bp length variant in intron 2 (it is to be noted that this female yielded a short sequence, which was excluded from further analyses). It thus seems that only the X-linked copy amplified from S. dioica. The Y copy may have been deleted in S. dioica. Alternatively, given the high X–Y divergence in S. latifolia (Table 1), the S. dioica Y-linked sequence may be too diverged for the primers to work.
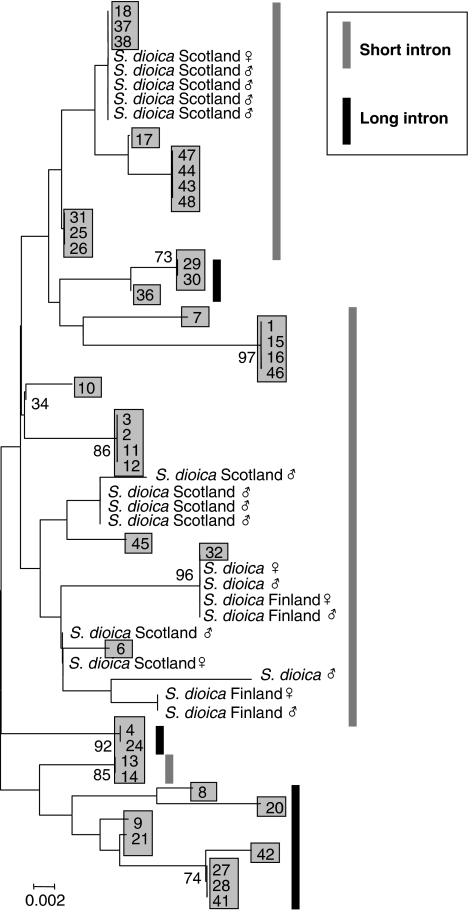
All 19 S. dioica X-linked sequences had the short haplotype (Figure 1). Diversity is slightly lower than for S. latifolia X-linked sequences (Table 1); for a region of 165 bp sequenced from both species, π=2.3%, versus 3.1% in S. latifolia. The shorter S. latifolia SlX9 haplotypes have sequences similar to the S. dioica sequences (silent site divergence 2.7%, with Jukes–Cantor correction, based on 314 sites), whereas the long ones differ slightly more (5.7%, based on 277 silent sites). In the NJ tree (Figure 2), several S. latifolia sequences cluster closely with S. dioica, and all these have the short intron 2. Several individuals from Northern France and England are very similar in sequence, or identical (plant 38), to Sd9 sequences, but the sequence from individual 6 is also similar to a group of SdX9 sequences (Figure 2), although it comes from Southern France, in which S. dioica is not found.
Figure 2.
Neighbour-joining tree of SlX9 and SdX9. The tree was constructed in MEGA, based on all sites, with complete deletion. Bootstrap support values >70% are shown at the branches. The S. latifolia male individuals (blue shading) are identified by their numbers in Table 1, and the S. dioica plants are identified by their populations of origin when known.
The part of the gene in which we have at least 2 X-linked sequences from each species contains 138 variable sites (including both species), of which 33 are in the intron 2 region that is absent from the short sequences. Our results from these sites suggest introgression. SlX9 and SdX9 share 20 polymorphisms, 12 at sites in regions present in both S. latifolia haplotypes and 8 in the intron 2 region that is present only in the S. latifolia short haplotype group plus S. dioica (Supplementary Figure 2, summarized in Supplementary Table 2). There were no fixed differences between the sequences of the two species, but 15 sites have polymorphisms exclusive to S. dioica.
Linkage disequilibrium among sites polymorphic in the X-linked alleles
As expected, recombination is detected in the S. latifolia SlX9 alleles (Table 3). Among the 37 single-nucleotide polymorphism sites, 9 pairs have significant linkage disequilibrium in our sample of alleles (Fisher's exact test; P<0.05, after Bonferroni correction for multiple comparisons). Eight of these pairs were <100 bp apart from each other, in or near exon 3, whereas one pair of sites is separated by 602 bp (Supplementary Table 5, Supplementary Figure 2). The LD results do not strongly indicate introgression, as opposed to ancestral polymorphism.
Table 3. Tests of neutrality and recombination for SlX9 and SlY9.
| Gene | Tajima's D | Fu and Li's D* | Fu and Li's F* | Fu's F | Strobeck's S statistic | Fay and Wu's H | R=3Ner (adjacent sites) |
|---|---|---|---|---|---|---|---|
| SlX9 | −0.32 | −0.25 | −0.32 | −7.63 | 1.000 | −1.23 | 0.14 |
| NS | NS | NS | P<0.05 | NS | |||
| SlY9 | −1.19307 | −1.74431 | −1.84007 | −0.515 | 0.836 | 0.46377 | — |
| P<0.1 | P<0.1 | P<0.1 | NS | NS |
Abbreviation: NS, nonsignificant.
However, ancestral polymorphism leaves the high diversity unexplained. Balancing selection in the common ancestor of the two species, and maintained in S. latifolia could explain this, but predicts that many variants should be at higher frequencies than expected under neutrality, resulting in a positive Tajima's D statistic. However, in our sample, Tajima's D for SlX9 was negative, although nonsignificant, as are other tests of selection on SlX9 (Table 3), except for Fu's F, which is very sensitive to a frequency spectrum bias towards rare polymorphisms.
Discussion
Causes of low Y diversity relative to the X
Like other known genes on the S. latifolia sex chromosomes, the SlY9 diversity is lower than expected purely from the lower Y effective population size assuming a 1:1 sex ratio and equal variance of offspring numbers in the two sexes. A reduced mutation rate on the Y cannot account for the diversity difference, because rates in S. latifolia either do not differ between the X and Y copies (as we found here), or are significantly higher for the Y-linked copies (Filatov and Charlesworth, 2002; Filatov, 2005; Nicolas et al., 2005).
Population structure can increase species-wide diversity, but subdivision should affect diversity on the Y chromosome more than that of the X, even if pollen and seed dispersal rates are the same (Laporte and Charlesworth, 2002). Strong population structure has been found in S. latifolia for the SlX4/Y4 and DD44-X/Y genes (Laporte et al., 2005) and SlX1/Y1 (Atanassov et al., 2001; Ironside and Filatov, 2005), in all cases much more markedly for the Y than the X, because of the low Y diversity. Our small within-population samples are not suitable for tests for subdivision, but we found modest subdivision at a larger geographic scale for the X, consistent with results for other X-linked loci. We conclude that diversity on the Y is reduced, rather than elevated on the X, and that subdivision or local adaptation probably cannot explain the high X diversity.
The low diversity is therefore probably because of hitch-hiking processes in the S. latifolia Y chromosome's large non-recombining region. This chromosome probably contains many active genes. There is as yet no adequate estimate of the proportion of X-linked genes that have copies on the Y, or of the absolute number of functional genes on the Y. Only 1 of the 11 genes so far identified on the X, the recently added SlCyt gene (Kaiser et al., 2009), has a missing or truncated Y copy, and none has a Y-linked allele with any evident loss-of-function mutation, although several of these genes have lower expression than their X-linked alleles, and molecular evolutionary analysis suggests some loss of adaptation, relative to the X-linked alleles (Marais et al., 2008). However, many of these genes were ascertained by Y-linkage of genes discovered from EST sequences, which will preferentially detect functional Y-linked genes. At present, therefore, one can conclude only that the S. latifolia Y carries several functional genes.
Hitch-hiking should produce an excess of low-frequency variants. However, with the small sample of genes whose diversity has been studied in this species, and the small number of variants in the Y-linked alleles, it is difficult to compare the frequency spectra for X- and Y-linked genes, or even to estimate Tajima's D for Y-linked genes. The X-linked genes so far studied, SlssX, SlCypX and SlCyt, have negative Tajima's D values (Bergero et al., 2008; Kaiser et al., 2009), so it is likely that recent demographic history may have produced a genome-wide excess of low-frequency variants, which will impede interpretation of the frequency spectrum of Y-linked variants. Previous studies of three Y-linked genes (SlY1, SlY4 and DD44Y) did not find significantly negative Tajima's D values, but pointed out that population subdivision, with local fixation of different Y haplotypes, could cause a deficit of rare variants, obscuring the effects of sweeps (Atanassov et al., 2001; Laporte et al., 2005). However, strong Y chromosome differentiation among populations (Ironside and Filatov, 2005) argues against recent species-wide selective sweeps, but is consistent with background selection, or geographically localized sweeps (Ironside and Filatov, 2005). The positive (though nonsignificant) Fay and Wu's H for SlY9 also argues against a selective sweep on the Y chromosome.
Our results are, however, consistent with purifying selection, and thus with the suggestion that Muller's ratchet and/or background selection should be most important early in Y chromosome evolution, when the number of functional genes that can undergo detrimental mutations is still very large (Bachtrog, 2008). Selective sweeps may become more important after the Y has lost many genes; there is then a higher chance that beneficial mutations can occur on chromosomes not carrying many deleterious mutations.
Causes of high X diversity in S. latifolia
To estimate the diversity for X-linked genes relative to homologues on the Y (or to estimate X/autosome diversity ratio), it is essential to have reliable diversity estimates for genes on the different chromosomes, and introgression from a different species will make this difficult and could increase the estimated diversity (Sweigart and Willis, 2003). We argued above that subdivision is not extreme for SlX9, and does not explain the high diversity observed.
The shared short intron 2 structure, and shared single-nucleotide polymorphisms are consistent with introgression from S. dioica. Evidence for introgression between S. dioica and S. latifolia has also been reported for DD44X and SlX4 (Laporte et al., 2005). For SlX4, a size variant in a S. latifolia intron matched an intron size variant in S. dioica, similar to our observation for SlX9 (Laporte et al., 2005); for SlX1, almost all shared polymorphisms were within the first 1755 bp, and fixed differences were found only at the 3′ end of the gene (Atanassov et al., 2001), whereas, for DD44X, the sequences sampled from the two species shared 14 polymorphic sites and had no fixed differences (Laporte et al., 2005). These results suggest that the introgressed regions can be very localized (Laporte et al., 2005), and thus that introgression may be infrequent, and that introgressed regions are often eliminated after recombination (presumably because of selection). For the diversity analysis of SlX9, exon 1 and intron 1 were not included, so that our diversity value might be an over-estimate if the region surveyed coincides with a region of the gene in which SdX9 sequence has been introgressed.
Introgression between S. latifolia and S. dioica has also been detected using AFLP markers and (maternally inherited) chloroplast markers: out of 209 markers studied by Minder et al. (2007), only 7 were species-specific, and 5 out of 7 chloroplast haplotypes in S. latifolia were also present in S. dioica (Prentice et al., 2008). Allopatric populations of S. dioica and S. latifolia in Switzerland separated by small distances were found to be more distinct than sympatric ones (Minder and Widmer, 2008), as expected if hybridization occurs locally, but the introgressed regions are usually eliminated, rather than persisting.
We find both of the SlX9 haplotypes in plants from the Mediterranean region, in which S. dioica is absent. This argues against a hypothesis of recent introgression creating the S. latifolia short intron 2 X-linked haplotypes. As S. latifolia sequences with the longer X-linked intron 2 also share polymorphisms with S. dioica, recombination must probably have occurred after introgression, to yield a group of haplotypes with the short intron 2 characteristic of the S. dioica X, but with variants derived from the long S. latifolia haplotype.
We find slightly higher diversity in SlX9 than in SdX9, suggesting that introgression of X-linked sequences occurred mainly from S. dioica into S. latifolia. This is consistent with the distribution of chloroplast versus genomic markers, which suggested that hybridization events mainly involve S. dioica as the pollen donor (Minder et al., 2007). However, this conclusion is not definitive, in the absence of extensive sampling of the chloroplast genome and its variants in both species, because ancestral polymorphisms may exist. Moreover, experiments with equal amounts of pollen from the two species yielded progeny from S. latifolia recipients in which <20% were hybrids, compared with 50% with S. dioica recipients (Rahme et al., 2009). We can detect no S. dioica Y-linked allele of SdX9, which implies that SlY9 does not introgress into S. dioica. Our results also exclude S. dioica Y chromosome introgression into S. latifolia. If such introgression occurs, and if the S. dioica Y has no copy of the gene, we should find individuals within S. latifolia lacking Y9, but all 46 S. latifolia males yielded SlY9 sequences. This lack of Y chromosome introgression supports results from other Y-linked genes (Laporte et al., 2005).
Is introgression the sole cause of the unexpectedly high X diversity? If introgression of the X occurred recently, it might be possible to remove the introgressed alleles and estimate X diversity for comparison with the Y. However, if introgression is not recent, introgressed alleles cannot be recognized. In an attempt to analyze diversity in S. latifolia that is unlikely to be due to introgression, we excluded from the SlX9 data set all polymorphisms that are shared with our S. dioica sequences. The X–Y difference in diversity remained unchanged, suggesting that introgression is not the sole factor causing higher X than Y diversity (HKA test: χ2=4.19, P<0.05). If most introgressed sequences are selectively eliminated, and only certain small introgressed regions remain, these will be difficult to distinguish from shared ancestral polymorphisms for such closely related species. In Mimulus guttatus, recent introgression from Mimulus nasutus was detectable because high diversity was found only in sympatric M. guttatus populations, and also because M. nasutus sequences were clearly distinguishable from M. guttatus (Sweigart and Willis, 2003), unlike the situation for our species, and so we cannot rule out that ancestral polymorphism has contributed to some of the observed pattern in S. latifolia.
Acknowledgments
We thank Helen Borthwick for assistance in the lab. VBK was funded by a postgraduate scholarship from the University of Edinburgh, and RB and DC by grant BB/E020909/1 from the Biotechnology and Biological Sciences Research Council. We are grateful to Dmitry Filatov and Graham Muir (Oxford University) for S. latifolia samples, and Christoph Haag (University of Fribourg) for S. dioica samples.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Atanassov I, Delichere C, Filatov DA, Charlesworth D, Negrutiu I, Moneger F. Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol Biol Evol. 2001;18:2162–2168. doi: 10.1093/oxfordjournals.molbev.a003762. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker HG. Accounts of Melandrium, M. dioicum and M. album for the biological flora of the British Isles sponsored by the British Ecological Society. J Ecol. 1947;35:271–292. [Google Scholar]
- Baker HG. Stages in invasion and replacement demonstrated by species of Melandrium. J Ecol. 1948;36:96–119. [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D, Filatov DA, Moore RC. Defining regions and rearrangements of the Silene latifolia Y chromosome. Genetics. 2008;178:2045–2053. doi: 10.1534/genetics.107.084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp S, Gottsberger G. Importance of Silene latifolia ssp. alba and S. dioica (Caryophyllaceae) as host plants of the parasitic pollinator Hadena bicruris (Lepidoptera, Noctuidae) Oikos. 2004;105:221–228. [Google Scholar]
- Cermak T, Kubat Z, Hobza R, Koblizkova A, Widmer A, Macas J, et al. Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Res. 2008;16:961–976. doi: 10.1007/s10577-008-1254-2. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. The Hill-Robertson effect: evolutionary consequences of weak selection and linkage in finite populations. Heredity. 2008;100:19–31. doi: 10.1038/sj.hdy.6801059. [DOI] [PubMed] [Google Scholar]
- Desfeux C, Maurice S, Henry JP, Lejeune B, Gouyon PH. Evolution of reproductive systems in the genus Silene. Proc R Soc London, Ser B Biol Sci. 1996;263:409–414. doi: 10.1098/rspb.1996.0062. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA. Evolutionary history of Silene latifola sex chromosomes revealed by genetic mapping of four genes. Genetics. 2005;170:975–979. doi: 10.1534/genetics.104.037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA. A selective sweep in or near the Silene latifolia X-linked gene SlssX. Genet Res. 2008;90:85–95. doi: 10.1017/S0016672307009056. [DOI] [PubMed] [Google Scholar]
- Filatov DA, Charlesworth D. Substitution rates in the X- and Y-linked genes of the plants, Silene latifolia and S. dioica. Mol Biol Evol. 2002;19:898–907. doi: 10.1093/oxfordjournals.molbev.a004147. [DOI] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I, Navarro A, Charlesworth B. Muller's ratchet and the pattern of variation at a neutral locus. Genetics. 2002;161:835–848. doi: 10.1093/genetics/161.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hobza R, Lengerova M, Svoboda J, Kubekova H, Kejnovsky E, Vyskot B. An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma. 2006;115:376–382. doi: 10.1007/s00412-006-0065-5. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ironside JE, Filatov DA. Extreme population structure and high interspecific divergence of the Silene Y chromosome. Genetics. 2005;171:705–713. doi: 10.1534/genetics.105.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Bergero R, Charlesworth D. SlCyt, a newly identified sex-linked gene, has recently moved onto the X chromosome in Silene latifolia (Caryophyllaceae) Mol Biol Evol. 2009;26:2343–2351. doi: 10.1093/molbev/msp141. [DOI] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The hitchhiking effect revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrenberg S, Favre A. Genetic and ecological differentiation in the hybridizing campions Silene dioica and S. latifolia. Evolution. 2008;62:763–773. doi: 10.1111/j.1558-5646.2008.00330.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laporte V, Charlesworth B. Effective population size and population subdivision in demographically structured populations. Genetics. 2002;162:501–519. doi: 10.1093/genetics/162.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte V, Filatov DA, Kamau E, Charlesworth D. Indirect evidence from DNA sequence diversity for genetic degeneration of the Y-chromosome in dioecious species of the plant Silene: the SlY4/SlX4 and DD44-X/DD44-Y gene pairs. J Evol Biol. 2005;18:337–347. doi: 10.1111/j.1420-9101.2004.00833.x. [DOI] [PubMed] [Google Scholar]
- Liu ZY, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu QY, et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- Marais GAB, Nicolas M, Bergero R, Chambrier P, Kejnovsky E, Moneger F, et al. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr Biol. 2008;18:545–549. doi: 10.1016/j.cub.2008.03.023. [DOI] [PubMed] [Google Scholar]
- Matsunaga S, Isono E, Kejnovsky E, Vyskot B, Dolezel J, Kawano S, et al. Duplicative transfer of a MADS box gene to a plant Y chromosome. Mol Biol Evol. 2003;20:1062–1069. doi: 10.1093/molbev/msg114. [DOI] [PubMed] [Google Scholar]
- Maynard Smith JM, Haigh J. Hitch-hiking effect of a favorable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McVean GAT, Charlesworth B. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minder AM, Rothenbuehler C, Widmer A. Genetic structure of hybrid zones between Silene latifolia and Silene dioica (Caryophyllaceae): evidence for introgressive hybridization. Mol Ecol. 2007;16:2504–2516. doi: 10.1111/j.1365-294X.2007.03292.x. [DOI] [PubMed] [Google Scholar]
- Minder AM, Widmer A. A population genomic analysis of species boundaries: neutral processes, adaptive divergence and introgression between two hybridizing plant species. Mol Ecol. 2008;17:1552–1563. doi: 10.1111/j.1365-294X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Marais G, Hykelova V, Janousek B, Laporte V, Vyskot B, et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 2005;3:47–56. doi: 10.1371/journal.pbio.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice HC, Malm JU, Hathaway L. Chloroplast DNA variation in the European herb Silene dioica (red campion): postglacial migration and interspecific introgression. Plant System Evol. 2008;272:23–37. [Google Scholar]
- Rahme J, Widmer A, Karrenberg S. Pollen competition as an asymmetric reproductive barrier between two closely related Silene species. J Evol Biol. 2009;22:1937–1943. doi: 10.1111/j.1420-9101.2009.01803.x. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Strobeck C. Average number of nucleotide differences in a sample froma single subpopulation: a test for population subdivision. Genetics. 1987;117:149–153. doi: 10.1093/genetics/117.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart AL, Willis JH. Patterns of nucleotide diversity in two species of Mimulus are affected by mating system and asymmetric introgression. Evolution. 2003;57:2490–2506. doi: 10.1111/j.0014-3820.2003.tb01494.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD. Recombination and the power of statistical tests of neutrality. Genet Res. 1999;74:65–79. [Google Scholar]
- Zluvova J, Georgiev S, Janousek B, Charlesworth D, Vyskot B, Negrutin I. Early events in the evolution of the Silene latifolia Y chromosome: male specialization and recombination arrest. Genetics. 2007;177:375–386. doi: 10.1534/genetics.107.071175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova J, Janousek B, Negrutiu I, Vyskot B. Comparison of the X and Y chromosome organization in Silene latifolia. Genetics. 2005;170:1431–1434. doi: 10.1534/genetics.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.