Abstract
Gene expression mediated by viral vectors is subject to cell-to-cell variability, which limits the accuracy of gene delivery. When coupled with single-cell measurements, however, such variability provides an efficient means to quantify signaling dynamics in mammalian cells. Here, we illustrate the utility of this approach by mapping the E2f1 response to MYC, serum stimulation, or both. Our results revealed an underappreciated mode of gene regulation: E2f1 expression first increased then decreased as MYC input increased. This biphasic pattern was also reflected in other nodes of the network including the miR-17-92 micro RNA cluster and p19Arf. A mathematical model of the network successfully predicted modulation of the biphasic E2F response by serum and a CDK inhibitor. In addition to demonstrating how noise can be exploited to probe signaling dynamics, our results reveal how coordination of the MYC/RB/E2F pathway enables dynamic discrimination of aberrant and normal levels of growth stimulation.
Introduction
Expression of MYC is deregulated in a wide spectrum of cancers and MYC levels show a strong association with clinicopathological markers of disease progression (Zeller et al., 2003). c-Myc encodes a nuclear protein that mediates extracellular growth signals by coordinating events related to metabolism, protein synthesis, and DNA replication during cell-cycle progression (Dang et al., 2006; Ren et al., 2002). Rat fibroblasts engineered to constitutively express high levels of c-Myc along with activated versions of the Ras proto-oncogene are readily transformed (Land et al., 1983). Similarly, increases in MYC expression and activity are amongst the minimal subset of genetic alterations required to transform human fibroblasts (Junttila et al., 2007; Rangarajan et al., 2004; Yeh et al., 2004). However, expression of ectopic MYC in the absence of additional oncogenes in normal cells is followed by cell-cycle arrest (Felsher et al., 2000; Leone et al., 1997), senescence (Grandori et al., 2003), and in some cases, apoptosis (Evan et al., 1992). The paradox that MYC mediates growth signals and triggers growth arrest is reconciled by the notion that anti-proliferative responses represent a safeguard activated in the presence of potentially oncogenic MYC signals (Lowe et al., 2004). Thus, a fundamental issue concerns how cells distinguish normal and aberrant growth signals.
MYC contains protein domains that are analogous to other classic sequence-specific transcription factors (Cowling and Cole, 2006). MYC heterodimerizes with the protein MAX via shared carboxy-terminal helix-loop-helix leucine zipper motifs – an interaction that is obligatory for MYC to associate with DNA (Blackwood and Eisenman, 1991). Mutations that disrupt heterodimerization and DNA binding also disable MYC-mediated transcriptional activation and its ability to promote proliferation, apoptosis, and transformation (Amati et al., 1993a; Amati et al., 1992; Amati et al., 1993b). MYC can interact with a large collection of chromatin modifying complexes that positively (Frank et al., 2001) and negatively (Wanzel et al., 2003) influence the accessibility of gene regulatory sequences to transcription factors. Genome-wide profiling studies have led to the notion that MYC may be involved in regulating a large number of genes – perhaps as much as 15% of the human genome (Fernandez et al., 2003; Guccione et al., 2006). This far-ranging influence on gene expression suggests that the effects of MYC are in large part a function of its transcriptional network.
One of the most intensively studied aspects of MYC is its role in coordinating cell-cycle progression. In quiescent cells, genes required for DNA synthesis are silenced by the Retinoblastoma (RB) family of pocket proteins (RB, p107, and p130) tethered to DNA via repressive E2F family members (E2F4/5) (Rayman et al., 2002; Takahashi et al., 2000). Upon growth factor stimulation, increases in MYC lead to activation of E2F-regulated genes through two routes. First, MYC regulates expression of Cyclin D (CYCD) which serves as the regulatory component of kinases that phosphorylate pocket proteins and disrupt their inhibitory activity (Ewen et al., 1993; Tedesco et al., 2002). Second, MYC facilitates transcriptional induction of activator E2Fs (E2F1-3) (Leung et al., 2008) which activate the transcription of genes required for S-phase. Expression of activator E2Fs is reinforced by two positive feedback loops. First, activator E2Fs can directly bind to their own regulatory sequences at or near those sites vacated by repressive E2Fs, and help to maintain an active transcriptional state (Adams et al., 2000; Johnson et al., 1994; Sears et al., 1997). Second, activator E2Fs transcriptionally upregulate CYCE which stimulates additional phosphorylation of pocket proteins and prevents them from sequestering activator E2Fs (Weintraub et al., 1992). Previous work demonstrated that this RB-E2F pathway functions as a bistable-switch that governs an all-or-none E2F response to serum (Yao et al., 2008). Like MYC, deregulation of the RB-E2F pathway is common in human cancers and is believed to play a role in the unrestrained proliferation of tumor cells (Nevins, 2001).
Tumor-related alterations often manifest in increased levels of MYC stemming from deregulation of the c-Myc locus (Leder et al., 1983; Zeller et al., 2003) or alterations to genes responsible for the rapid turnover of the protein (Yeh et al., 2004). An elegant study by Murphy et al. (Murphy et al., 2008) demonstrated that only two additional copies of a transgene directing persistent low levels of c-Myc was necessary to drive ectopic proliferation whereas a further increase was necessary to induce apoptosis. These results demonstrate that distinct thresholds of MYC govern the decision to engage different MYC-related responses. Given the central role of MYC in regulating transcription, it is likely that differences in MYC concentration have profound impact on the expression of its targets. This is consistent with the finding that MYC binds to an increasing number of promoters in a concentration-dependent fashion (Fernandez et al., 2003). While the effects of MYC overexpression often depends on the presence wild-type E2f1 (Baudino et al., 2003; Leone et al., 2001; Rounbehler et al., 2002), the quantitative relationship between E2f1 expression and MYC remains undefined in this context (Figure 1). Here, we set out to quantify the E2F response to increasing levels of MYC stimulation.
Figure 1. The MYC/E2F pathway in cell-cycle progression.
Growth factor stimulation leads to induction of MYC that facilitates expression of the activator subclass of E2Fs. Despite the dependence of MYC on E2F for cell-cycle progression, survival, and death, the quantitative relationship between deregulated MYC and E2F is poorly understood.
Results
Viral-mediated noisy gene expression generates a broad range of MYC input
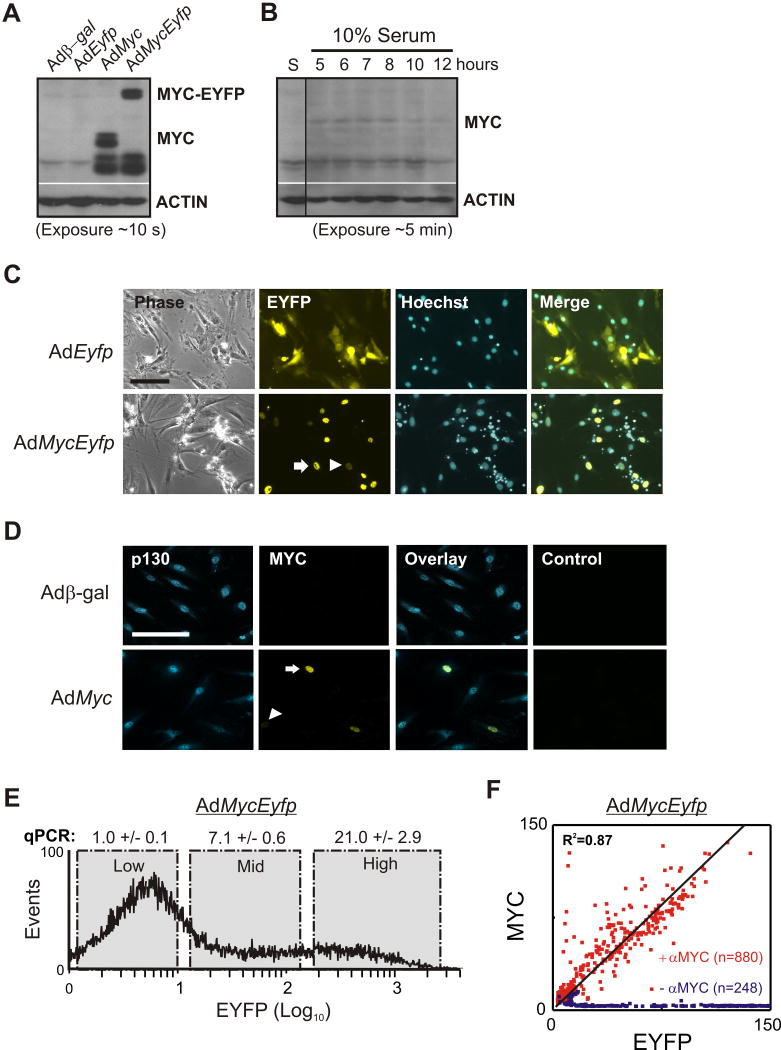
To introduce and quantify MYC in single cells, we generated a replication-defective recombinant adenovirus expressing MYC tagged with yellow fluorescent protein (AdMycEyfp). An adenovirus expressing native MYC (AdMyc) was also generated to calibrate effects of the corresponding MYC-EYFP fusion. REF52 cells infected with virus expressing native MYC produced a 65 kDa species while the virus encoding MYC-EYFP generated a super-shifted species, consistent with its fusion to YFP (Figure 2A). Adenoviral-mediated MYC protein expression was persistent and sustained at higher levels than that resulting from serum-stimulation (Figure 2B).
Figure 2. Adenoviral-mediated variable MYC expression.
(A and B) Western blots of protein from REF52 cells that were first starved in 0.01% serum for 48 hours then (A) infected with the indicated adenovirus at a MOI of 1 000 for 42 hours or (B) stimulated with 10% serum for various times then probed with a MYC antibody. Blots were stripped and re-probed for β-ACTIN. Native MYC has an apparent molecular weight of 67 kDa and the reactive species in AdMycEyfp extracts runs at approximately 90 kDa. S - cells starved in 0.01% serum.
(C) Microscopy of cells infected as in (A) and stained with Hoechst 33258. Merge shows overlay of yellow fluorescence and Hoechst. Arrows highlight cell-to-cell variability in MYC-EYFP. Scale bar: 100 μm.
(D) Fluorescence microscopy of infected cells incubated with antibodies against p130 and MYC, or IgG (Control). Arrow head and arrow highlight cells expressing low and high levels of MYC, respectively. Scale bar: 200 μm.
(E) Flow cytometry of MYC-EYFP (EYFP) levels in cells infected with AdMycEyfp. Representative real-time PCR (qPCR) results of c-Myc mRNA levels normalized to β-Actin in sorted subpopulations expressed relative to Low fraction. Errors indicate the standard deviation of four technical replicates.
(F) MYC-EYFP fluorescence (x-axis) correlated approximately linearly with MYC levels measured by fluorescent immunolabeling (y-axis). Infections were performed with AdMycEyfp at an MOI of 1 000 for 36 hours prior to indirect staining with antibody against c-MYC (red scatter; +αMYC) or without MYC antibody (blue scatter; -αMYC) and fluorescent-labelled secondary antibody. Fluorescence images were processed as described in Experimental Procedures. See additional data in Figures S1D and S2G.
MYC has previously been shown to regulate a number genes involved in ribosomal biogenesis (Arabi et al., 2005; Grandori et al., 2005) and overexpression of MYC results in nucleolar expansion (Kim et al., 2000). Consistent with this notion, expression of MYC and MYC-EYFP for two days resulted in cells with pronounced nucleoli that were not apparent in serum-starved controls (Adβ-gal or AdEyfp) (Figure S1A). Increased expression of MYC in serum-limiting conditions has also been shown to induce cell death (Harrington et al., 1994). Likewise, introduction of MYC and MYC-EYFP for three days induced rounding and detachment across a vast majority of serum-starved cells and was partially suppressed by high serum (Figure S1B). Overall, these observations indicate that the MYC-EYFP fusion protein and native MYC had a similar impact on cell morphology.
We also observed that fusion between MYC and YFP did not interfere with subcellular localization: Unlike EYFP, fluorescence from MYC-EYFP was restricted to regions coincident with nuclei (Figure 2C). Fluorescence microscopy shows that the number of MYC-EYFP expressing cells increased with the adenoviral multiplicity of infection (MOI) (Figure S1C). Importantly, microscopy revealed drastic cell-to-cell variability in fluorescence after infection with adenoviruses expressing either EYFP or MYC-EYFP. Immunolabeling experiments confirmed that variable amounts of native MYC were also detected in nuclei (indicated by p130 staining) in cells infected with AdMyc (Figure 2D). Flow cytometry indicated that cells infected with AdMycEyfp generated fluorescence that spanned more than three orders of magnitude and was largely bimodal (Figure 2E). Quantitative PCR measurements showed that part of the increase in MYC-EYFP was reflected by the abundance of its mRNA. The range in Myc mRNA (∼20-fold) is comparable to the differences observed between normal and cancerous breast tissue (Bieche et al., 1999). Amplification of Myc in human cancers has been observed to range between 10-fold to 300-fold, supporting the notion that a large range in MYC is of biological relevance (Dalla-Favera et al., 1982; Lee et al., 1984; Schwab et al., 1984). Data from fluorescence microscopy revealed approximately linear correlation between the level of fluorescence resulting from MYC-EYFP and the extent of MYC immunolabeling in individual cells (Figure 2F and Figure S1D). Variability in gene expression is not restricted to adenoviral vectors as infection with a retroviral vector reproduced the same degree of cell-to-cell variability (Figure S1E). Variability in viral-mediated gene expression could arise as a result of multiple factors. Viral transduction could vary with the particular ‘population context’ (e.g. cell size, position within a colony, and local density) in which cells exist (Snijder et al., 2009). In addition, variability could arise from the stochastic dynamics intrinsic to gene expression and protein modification. Regardless of the underlying mechanisms, the tremendous variability in viral-mediated expression represents an efficient means to introduce an input with broad dynamic range.
E2f1 is biphasic with respect to MYC
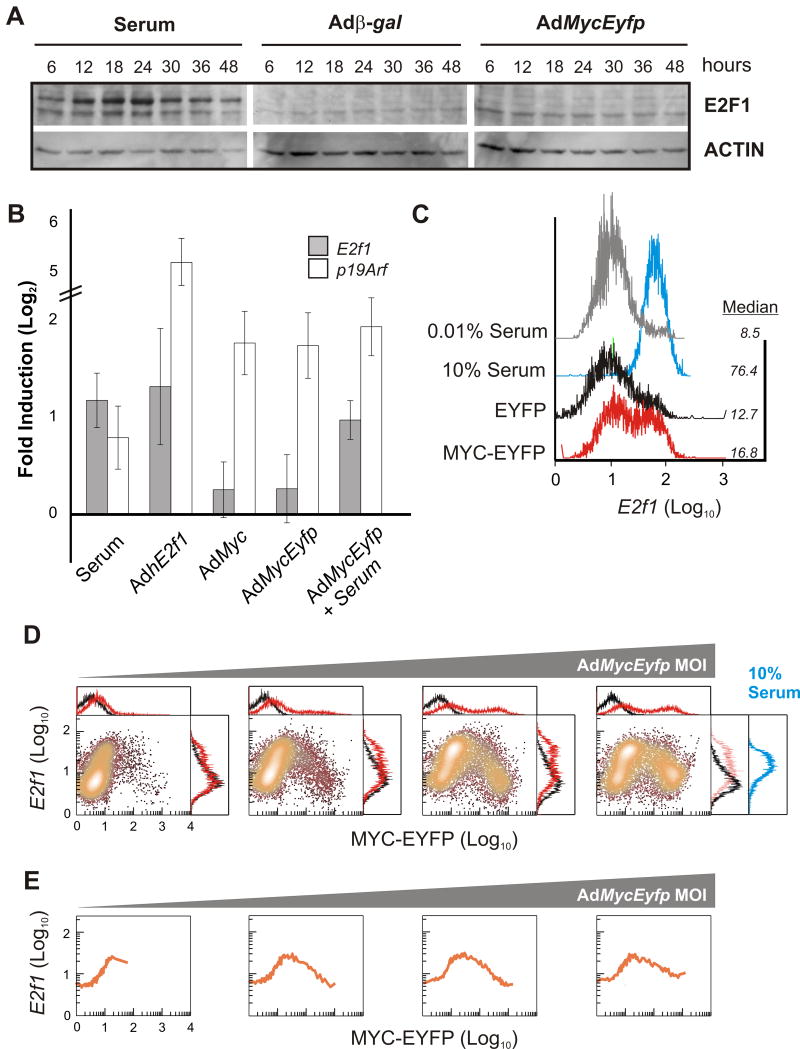
We compared the regulation of E2f1 in response to serum and deregulated MYC. In line with previous observations, Western blots showed that accumulation of endogenous E2F1 peaked at 18 hours after serum-stimulation (Figure 3A) and real-time PCR experiments confirmed that this pattern was mirrored by changes in mRNA (Figure S2A). In stark contrast, overexpression of MYC did not result in a detectable increase in E2F1 protein relative to controls. Real-time PCR experiments show that unlike serum and ectopic human E2F1 (hE2F1) (Johnson et al., 1994), native MYC and MYC-EYFP did not strongly increase E2f1 mRNA (Figure 3B). On the other hand, both species of MYC and hE2F1 were capable of inducing p19Arf (Bates et al., 1998; Zindy et al., 1998), supporting the notion that each is fully competent in upregulating gene expression in this context.
Figure 3. Comparison of E2f1 response to normal and aberrant growth signals.
(A) Western blots of protein extracts from starved REF52 cells collected various times after treatment with 10% serum or infection with the indicated adenovirus at an MOI of 1 000.
(B) Real-time PCR results. mRNA was measured 42 hours after treatment. Signals from cells stimulated with 10% BGS and adenoviral-infected samples were normalized to β-Actin and expressed relative to those in serum-starved and AdEyfp infected samples, respectively. Each error bar represents the standard error in the mean (n=3).
(C) Flow cytometry of E2f1 reporter expression (GFP fluorescence) in starved cells treated for 36 hours as indicated. Cells expressing EYFP and MYC-EYFP were infected with adenovirus at an MOI of 50, 100, 200, 300, 400, 500, 600, 800, and 1 000 in 0.01% serum and subsequently pooled before analysis. Median fluorescence is indicated.
(D) Flow cytometry results from cells infected at increasing MOI (200, 400, 800, and 1 000) for 42 hours. Black histograms - Adβ-gal infection; blue - 10% serum. (E) Moving median E2f1 values for data in (D) calculated from EYFP values that were ordered and grouped into contiguous bins of size 100.
To examine and quantify E2f1 in individual cells, we employed a REF52 cell line harbouring a stable GFP reporter that had been previously established (Yao et al., 2008). Combined with the variability inherent in adenoviral-mediated expression, these cells enabled us to perform high-throughput, single-cell measurements of dose responses to serum and deregulated MYC. Flow cytometry measurements used to detect E2f1 reporter activity confirmed that the underlying temporal expression pattern of E2f1 in response to serum was bimodal (Figure S2B) - characteristic of the ‘all-or-none’ response (Yao et al., 2008). Indeed, serum-stimulation for 36 hours increased median E2f1 over 8-fold as a result of a population-wide shift in expression (Figure 3C).
Analogous to the behaviour of E2F1 protein, introduction of MYC resulted in a modest increase in median E2f1 expression relative to a control expressing EYFP (compare red and black histograms in Figure 3C). However, the distribution of E2f1 across the population of cells expressing MYC was considerably broad, with a proportion of cells expressing E2f1 at levels comparable with serum. To directly establish how a particular level of MYC impacts E2f1, we analysed E2f1 output as a function of MYC input in single cells. The first scatter plot in Figure 3D shows that MYC induced E2f1 to levels at least as high as serum. Median values calculated from flow cytometry data indicate that a 4-fold increase in E2f1 levels resulted from a 7-fold change in MYC levels (Figure 3E). This steep increase in E2f1 is reminiscent of the switch-like response to serum generated in part through at least two modes of positive autoregulation on E2f1. However, in cells that express higher levels of MYC, E2f1 was gradually suppressed to near-baseline levels. This suppression is not simply a result of viral load as co-incubation with a control virus (Adβ-gal) at a high MOI had negligible effect on the ability of MYC to induce E2f1 (Figure S2C). We did observe a subtle increase of E2f1 in cells expressing elevated levels of EYFP alone (Figures S2D). However, this induction was monotonic and much lower than that induced by MYC, confirming that MYC was responsible for E2f1 induction and suppression. Furthermore, the overall shape of the E2f1 dose-response to MYC was insensitive to the amount of virus applied: increasing the MOI served only to expand the range of the MYC domain surveyed. Time-course experiments showed that once established, the biphasic E2f1 pattern remained largely invariant, despite the fact that MYC-EYFP expression continued to increase (Figures S2E and S2F). Finally, we observed that endogenous E2F1 protein levels followed a biphasic pattern in response to MYC-EYFP, reminiscent of the changes in promoter activity (Figure S2G). This observation is consistent with Western blots showing the absence of E2F1 in cells overexpressing MYC-EYFP (Figure 3A).
The biphasic response underscores a critical, underappreciated challenge in analysing phenotypic consequences of stimulating cells by over-expressing MYC. Our results show that biphasic behaviour allows full activation of E2f1 when MYC occurs within a narrow concentration range. Taken another way, the absence of E2f1 expression could result from either very low or high MYC. Indeed, flow cytometry measurements revealed that a relatively low MOI of virus expressing native MYC generated a broad distribution of E2f1 that was narrowed and reduced upon further increase in the MOI (Figure S2H). This is reminiscent of low MYC levels increasing E2f1 in a subpopulation of cells (Figure 3D; first panel) and subsequently suppressing output when cells express uniformly high MYC-EYFP (Figure S2E; last panel). Thus, suppression of E2f1 transcription by overexpression of MYC correlates with negligible increases in average mRNA and protein.
Expression of miR-17-92 miRNA and p19Arf are biphasic in response to MYC
The MYC/RB/E2F network is subject to multiple levels of regulatory control. MYC and E2F have both been implicated in the induction of the p19Arf (Arf) tumor suppressor (Bates et al., 1998; Zindy et al., 1998) and the miR-17-92 microRNA (miRNA) cluster (O'Donnell et al., 2005; Woods et al., 2007) (Figure S3A). Apart from its role in p53 stabilization, ARF can physically associate with and suppress aspects of MYC (Qi et al., 2004) and E2F (Datta et al., 2005) activity. The miR-17-92 cluster is an oncogene that cooperates with MYC in tumorigenesis (Mendell, 2008). miR-17-1 and miR-20a temper E2F1 protein levels and precocious induction during cell-cycle entry (Pickering et al., 2008). Transgenic analyses have demonstrated that miR-19a and miR-19b-1 are oncogenic components of miR-17-92 that downregulate PTEN and buffer apoptosis (Mu et al., 2009; Olive et al., 2009).
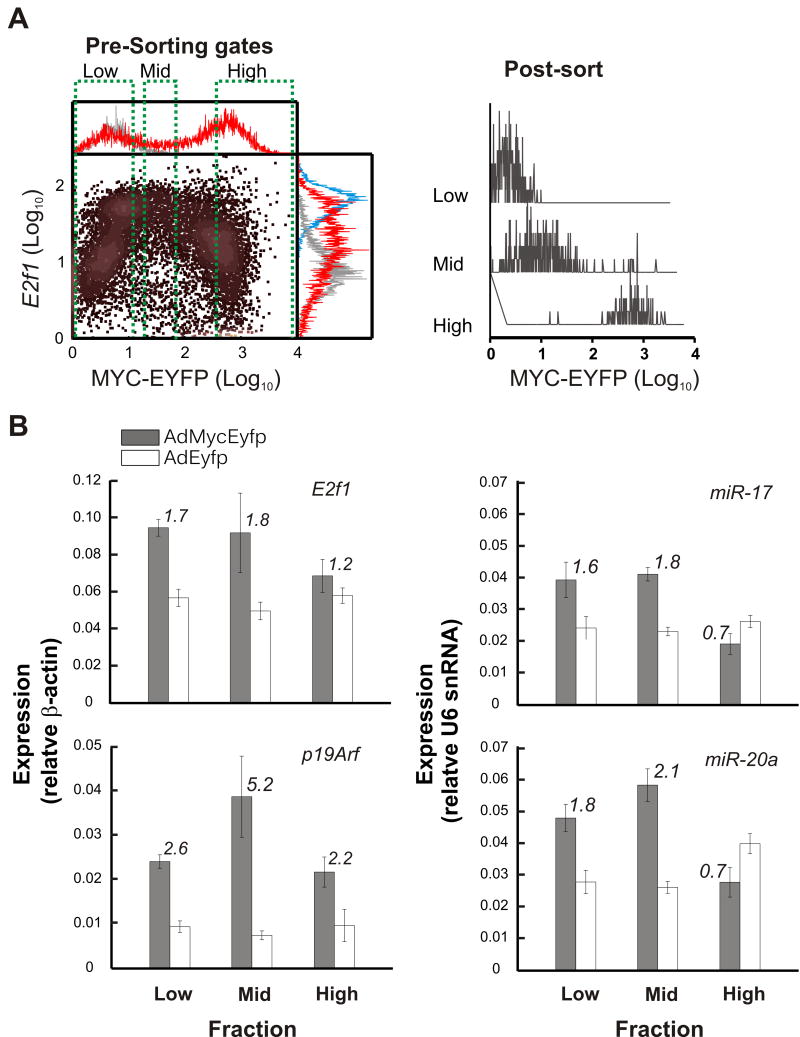
To provide further validation of our E2f1 reporter observations and gain initial insight into the broader ramifications of increasing MYC signaling, we analysed the expression of p19Arf and miRNA within the miR-17-92 cluster. Cells infected with AdMycEyfp were sorted into three subpopulations on the basis of their yellow fluorescence (Figure 4A) and mRNA levels were surveyed with quantitative real-time PCR (Figure 4B and Figure S3B). In line with previous observations, E2f1 expression levels demonstrated a biphasic trend. Surprisingly, expression of p19Arf and miRNA also demonstrated a qualitatively biphasic trajectory in which levels initially increase relative to controls then decrease in successive MYC-expressing fractions. However, subtle differences in E2f1, p19Arf, and miRNA expression were apparent such as the extent of initial increase in Low fractions, the fraction in which peak expression occurs, and the extent of suppression achieved in HI fraction (e.g. p19Arf remains elevated above controls). An overall biphasic trend was also observed when the population was sorted on the basis of both MYC-EYFP and E2f1 levels (Figure S3C and S3D). Thus, increasing MYC generates biphasic expression in multiple nodes within the wider MYC/RB/E2F network.
Figure 4. Biphasic expression of MYC/E2F-regulated genes.
(A) Fluorescence-activated cell sorting (FACS) results showing E2f1 reporter fluorescence as a function of MYC-EYFP input for cells treated as described in Figure 3C. Cells were sorted into three subpopulations based on yellow fluorescence as indicated by green dotted brackets. Cells expressing EYFP were sorted in a similar fashion (not shown). Red histograms - AdMycEyfp; grey - 0.02% serum; blue - 10% serum.
(B) Representative real-time PCR results from subpopulations sorted as described in (A). RNA levels in serum starved cells infected with AdEyfp and AdMycEyfp are expressed relative to β-Actin (for E2f1 and p19Arf) or U6 snRNA (for miRNA). Numbers above each bar indicate fold difference expression in MYC-EYFP expressing cells compared to EYFP. Each error bar represents the standard deviation of 4 technical replicates.
Origins of the biphasic response to MYC
A variety of mechanisms may underlie biphasic behaviour. A simple example involves the Drosophila transcription factor Kruppel, which activates expression at low levels whereas at elevated concentrations, the protein homodimerizes and functions as a repressor at the identical DNA site (Sauer and Jäckle, 1993). In another case, the ‘bell-shaped’ response of a synthetic gene construct results from complex antagonistic interactions between upstream and downstream transcriptional activators (Buetti-Dinh et al., 2009). The incoherent feed-forward motif, whereby an input signal both activates and regulates a repressor of an output, has been shown to underlie biphasic biological responses (Levchenko et al., 2004; Ma et al., 2009). Biphasic induction of the adenovirus-2 major late promoter has been shown to result from direct induction and suppression by MYC (Li et al., 1994; Peukert et al., 1997). Regardless of its implementation, a universal theme in these mechanisms is the interplay between activation and repression mediated by a common input.
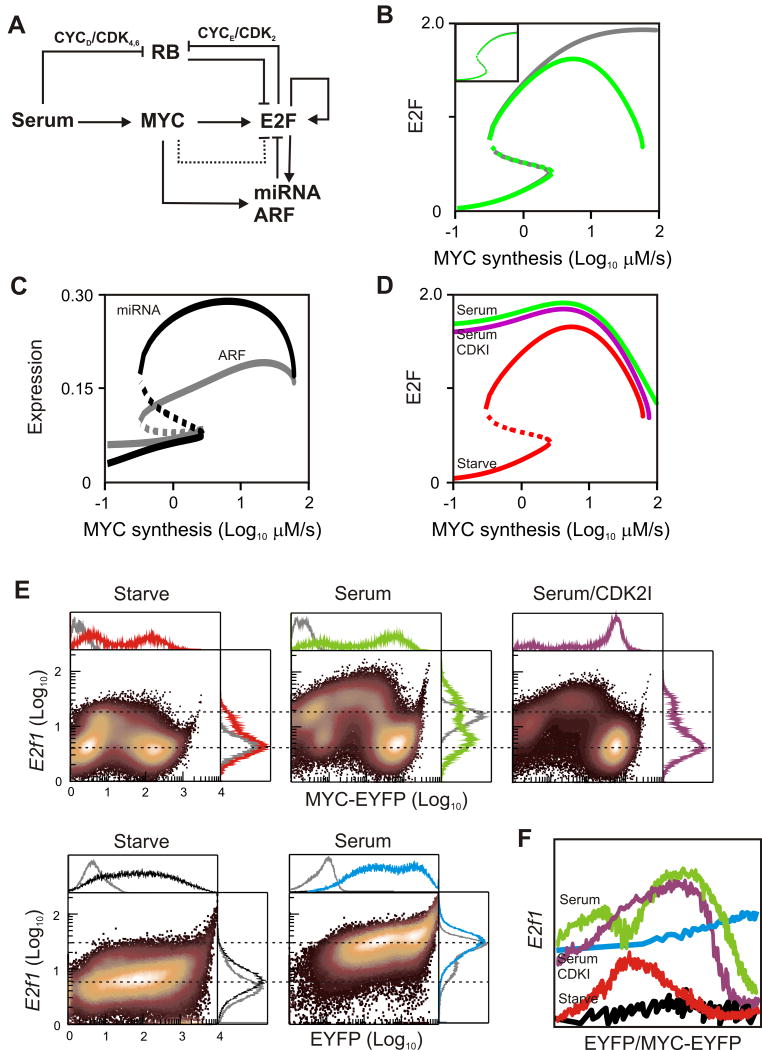
We used a previously developed mathematical model of the MYC/RB/E2F network (Yao et al., 2008) to address three main questions. First, we attempted to determine the minimal conditions for biphasic E2F expression. Second, we attempted to reconcile the involvement of E2F suppression by MYC with the strict increase in E2F observed after serum-stimulation. Third, we explored how serum-stimulation might impact the response of E2F to increasing levels of deregulated MYC. We examined different potential system dynamics by bifurcation analysis.
What are the minimal requirements for biphasic E2F expression by MYC? We reasoned that increased E2F resulting from MYC, E2F-autoregulation, and RB inactivation must be counteracted in some fashion when MYC levels become elevated. We extended the MYC/RB/E2F model to include contributions from miRNA and Arf (Supplementary Data). Within this framework, our model simulations revealed that concurrent biphasic E2F, miRNA, and Arf could not be generated with physiologically relevant parameters (data not shown). This result points to the existence of a source of E2F suppression downstream of MYC, in addition to miRNA and Arf. In light of these findings, we took a parsimonious and general approach in attempting to describe MYC-mediated E2F suppression that encapsulates possible contributions from miRNA, p19Arf, as well as additional, possibly unknown, sources (dotted edge in Figure 5A). Under these conditions, simulations of the model indeed captured the biphasic E2F response with respect to MYC (Figure 5B). In each case bistability is maintained, which is depicted as overlapping steady-states within the same region of the domain (MYC synthesis). This predicted bistability may underlie the observed bimodal activation of E2F in response to moderate MYC stimulation. However, we note that further analysis is needed to conclusively establish this predicted bistable E2F response. In line with real-time PCR experiments in cells expressing increasing MYC, model simulations reproduced the biphasic nature of both miRNA and ARF (Figure 5C).
Figure 5. Modelling biphasic E2F response.
(A) Signaling network underlying biphasic E2F response. Dotted line indicates postulated repression of E2F by MYC. Abbreviations: RB – Pocket proteins (Retinoblastoma, p107, p130); CYC – CYCLIN; CDK – CYCLIN-dependent kinase; miRNA – micro RNA within the miR-17-92 cluster; ARF – p19ARF. Inhibition of RB is mediated via phosphorylation by CYC/CDK complexes. Not depicted is weak activation of CYCD by MYC and explicit description of E2F mRNA and protein species.
(B) Bifurcation diagram showing steady-state trajectories of E2F when MYC-mediated suppression is negligible (grey; KR=104 μM) or significant (green; KR=102 μM). Dotted regions indicate unstable steady-states. Inset: Simulated E2F response to serum in the presence and absence of suppression (x-axis: serum between 0.01% to 10%).
(C) Simulated response of miRNA and ARF.
(D) Simulated E2F response to MYC under serum-starvation (Starve; parameter S=0.01%), high serum (Serum; S=10%) or high serum with inhibitor of CYCE/CDK2 phosphorylation of RB (CDK2I; parameter kRBP2=0.18 μM*h-1).
(E) Flow cytometry results showing E2f1 expression in the presence and absence of serum. Top row: Cells infected with virus expressing MYC-EYFP or β-gal control (grey histograms) as described in Figure 3C. Bottom row: Cells infected with EYFP virus or uninfected (grey histograms). CDK2 Inhibitor (CVT-313) was added at 10 μM.
(F) Moving median values corresponding to panels in (E).
Modulation of E2F response to MYC
In contrast to deregulated MYC, increasing serum alone is predicted to generate monotonic induction of E2F whose overall levels are only slightly reduced in the presence of suppression (inset Figure 5B). Unlike deregulated MYC, serum can stimulate RB phosphorylation via its more potent influence on CYCD/CDK4,6 activity (Leone et al., 1997). However, monotonic response to serum is largely a result of the relatively modest levels of MYC. Our model predictions underscore the importance of MYC concentration, rather than MYC deregulation per se (i.e. the absence of parallel pathways triggered by growth factors), in the activation of anti-proliferative responses (Murphy et al., 2008).
We have noted that serum partially counteracted the negative effects of elevated MYC on viability and we wished to determine how it might affect MYC-mediated expression of E2F. Our model predicted that serum stimulation would shift the E2F dose response upwards with its biphasic character maintained (Figure 5D). Specifically, initially high levels of E2F induced by serum are increased further and subsequently reduced with increasing levels of deregulated MYC. The model further predicted that the serum effect is attenuated by inhibiting the activity of CYCE/CDK2, which mediates the overall strength of E2F positive feedback by phosphorylation of RB. Experimentally, this can be realized by using the small molecule CVT-313, a specific inhibitor of CDK2 (Brooks et al., 1997). These simulations suggest that by reducing RB activity, serum signals greatly expand the window of deregulated MYC that propels E2F into a supraphysiological range.
Consistent with model predictions, addition of serum led to a shift in the biphasic pattern: except for very high MYC levels, E2f1 was induced in all MYC-expressing cells to a greater extent than that observed with MYC alone (upper middle panel of Figure 5E, Figure 5F). These observations were consistent with quantitative PCR results showing that serum induced E2f1 mRNA regardless of the MOI of MYC virus used to infect them (Figure S4A and Figure 3B). On the other hand, expression of EYFP control failed to reproduce any of these effects, underscoring the notion that these biphasic patterns are a function of MYC rather than viral load (bottom row of Figure 5E). Also predicted by simulations, reduced inhibition by RB via small molecule inhibition of CDK2 resulted in partial suppression of the serum effect (upper right panel in Figure 5E, Figure 5F). These experimental results support the notion that stringent restrictions imposed on MYC via the biphasic response are attenuated by serum.
Discussion
Our analysis revealed a hitherto unknown biphasic induction of E2f1 by MYC. The biphasic E2F response provides an intuitive explanation for the variable results in previous studies regarding MYC-mediated induction of E2f1. These reports, listed in Table 1, share two characteristics: (1) all employ Northern and/or Western blotting, which provide information about average levels of E2f1 expression; and (2) none provides quantitative information regarding MYC input levels. Specifically, Leone et al. (Leone et al., 1997) showed that adenoviral-MYC induced modest accumulation of E2f1 mRNA and DNA synthesis in REF52 cells compared to serum-stimulation. This is consistent with our observations that variability in adenoviral expression can result in a small fraction of a cell population with MYC levels within the appropriate window for E2f1 induction. This concept may also explain why adenoviral expression of MYC in mouse liver (where most virus is sequestered after intravenous injection) failed to increase E2F1 and Ki-67 staining (Kim et al., 2000) whereas abundant Ki-67 staining was apparent in livers of transgenic mice with only two additional copies of c-Myc (Murphy et al., 2008). A particularly striking example involves primary mouse embryonic fibroblasts: large amounts of E2F1 accumulated in response to small-molecule activation of MYC fused to the oestrogen receptor (MYC-ER) (Baudino et al., 2003) but not when native MYC was constitutively expressed from a retrovirus (Zindy et al., 1998). While the level of MYC activity in these two systems awaits a direct quantitative comparison, small-molecule protein activation might allow for more modest and uniform increases across a population. In contrast, our results indicate that retroviral-mediated gene expression demonstrates large cell-to-cell variability comparable to that observed with adenoviruses. Thus, considering the biphasic E2F response, these apparently conflicting conclusions surrounding E2f1 may be reconciled by taking into account the different amounts of MYC present in target cells. Furthermore, it is conceivable that the degree of MYC-mediated E2f1 suppression may vary in a cell and context-specific manner.
Table 1. Literature on E2f1 regulation by MYC.
| MYC Input | Target cells | Species (Assay) | aE2Fl Induction | Reference | |
|---|---|---|---|---|---|
| MYC | Serum | ||||
| adenovirus | REF52 (rat fibroblast) | mRNA (Northern) | + | +++ | (Leone et al., 1997) |
| adenovirus | SkBr3 (breast cancer) | Protein (Western) | - | N/A | (Mitchell and El-Deiry, 1999) |
| adenovirus | mouse liver (in vivo) | Protein (Western) | - | N/A | (Kim et al., 2000) |
| bretrovirus (constitutive) | MEF(WT/ARF-/-/p53-/-) | Protein (Western) | - | N/A | (Zindy et al., 1998) |
| transgenic c(MT) | mouse liver (in vivo) | mRNA/Protein (Northern/Western) | - | N/A | (Santoni-Rugiu et al., 1998) |
| +++ (tumor) | |||||
| transgenic d(MMTV) | mouse breast (in vivo) | mRNA/Protein (Northern/Western) | - | N/A | (Liao et al., 2000) |
| +++ (tumor) | |||||
| eretrovirus (MYC-ER) | MEF | Protein (Western) | +++ | N/A | (Baudino et al., 2003) |
| tet-Off | P493-6 (human B-cell) | RNA (Northern) | +++ | N/A | (O'Donnell et al., 2005) |
| Protein (Western) | + | ||||
Indicates change in E2F1 levels. Scored as undetectable change (-) or induction ranging from low (+) to high (+++)
unselected MEFs from day 13.5 embryos constitutively expressing MYC from retrovirus
c-Myc under the control of regulatory sequences from the metallothionein gene
c-Myc under the control of the mouse mammary tumor virus long terminal repeat
drug-selected MEFs from day 14.5 embryos expressing MYC-ER from retrovirus
Equally important, the biphasic E2f1 response to MYC represents an intrinsic safeguard to curtail uncontrolled proliferation. Consistent with previous observations (Murphy et al., 2008), we observed that low levels of MYC induced by serum can potently induce E2f1 with modest activation of Arf (Figure 3B) allowing cells to proceed with S-phase entry and proliferation. In response to direct MYC stimulation, this ability is moderated by upregulation of miRNAs and Arf at modest levels of MYC (Figure 4). That high MYC leads to downregulation of miRNAs and Arf is consistent with this conceptual framework. For instance, perhaps even lower levels of Arf achieved at high MYC inputs are sufficiently potent to trigger downstream pathways. As such, Arf induction and E2F downregulation would combine to promote cell cycle arrest or apoptosis. Alternatively, the reduced Arf levels may be too low (due to lack of sufficient E2F1 induction (Figure 5A)) to induce further growth suppression. In this case, E2F downregulation alone would still be sufficient to prevent cell cycle progression, obviating the need for potent Arf activity. Indeed, Ad-mediated expression of MYC in the livers of mice resulted in low E2F1 and was unable to induce proliferation and apoptosis (Kim et al., 2000). To conclusively establish the interplay among these genes, however, single cell analyses like those used to interrogate E2f1 are required to quantitatively determine how they are correlated in individual cells.
Our model successfully predicted that serum-mediated suppression of RB phosphorylation expands the range of MYC capable of inducing E2f1. The cooperation between serum and deregulated MYC is reminiscent of the ability of some physiological growth signals to exacerbate tumorigenesis. In particular, serum stimulation of fibroblasts is typically involved in the activation of an inflammatory or ‘wound healing’ response. Interestingly, inflammation is also a driving force in some types of tumors and likely reflects the ability of serum to promote survival, proliferation, and invasion (Bissell and Radisky, 2001). According to our results, part of the insidious nature of inflammatory signals may lie in the supraphysiological induction of E2f1 in cell variants that possess sustained increases in MYC. In this context, serum may potentially overcome MYC-mediated E2f1 suppression while muting the apoptotic program downstream of E2f1 (Hallstrom et al., 2008).
It is now well appreciated that intracellular processes are noisy due to small numbers of interacting molecules and environmental perturbations (Kaern et al., 2005; Longo and Hasty, 2006; Raj and van Oudenaarden, 2008; Raser and O'Shea, 2005). Studies to date have primarily focused on two aspects of cellular noise: (1) propagation and regulation of noise by intracellular and communication based mechanisms, and (2) implications of noise in cell-fate decisions or cellular adaptation to changing environments. With respect to variability in gene expression generated by viral vectors, on one hand it may undermine interpretation of increasing levels of a gene of interest and limit the accuracy of viral-mediated therapeutic gene delivery. Here, our study underscores an underappreciated application of noise that is particularly well-suited for the use of viral vectors: when coupled with quantitative single-cell measurements, drastic cell-to-cell variability associated with viral-mediated gene expression can provide an input with a wide dynamic range that facilitates analysis of cell signaling dynamics in a high-throughput manner. In addition to providing basic insights into dynamics of signaling pathways, this efficient mapping may provide a powerful approach to define noise-based phenotypic signatures of cell physiology under normal or pathological states.
Experimental Procedures
Construction of viral vectors
For adenoviruses expressing MYC, the full-length cDNA for the murine myelocytomatosis proto-oncogene (c-Myc; Accession No. NM_010849) contained in the plasmid pRc/CMV-cmyc (Hann et al., 1994) was amplified by PCR with primers cMYC KOZAK FORWARD (5′-ACC ATG GCC CTC AAC GTG AAC TTC ACC-3′) and cMYC REVERSE (5′-ACC GGT TGC ACC AGA GTT TCG AAG-3′). The enhanced yellow fluorescent protein (EYFP) sequence contained in pLuxR-EYFP was amplified with primers EYFP FORWARD (5′-ACC GGT ATG GTG AGC AAG GGC GAG-3′) and EYFP REVERSE (5′-TTA CTT GTA CAG CTC GTC CAT GCC-3′). PCR products were cloned into the pCR2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen) to form MYC-TOPO and EYFP-TOPO and subsequently sequenced. An AgeI/XhoI fragment containing the EYFP sequence in EYFP-TOPO was cloned into the corresponding sites in MYC-TOPO to generate MYC-EYFP-TOPO. A BamHI/XhoI fragment from either EYFP-TOPO or MYC-EYFP-TOPO was subcloned into the corresponding sites of the Gateway pENTR1A vector (Cat. no. 11813-011; Invitrogen). pENTR1A construct containing native c-Myc was generated by XhoI/AgeI digestion of MYC-EYFP-TOPO followed by blunt-ending with Klenow and ligation of the formerly cohesive ends. For construction of E2f1 based adenoviral vectors, a BamHI/EcoRI fragment from pS65LHA-E2F1 (Addgene plasmid 10736) (Sellers et al., 1998) was subcloned into pENTR1A. Adenoviral plasmids were generated using the destination vector pAd/CMV/V5-DEST (Cat. no. V493-20; Invitrogen). Adenoviral construction using the ViraPower Adenoviral Gateway Expression Kit (Cat. no. K4930-00) was performed according to the manufacturer's instructions. Destination plasmid coding for Adβ-galactosidase was supplied in the same kit. Growth and maintenance of replication incompetent adenoviral vectors has been described previously (Nevins, 1980). For retrovirus vector constitutively expressing mCherry, the complete coding sequence was excised from pCMV-mCherry by restriction digest and cloned into the multiple cloning region of the pQCXIN retroviral vector (Cat. No. 631514; Clontech) downstream of the CMV promoter.
Cell culture and viral infection
Construction of stable puromycin resistant REF52 cells containing a cassette with the d2GFP reporter driven by the E2F1 promoter (E2F1p) has been described previously (Yao et al., 2008). REF52 cells containing retroviral vectors were passaged in α-MEM (Cat. No.1257-063; Gibco) supplemented with Bovine Growth Serum (i.e. ‘BGS’; Cat. No. SH30541.03; Hyclone). Maintenance of E2f1 reporter was accomplished by supplementing growth media with 2.5 μg/mL Puromycin (Cat. No. P8833; Sigma) while mCherry retroviral vector was maintained with Geneticin at 400 μg/mL (Cat. No. 10131-035; Gibco). For typical flow cytometry experiments, cells were plated in 6-well dishes at a density of 2- 105 per well and serum starved in α-MEM supplemented with 0.01% BGS for 48 hours. Uninfected cells were either supplemented with 2 mL serum-starvation media or serum-stimulated in 2 mL α-MEM with 10% BGS. For adenoviral infection, adenoviruses were diluted into 250 μL α-MEM supplemented with 25 mM HEPES, added drop-wise onto cells and incubated for 90 minutes at 37°C with periodic rocking. Post-infection, cells were then supplemented with 2 mL of the appropriate culture media. For pharmacological inhibition of CDK2, CVT-313 (2(bis-(Hydroxyethyl)amino)-6-(4-methoxybenzylamino)-9-isopropyl-purine, Cat. No. 238803; EMD Biosciences) was diluted in DMSO to 25 mM and added directly to media.
Western blotting
Cells were lysed with RIPA lysis buffer, scraped into microfuge tubes and agitated at 4°C for 30 minutes before centrifugation. Supernatants were rescued and stored at -80°C. Concentration of protein in cell extracts was determined using the Microplate BCA Protein Assay Kit - Reducing Agent Compatible (Cat. No. 23252, Thermo Scientific, Rockfort IL). Approximately 40 μg of protein were loaded onto 10% polyacrylamide gels and subjected to SDS-PAGE and Western blotting onto Immuno-Blot PVDF membranes (BIO-RAD, Hercules, CA). Membranes were blotted using standard procedures with primary antibodies against MYC (Cat. No. C-33, Santa Cruz Biotech), E2F-1 (Cat. No. KH95, Santa Cruz Biotech) and β-ACTIN (Cat. No. C-2, Santa Cruz Biotech). Secondary antibodies employed were either an ECL Anti-Mouse IgG HRP-linked whole antibody (Cat. No. NA931, GE Healthcare) or ECL Anti-Rabbit IgG HRP-linked species specific F(ab') fragment (Cat. No. NA9340, GE Healthcare). Western blots were visualized using the ECL Plus Western Blotting Detection Kit (Cat. No. 2132, GE Healthcare) and HyBlot CL autoradiography film (Cat. No. E3018, Denville Scientific, Metuchen, NJ) according to the manufacturer's instructions.
Fluorescence microscopy
For fluorescence microscopy, cells were fixed with 3.7% formaldehyde in PBS for 5 minutes at room temperature. For nuclear staining, cells were incubated in Hoechst 33342 (Cat. No. H3570, Invitrogen) diluted in PBS to 10 μg/mL for 5 minutes at room temperature and washed twice with PBS. Cells cultured in 6-well dishes were imaged using a Leica DMI 6000 B inverted fluorescent microscope with Semrock Brightline filters. Images were taken using a Hamamatsu 1394 ORCA-ERA camera using the SimplePCI software (version 6.1.2.020107; Compix Inc.). For phase contrast, software settings were: Offset = 255, Gain=0, and variable exposure times. For assaying YFP software settings were: Offset = 255, Gain=0, Exposure=0.50 seconds. For indirect immunofluorescence microscopy, cells were grown on cover slips (SecureSlip Cat. No. S1815; Sigma) coated with 0.01% gelatin. Treated cells were fixed with ice-cold methanol for 10 minutes at -20°C, permeabilized with PBS/0.25% Triton X-100/1% BSA for 10 minutes, and blocked with PBS/3% BSA/0.02% Tween-20 for 30 minutes. Cells were then incubated with primary antibodies overnight at a dilution of 1:100 in PBS/1% BSA: p130 (Cat. No. sc-317; Santa Cruz); c-MYC (Cat. No. sc-42; Santa Cruz); and E2F1 (Cat. No. sc-251). Cells were then washed with PBS, incubated with goat anti-rabbit AlexaFluor 405 (Invitrogen) and goat anti-mouse AlexaFluor 594 (Invitrogen) secondary antibodies in PBS/1% BSA for 1 hour prior to mounting with SlowFade Gold antifade solution (Cat. No. S36936; Invitrogen). Slides were imaged on a Zeiss LSM 510 inverted confocal microscope located at the Duke University light microscopy core facility.
Flow cytometry and fluorescence activated cell sorting
For flow cytometry without sorting, cells were harvested by trypsinization and resuspended in phosphate buffered saline (PBS) supplemented with 3.7% formaldehyde and analyzed with a BD FACstar flow cytometer (BD Biosciences). Typically, 10 000 events were measured for each flow cytometry experiment. For flow cytometry with sorting, cells were resuspended in PBS with 1% bovine serum albumin and processed with a BD FACSVANTAGE with DIVA sorter. Typically, at least 100 000 cells were obtained for each sorted subpopulation.
Quantitative real-time PCR
RNA extracts were prepared from cells using the RNeasy Protect Cell Mini Kit (Qiagen) and miRNA was extracted with the miRVana miRNA Isolation Kit (Ambion) according to the manufacturer's protocol. For E2f1 and Arf, RNA was interrogated by real-time PCR using the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems). Gene specific primers used: for rat E2f1 (5′-ttgacccctctggatttctg-3′) and (5′-ccctttggtctgctcaatgt-3′); for rat p19Arf (5′-ccttggtgttgaggccagagag-3′) and (5′-ggtcctcgcagttcgaatctgc-3′); for rat β-Actin (5′-gtcgtaccactggcattgtg-3′) and (5′-ctctcagctgtggtggtgaa-3′) (IDT technologies). Gene specific primers and probes for murine c-Myc (Assay No. Mm00487804_m1) and rat β-Actin (Assay No. Rn00667869_m1) were purchased from Applied Biosystems and amplified using the TaqMan® RNA-to-CT™ 1-Step Kit (Applied Biosystems Part No. 4392938). Gene-specific primers and probes were purchased from Applied Biosystems in order to detect rat miR-17-1 (Assay ID 002308), rat miR-20a (Assay ID 000580), rat miR-19a (Assay ID 000395), rat miR-19-b-1 (Assay ID 000396), and U6 snRNA (Assay ID 001973). cDNA from miRNA was generated using the TaqMan® MicroRNA RT Kit (Applied Biosystems Part No. 4366596) and amplified using the TaqMan® Universal PCR Master Mix, No AmpErase® UNG (Applied Biosystems Part No. 4324018). All samples were run on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems) according to manufacturer's protocol.
Computational modelling and data analysis
Data analysis was performed using Matlab (Mathworks, Natick, Massachusetts). For analysis of images from fluorescence microscopy, a mask for each image was created by first removing background (global minimal pixel intensity = 12) and then removing areas smaller than a typical nucleus (minimal number of pixel = 500). Mean signal intensity from each channel was then extracted. Bifurcation analysis was performed using XPP-AUTO software (http://www.math.pitt.edu/∼bard/xpp/xpp.html).
Supplementary Material
Acknowledgments
We thank Mike Cook (Flow Cytometry Resource, Duke Comprehensive Cancer Center), Dawn Chasse for help with adenoviral stocks, Tae Jun Lee for help with development of the mathematical model, Quanli Wang for help with image data analysis (Department of Statistical Science, Duke University), and Yu Tanouchi for assistance with adenovirus construction. This work was partially supported by the National Institutes of Health (1P50GM081883), a DuPont Young Professorship (LY), and a David and Lucile Packard Fellowship (LY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MR, Sears R, Nuckolls F, Leone G, Nevins JR. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol Cell Biol. 2000;20:3633–3639. doi: 10.1128/mcb.20.10.3633-3639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993a;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- Amati B, Littlewood TD, Evan GI, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993b;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Baudino TA, Maclean KH, Brennan J, Parganas E, Yang C, Aslanian A, Lees JA, Sherr CJ, Roussel MF, Cleveland JL. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol Cell. 2003;11:905–914. doi: 10.1016/s1097-2765(03)00102-3. [DOI] [PubMed] [Google Scholar]
- Bieche I, Laurendeau I, Tozlu S, Olivi M, Vidaud D, Lidereau R, Vidaud M. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 1999;59:2759–2765. [PubMed] [Google Scholar]
- Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Brooks EE, Gray NS, Joly A, Kerwar SS, Lum R, Mackman RL, Norman TC, Rosete J, Rowe M, Schow SR, et al. CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. J Biol Chem. 1997;272:29207–29211. doi: 10.1074/jbc.272.46.29207. [DOI] [PubMed] [Google Scholar]
- Buetti-Dinh A, Ungricht R, Kelemen JZ, Shetty C, Ratna P, Becskei A. Control and signal processing by transcriptional interference. Mol Syst Biol. 2009;5:300. doi: 10.1038/msb.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Datta A, Sen J, Hagen J, Korgaonkar CK, Caffrey M, Quelle DE, Hughes DE, Ackerson TJ, Costa RH, Raychaudhuri P. ARF directly binds DP1: interaction with DP1 coincides with the G1 arrest function of ARF. Mol Cell Biol. 2005;25:8024–8036. doi: 10.1128/MCB.25.18.8024-8036.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato J, Livingston DM. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Zetterberg A, Zhu J, Tlsty T, Bishop JM. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci U S A. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, et al. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 2003;17:1569–1574. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR, Dixit M, Sears RC, Sealy L. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- Harrington EA, Bennett MR, Fanidi A, Evan GI. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- Junttila MR, Puustinen P, Niemela M, Ahola R, Arnold H, Bottzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130:51–62. doi: 10.1016/j.cell.2007.04.044. [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci U S A. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Cellular oncogenes and multistep carcinogenesis. Science. 1983;222:771–778. doi: 10.1126/science.6356358. [DOI] [PubMed] [Google Scholar]
- Leder P, Battey J, Lenoir G, Moulding C, Murphy W, Potter H, Stewart T, Taub R. Translocations among antibody genes in human cancer. Science. 1983;222:765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Lee WH, Murphree AL, Benedict WF. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature. 1984;309:458–460. doi: 10.1038/309458a0. [DOI] [PubMed] [Google Scholar]
- Leone G, DeGregori J, Sears R, Jakoi L, Nevins JR. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell. 2001;8:105–113. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- Leung JY, Ehmann GL, Giangrande PH, Nevins JR. A role for Myc in facilitating transcription activation by E2F1. Oncogene. 2008 doi: 10.1038/onc.2008.55. [DOI] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, Sternberg PW. Regulatory modules that generate biphasic signal response in biological systems. Syst Biol (Stevenage) 2004;1:139–148. doi: 10.1049/sb:20045014. [DOI] [PubMed] [Google Scholar]
- Li LH, Nerlov C, Prendergast G, MacGregor D, Ziff EB. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao DJ, Natarajan G, Deming SL, Jamerson MH, Johnson M, Chepko G, Dickson RB. Cell cycle basis for the onset and progression of c-Myc-induced, TGFalpha-enhanced mouse mammary gland carcinogenesis. Oncogene. 2000;19:1307–1317. doi: 10.1038/sj.onc.1203430. [DOI] [PubMed] [Google Scholar]
- Longo D, Hasty J. Dynamics of single-cell gene expression. Mol Syst Biol. 2006;2:64. doi: 10.1038/msb4100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KO, El-Deiry WS. Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ. 1999;10:223–230. [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR. Definition and mapping of adenovirus 2 nuclear transcription. Methods Enzymol. 1980;65:768–785. doi: 10.1016/s0076-6879(80)65072-1. [DOI] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peukert K, Staller P, Schneider A, Carmichael G, Hanel F, Eilers M. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2008 doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounbehler RJ, Rogers PM, Conti CJ, Johnson DG. Inactivation of E2f1 enhances tumorigenesis in a Myc transgenic model. Cancer Res. 2002;62:3276–3281. [PubMed] [Google Scholar]
- Santoni-Rugiu E, Jensen MR, Thorgeirsson SS. Disruption of the pRb/E2F pathway and inhibition of apoptosis are major oncogenic events in liver constitutively expressing c-myc and transforming growth factor alpha. Cancer Res. 1998;58:123–134. [PubMed] [Google Scholar]
- Sauer F, Jäckle H. Dimerization and the control of transcription by Krüppel. Nature. 1993;364:454–457. doi: 10.1038/364454a0. [DOI] [PubMed] [Google Scholar]
- Schwab M, Varmus HE, Bishop JM, Grzeschik KH, Naylor SL, Sakaguchi AY, Brodeur G, Trent J. Chromosome localization in normal human cells and neuroblastomas of a gene related to c-myc. Nature. 1984;308:288–291. doi: 10.1038/308288a0. [DOI] [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, Lassar AB, Kaelin WG. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder B, Sacher R, Ramo P, Damm EM, Liberali P, Pelkmans L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009 doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2) Genes Dev. 2002;16:2946–2957. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Prater CA, Dean DC. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.