Abstract
Krüppel-like factor 4 (KLF4), a zinc finger-containing transcriptional factor, regulates a variety of biological processes, including cell proliferation, differentiation, apoptosis, and stem cell reprogramming. Post-translational modifications of KLF4, including phosphorylation, acetylation, and sumoylation, regulate its transcriptional activity. Most recent studies also demonstrate that KLF4 is targeted for ubiquitin-dependent proteolysis during cell cycle progression. However, the underlying mechanism remains largely unknown. In this study, we demonstrated that KLF4 is profoundly degraded in response to TGF-β signaling. We have identified the Cdh1-anaphase promoting complex as a putative E3 ligase that governs TGF-β-induced KLF4 degradation. The TGF-β-induced KLF4 degradation is mediated by the destruction box on the KLF4. Either depletion of Cdh1 by RNA interference or stabilization of KLF4 by disruption of its destruction box significantly attenuates TGF-β-induced ubiquitylation and degradation. In addition, depletion of Cdh1 or stabilization of KLF4 antagonizes TGF-β-induced activation of transcription. Determining the role of KLF4 proteolysis in response to TGF-β signaling has opened a new perspective to understand the TGF-β signaling pathway.
Keywords: Cell Cycle, Cytokine, E3 Ubiquitin Ligase, Protein Turnover, Ubiquitylation
Introduction
Krüppel-like factor 4 (KLF4),2 a zinc-finger containing transcription factor, regulates a variety of cellular processes, including cell proliferation, differentiation, apoptosis, as well as maintenance of tissue homeostasis (1). Recent studies have demonstrated that KLF4 is one of four critical transcriptional factors (Oct3/4, Sox2, KLF4, and c-Myc) that orchestrate the reprogramming of differentiated cells into pluripotent stem cells (2). In addition, current studies in carcinogenesis have revealed an interesting feature of KLF4 in that it could function as both tumor suppressor and tumor promoter depending on tissue type and cellular context (1). The tumor-suppressive effect of KLF4 has been characterized in various types of cancer, including gastrointestinal cancer (3), esophageal cancer (4, 5), bladder cancer (6), lung cancer (7), and lymphoma (8). In contrast, its oncogenic effect has been reported in breast and squamous cell carcinoma (9–13). Mechanistically, KLF4 exerts its tumor-suppressive function mainly by regulating cyclin-dependent kinase inhibitor (CKI) p21WAF1 (11, 14). Inactivation of p21Waf1 completely abolishes the cytostatic action of KLF4. Furthermore, KLF4 also up-regulates other inhibitors of proliferation such as p27Kip1 (15) and down-regulates the expression of Cyclin B (16) and Cyclin D1 (17), positive regulators of cell cycle progression. The oncogenic role for KLF4 is thought at this time to depend principally on its ability to inhibit apoptosis through the suppression of p53 expression (11). Depletion of Klf4 in breast cancer cells induces p53 accumulation, which in turn leads to p53-dependent apoptosis. In addition to p53 suppression, KLF4 could also inhibit the expression of Bax, a pro-apoptotic factor that is governed by p53 (18). Also, Rowland et al. (11) recently reported that KLF4 could override RasV12-induced senescence in primary fibroblasts and induce its transformation with regulation of p21WAF1 being pinpointed as the key to converting between oncogene and tumor suppressor. The dual and opposing roles of KLF4 in tumorigenesis and cell cycle arrest have attracted the attention in the field. However, thus far, the mechanism by which KLF4 switches from tumor suppressor to oncogene in different types of cancer remains largely unknown.
The function of KLF4 is highly regulated at both transcriptional and post-transcriptional levels. Previous studies revealed that KLF4 is down-regulated by promoter hypermethylation and loss-of-heterozygosity in many types of cancer (3). In addition, KLF4 can be induced by a variety of stimuli, including serum starvation (19, 20), oxidative stress (21), sodium butyrate (22), selenium (23), interferon-γ (IFN-γ) (24), and cAMP (25). Interestingly, KLF4 expression can be either elevated or repressed depending on the extent of DNA damage (26). Stimuli-elicited KLF4 regulation is known to be governed by multiple mechanisms including increased transcription, decreased mRNA stability, and increased protein stability. For example, severe DNA damage can trigger a rapid dissociation of Klf4 mRNA from the ubiquitous RNA-binding protein, HuR, which destabilizes Klf4 mRNA (26). In addition to its expression being regulated, KLF4 is also subjected to many kinds of post-translational modifications such as acetylation (27) and SUMOylation (28). Moreover, the addition of Sumo1 to KLF4 has been thought to be necessary to facilitate KLF4-mediated transactivation (28). A most recent study has implicated the importance of ubiquitin-proteasome system (UPS) in KLF4 regulation in response to serum stimulation (19). However, how KLF4 is regulated by UPS and which E3 ligase is involved in the serum-responsive KLF4 ubiquitylation remains unknown.
Transforming growth factor β (TGF-β) is a pluripotent cytokine involved in almost every aspect of cellular behavior. Perturbations of TGF-β signaling are central to tumorigenesis and tumor progression (29, 30). Transduction of the complex signaling starts on the cell surface where TGF-β binding induces the formation of the type I and II receptor complexes. Type II receptor phosphorylates and activates Type I receptor. Type I receptor then propagates the signal through phosphorylation of Smad2 or Smad3 (receptor-regulated Smad, R-Smad). Upon phosphorylation, Smad2 or Smad3 form an oligomeric complex with Smad4 (co-mediator Smad, Co-Smad) and translocates to the nucleus, where they regulate gene transcription in collaboration with DNA-binding co-factors such as forkhead family member FoxH1, co-activators such as p300, or co-repressors such as Ski (31). Like KLF4 and Runt-related transcription factors (Runx) (32), TGF-β has dual functions in carcinogenesis, where it acts as either a cytostatic factor in epithelial cells or in the early stage of cancer cells or a promoter of invasiveness and metastasis in late stage tumors (29, 30, 33, 34). As cytostatic regulator, TGF-β induces expression of several crucial CKIs, including p15INK4B, p21WAF1, and p57KIP2 (33), which in turn represses growth promoting transcription factor, c-Myc, and inhibitor of differentiation (31). On other hand, epithelial-mesenchymal transition (EMT) closely correlates to TGF-β-induced invasiveness and metastasis. Besides the above important features of the TGF-β pathway, how this pathway is regulated has greatly attracted our attention. It has been demonstrated that post-translational modification plays a critical role in regulating TGF-β signaling, including phosphorylation, acetylation, sumoylation, and ubiquitylation (35–39). Recent studies have demonstrated that the TGF-β signaling pathway is tightly regulated by UPS with several molecules in its signaling cascade being targeted by UPS for proteolysis, including TGF-β receptors, Smads, and co-suppressors of transcription (38). Several E3 ligases have been suggested to regulate the TGF-β signaling pathway, such as Smurfs (40, 41), Cdh1-anaphase promoting complex (APC) (42, 43), SCFFbxw7 (44), Arkadia (45), and CHIP (46). The list of targets by UPS in the TGF-β signaling circuitry is still expanding. Identification of new UPS targets in regulating TGF-β-mediated cytostatic and tumorigenic function will advance our knowledge in the molecular basis of TGF-β signaling pathway.
Our recent endeavor to search for new TGF-β-induced fast turnover proteins by large scale protein profiling led to our identification of KLF4 as a TGF-β-induced proteolytic component. In this study we have elucidated the mechanism by which KLF4 is degraded in response to TGF-β signaling and further determined the role of KLF4 proteolysis in TGF-β-mediated transactivation. Results from this work have opened a new avenue to understand the mechanism in the regulation of TGF-β signaling.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection (Manassas, VA). Mink lung epithelial cells Mv1Lu were generously provided by Dr. Xuedong Liu (University of Colorado, Boulder, CO). Mv1Lu cells stably expressing Cdh1 siRNA or control siRNA was generated as described previously (47). These cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1× antibiotic/antimycotic solution (100 units/ml of streptomycin, 100 units/ml of penicillin, and 0.25 μg/ml of amphotericin B), 100 μmol/liter of non-essential amino acids and 100 mmol/liter of HEPES buffer solution (all from Invitrogen). All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. For TGF-β stimulation experiments, 2 ng/ml of TGF-β (R&D systems) was used.
Plasmids and Transfection
The PAI-1 reporter plasmid was the gift from Dr. Xuedong Liu (University of Colorado, Boulder). Klf4 constructs were generated by PCR amplification of the Klf4 coding sequence from pcDNA3.1-Klf4 (the gift from Dr. Daniel S. Peeper, Netherlands Cancer Institute, Netherlands) and subsequent subcloning into pCS2-HA (where HA is hemagglutinin), a mammalian expression vector. FLAG tag was introduced into the primers and the sequences for the primers are as following: for F/H-KLF4, 5′-CCATCGATGCCATGGACTACAAGGACGACGATGACAAGGCTGTCAGCGACGCGCTGCTCCCA-3′ (forward primer) and 5′-TTGGCGCGCCAAAATGCCTCTTCATGTGTAAG-3′ (reverse primer); for FLAG-KLF4, 5′-CCATCGATGCCATGGACTCAAGGACGACGATGACAAGGCTGTCAGCGACGCGCTGCTCCCA-3′ (forward primer) and 5′-TTGGCGCGCCTTAAAAATGCCTCTTCATGTGTAAG-3′ (reverse primer). Klf4 constructs with mutations on destruction boxes (deletion of amino acids from 44–49 and 204–209) were engineered using the site-directed mutagenesis kit (Stratagene). The primer sequences are as following: for the D1 mutation, 5′-CCCGAATAACCGCTGGTCCCACATGAAGCGAC-3′ (forward primer) and 5′-GTCGCTTCATGTGGGACCAGCGGTTATTCGGG-3′ (reverse primer); for the D2 mutation, 5′-GGCCGAGCTCCTGGACCCGGTGTACA-3′ (forward primer) and 5′-TGTACACCGGGTCCAGGAGCTCGGCC-3′ (reverse primer). For transfection, cells were plated to form a 50–70% confluent culture. The HEK293T and Mv1Lu cells were transfected using Lipofectamine 2000 (Invitrogen).
To construct pLenti6-IRES-GFP-HA-Klf4 and pLenti6-IRES-GFP-HA-Klf4, the Klf4 coding sequence was first amplified from pCS2-F/H-KLF4 or pCS2-F/H-KLF4–2D and cloned into pMX-IRES-GFP, resulting in pMX-IRES-GFP-HA-KLF4 or pMX-IRES-GFP-HA-KLF4–2D. The sequences of used primers are: 5′-CCG CTC GAG ACC ATG TAC CCT TAT GAC GTG CCC GAT TAC-3′(forward), 5′-ATA AGA ATG CGG CCG CTT AAA AAT GCC TCT TCA TGT GTA AG-3′(reverse). Then the fragment encompassing HA-KLF4 or HA-KLF4–2D and IRES-GFP was amplified and cloned into pENTR/D-TOPO, an entry vector for the Gateway system. The sequences of used primers are: 5′-CAC CAT GTA CCC TTA TGA CGT G-3′ (forward), 5′-TTA CTT GTA CAG CTC GTC CAT GC-3′ (reverse). The resulting plasmids, ENTR-IRES-GFP-Klf4 or ENTR-IRES-GFP-Klf4–2D, were used for LR recombination reaction together with the destine vector Lenti6/V5-Dest, leading to the generation of pLenti6-IRES-GFP-HA-Klf4 or pLenti6-IRES-GFP-HA-Klf4–2D.
Lentiviral Infection
The pLenti6/V5-Dest plasmid was co-transfected with VSV-G, pRRE, and RSV-REV into HEK293T cells. Lipofectamine 2000 was used. The packaged lentiviral particle was collected, mixed with Polyprene, and added into target cells Mv1Lu. The GFP positive cells were sorted out by flow cytometry for further assay.
RNA Extraction, cDNA Synthesis, and Real Time PCR
Total RNA was isolated from various samples using TRIzol reagent (Invitrogen). 2 μg of RNA was primed by oligo(dT) (Promega) and reverse transcribed into cDNA with Moloney murine leukemia virus reverse transcriptase (M-MLV RT) (Promega). Real time PCR was carried out on a StepOne®Plus Real-time PCR system (Applied Biosystems) using Fast SYBR® Green master mix (Applied Biosystems). To pick up the primers for real time PCR detecting the expression of Klf4 and β-actin in Mv1Lu cells, the fragments of the mink Klf4 and β-actin gene were amplified from Mv1Lu cDNA and sequenced using the primers within the conserved regions as revealed by the nucleotide sequence alignment of Klf4 and the β-actin gene from different species. The primers for mink Klf4 and β-actin are as following: Klf4, 5′-GAGGGAGACGGAGGAGTTCAA-3′ (forward primer) and 5′-GGATGGGACAGCGAATTGG-3′ (reverse primer); β-actin, 5′-GGGAGATCGTGCGTGACAT-3′ (forward primer) and 5′-GCCATCTCCTGCTCGAAGTC-3′ (reverse primer).
Western Blotting and Immunoprecipitation Assay
Cells were harvested and lyzed in radioimmunoprecipitation assay (RIPA) lysis buffer (Upstate Biotechnology) containing protease inhibitor mixture (Sigma). The protein concentration was determined using the Bio-Rad Protein Assay Reagent (Bio-Rad). Western blotting was performed using anti-Cdk4 (Santa Cruz), Cdk6 (Santa Cruz), Cyclin D1 (Santa Cruz), p15INK4B (Santa Cruz), anti-SnoN (Santa Cruz), Skp2 (Santa Cruz), KLF4 (Santa Cruz), HA (Santa Cruz), p21 (BD Biosciences), ubiquitin (BD Biosciences), FLAG (Sigma), β-actin (Sigma), and horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit secondary antibody (Promega). Signals were detected with enhanced chemiluminescence reagents (Amersham Biosciences). Semi-quantification of data were performed using NIH Image. For the immunoprecipitation assay, cell lysate was incubated with anti-FLAG M2 gel (Sigma) or anti-Cdh1 (Calbiochem) antibody overnight at 4 °C on a rotator, following by addition of protein A/G plus-agarose (Pierce) to the reaction containing anti-Cdh1 antibody for 2 h at 4 °C. After five washes with RIPA lysis buffer supplemented with protease inhibitor mixture, complexes were released from the anti-FLAG M2 gel and protein A/G plus-agarose by boiling for 5 min in 2× SDS-PAGE loading buffer. Western blotting was used to detect Myc-Cdh1 and KLF4 with anti-Myc and anti-KLF4 antibody, respectively.
PAI-1 Reporter Assay
Cells were plated in 24-well plates. After 24 h, cells were co-transfected with the PAI-1 reporter plasmid and Klf4 constructs or the empty vector control using Lipofectamine 2000 according to the manufacturer's instruction. Firefly and Renilla luciferase activities were measured using a dual luciferase kit (Promega). The firefly luciferase data for each sample was normalized based on transfection efficiency as determined by Renilla luciferase activity. Each experiment was performed in triplicate and repeated at least three times.
Cell Viability Assay
2 × 103 of Mv1Lu cells were seeded into a 96-well plate and cultured overnight. Then the cells were treated with TGF-β (100 pm) for 48 h. Cell viability was determined using the CellTiter 96 AQuesous One Solution (Promega) according to the manufacturer's protocol.
RESULTS
KLF4 Protein Levels Are Drastically Down-regulated in Response to TGF-β Signaling
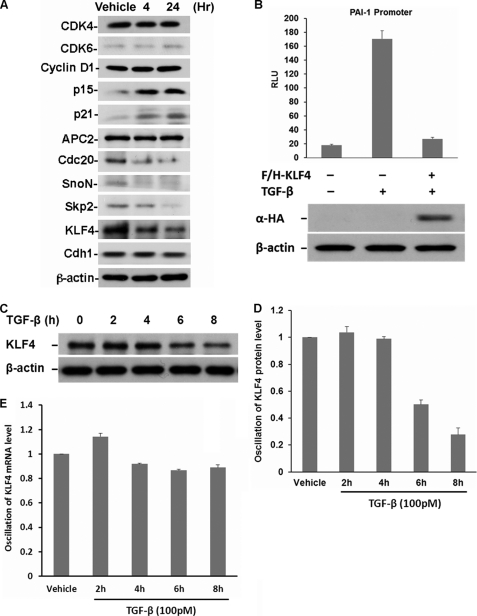
Previous studies have indicated that the TGF-β signaling pathway is tightly regulated by proteolysis. The role of UPS has been suggested to orchestrate TGF-β signaling by activation of TGF-β-induced transcription via removal of co-suppressors and resetting the signaling transducing system to the initial state by the degradation of phosphorylated components, such as Smad2 and receptors (37, 48). To identify potential novel proteolytic targets, we systematically analyzed the protein profile in response to TGF-β signaling. Mv1Lu cells were treated with TGF-β for 4 and 24 h. TGF-β-induced protein alteration was measured by immunoblotting. The results of TGF-β-induced alteration in protein expression can be divided into three groups. Although no obvious change in the concentrations of Cdk4, Cdk6, Cyclin D1, and APC was observed, the abundance of p15INK4B and p21WAF1 increased and the amounts of Cdc20, SnoN, and Skp2 decreased (Fig. 1A). Although down-regulation of SnoN and Skp2 by TGF-β has been described previously, we were nevertheless, surprised by our observation of the dramatic down-regulation of KLF4 in response to TGF-β signaling, which has not yet been reported. We decided to characterize the relevance of TGF-β-induced KLF4 down-regulation and further elucidate its underlying regulatory mechanism because KLF4 is critical to determine tumorigenesis and maintain stem cell pluripotency (49).
FIGURE 1.
KLF4 protein levels are drastically down-regulated in response to TGF-β signaling. A, profiling of TGF-β-induced protein alteration. Lysates were prepared from Mv1Lu cells treated with 100 pm TGF-β at the indicated time point. Alteration of various proteins involved in cell cycle progression, cellular differentiation, and transcriptional regulation were measured by immunoblotting. KLF4 protein levels dramatically dropped in response to TGF-β signaling. B, KLF4 profoundly affects TGF-β-initiated transactivation. TGF-β-induced PAI-1 transactivation was used as a readout. Mv1Lu cells were co-transfected with plasmid carrying KLF4, PAI-1 reporter plasmid, and a Renilla luciferase construct (pRL-TK). Transfected cells were then treated with TGF-β for 24 h and harvested for dual-luciferase activity assay. After normalization to Renilla, firefly luciferase activity was presented as relative light units (RLU), plotted against various treatments, and shown in the upper panel. Each bar represents the mean ± S.D. for triplicate samples. KLF4 expression in various treatments was estimated by immunoblotting using anti-hemagglutinin (HA) antibody, which is shown in the lower panel. C, time-dependent and TGF-β-induced KLF4 down-regulation. Mv1Lu cells were treated with TGF-β, collected at different time points as indicated. TGF-β-induced alteration of KLF4 protein levels was measured by immunoblotting using the antibody against KLF4. β-Actin was measured as loading control. D, quantification of TGF-β-induced down-regulation of KLF4. E, examination of Klf4 mRNA levels in response to TGF-β signaling. Total RNA was prepared from Mv1Lu cells treated with TGF-β at different time points. TGF-β-induced alteration of mRNA levels was measured by real time PCR. After normalization to the internal control β-actin, TGF-β-induced alteration of Klf4 mRNA levels were represented as the percentage of the vehicle control and plotted as indicated.
Given that KLF4 is an important regulator for transcription, we first explored its potential role involving TGF-β-mediated transcriptional regulation by conducting a plasminogen activator inhibitor-1 (PAI-1) reporter assay (50). The KLF4 expression construct and PAI-1 reporter plasmid were co-transfected into Mv1Lu cells and then the PAI-1 reporter activity was measured by the dual-luciferase reporter assay system. As shown in Fig. 1B, whereas an 8-fold increase in PAI-1 reporter activity was measured in response to TGF-β stimulation, expression of KLF4 profoundly attenuated the TGF-β-induced PAI-1 activity suggesting that KLK4 plays a suppressing role in TGF-β-induced transactivation and thus, antagonizes TGF-β effect.
To study the kinetics of the alteration in KLF4 protein concentration by TGF-β, we determined the expression profile of the KLF4 protein in response to TGF-β stimulation at different time points. Mv1Lu cells were stimulated with TGF-β and harvested at different time points. Lysates were subjected to immunoblotting. As shown in Fig. 1, C and D, a dramatic drop in KLF4 expression was observed after 6 h following TGF-β stimulation. To obtain further clues as to whether TGF-β-induced KLF4 alteration is due to transcriptional or post-transcriptional regulation, we assessed the dynamics of Klf4 mRNA at different time points after TGF-β stimulation. As shown in Fig. 1E, no significant alteration of mRNA was observed in response to TGF-β suggesting that TGF-β-induced down-regulation of KLF4 is via post-transcriptional regulation.
TGF-β-induced KLF4 Alteration Is Mediated by the Ubiquitin-Proteasome Pathway
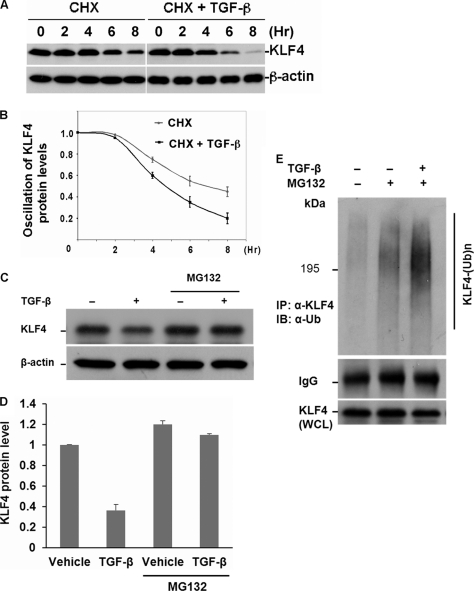
To investigate whether the TGF-β-induced drop of KLF4 protein levels is due to post-translational regulation, we measured the protein half-life of KLF4 in the absence and presence of TGF-β signaling. Mv1Lu cells were treated with the protein synthesis inhibitor cycloheximide (CHX) with or without TGF-β. Cells were harvested at different time points and the dynamic of KLF4 protein levels were measured by immunoblotting. As shown in Fig. 2, A and B, addition of TGF-β significantly enhanced the rate of drop for the abundance of KLF4 protein, suggesting that TGF-β enhances KLF4 turnover rate.
FIGURE 2.
TGF-β-induced KLF4 alteration is mediated by the ubiquitin-proteasome pathway. A, turnover rate of KLF4 protein in the absence and presence of TGF-β signaling. Mv1Lu cells were treated with 10 μg/ml of cycloheximide (CHX) only or CHX plus TGF-β. The cell pellet was collected at different time points as indicated. KLF4 protein levels were measured by immunoblotting. The KLF4 turnover rate was enhanced by stimulation with TGF-β. B, summary of A. C, TGF-β-induced KLF4 protein turnover is blocked by proteasomal inhibitor, MG132. Cells were collected 4 h after stimulation with TGF-β. The TGF-β-induced drop of KLF4 levels was attenuated by MG-132. D, quantification of C. E, TGF-β-induced KLF4 down-regulation is mediated by ubiquitylation. TGF-β-treated Mv1Lu cells were collected 4 h after cellular stimulation with TGF-β. The endogenous KLF4 protein complex was purified by immunoprecipitation using anti-KLF4 antibody coupled with protein A beads. The ubiquitin-conjugated KLF4 in the immunoprecipitation (IP) complex was detected by immunoblotting (IB) using anti-ubiquitin antibody. KLF4 protein levels in the whole cell lysates (WCL) were measured by immunoblotting. Obvious KLF4 ubiquitin conjugates were visualized in response to TGF-β signaling.
To test if the TGF-β-induced drop in KLF4 protein levels was mediated by proteasome, we then evaluated the effect of MG-132, a proteasomal inhibitor, on TGF-β-induced KLF4 down-regulation. Mv1Lu cells were treated with TGF-β in the absence and presence of the proteasome inhibitor MG132, and KLF4 protein expression was analyzed by immunoblotting. As shown in Fig. 2, C and D, whereas TGF-β stimulation induced a significant drop in KLF4 protein levels, the addition of MG132 completely abolished TGF-β-induced KLF4 down-regulation, suggesting that the TGF-β-induced KLF4 alteration is via a proteasomal pathway. To further examine if TGF-β-induced KLF4 down-regulation is mediated by ubiquitylation, we tested endogenous KLF4 ubiquitylation. Mv1Lu cells were treated with TGF-β and the ubiquitin conjugate of KLF4 was pulled down and detected by immunoblotting. As shown in Fig. 2E, whereas minor KLF4 ubiquitin conjugates were detected in the absence of TGF-β, a significant increase in KLF4 ubiquitin conjugates was measured in response to TGF-β stimulation. Taken together, results from the above experiments suggest that the TGF-β-induced KLF4 down-regulation is mediated by the ubiquitin-proteasome pathway.
Cdh1/APC Is a Putative E3 Ubiquitin Ligase Governing TGF-β-induced KLF4 Proteolysis
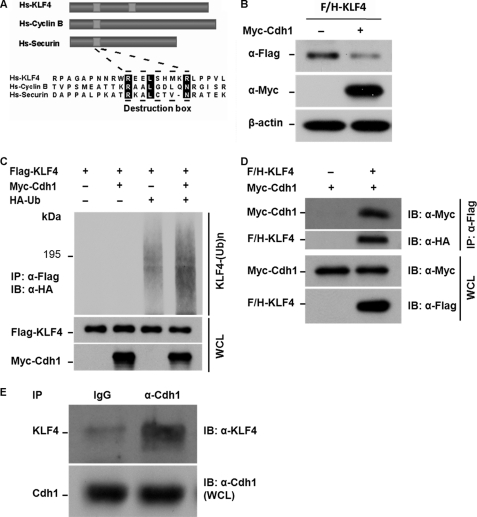
To identify the potential E3 ligase that governs TGF-β-induced KLF4 degradation, we have systematically searched for putative degron on KLF4. Like most of APC substrates, we found two conserved destruction boxes on KLF4, suggesting that APC could be a candidate E3 ligase for KLF4 degradation (Fig. 3A). In addition, previous studies have shown that the APC activity is responsive to TGF-β stimulation and Cdh1 is the substrate activator for APC to target the TGF-β-induced protein turnover such as SnoN and Skp2 (42, 43). We therefore hypothesize that Cdh1/APC is a putative E3 ligase that governs TGF-β-induced KLF4 degradation. To test this hypothesis, we tested the stability of the KLF4 protein in response to overexpression of Cdh1. As shown in Fig. 3B, co-transfection of Klf4 and Cdh1 in human embryonic kidney cells (HEK293T) resulted in significant down-regulation of KLF4 expression supporting the notion that KLF4 is regulated by Cdh1/APC. We then tested whether Cdh1/APC could facilitate KLF4 ubiquitylation. FLAG-KLF4, Myc-Cdh1, and HA-ubiquitin were cotransfected into HEK293T cells. KLF4 ubiquitin conjugates were purified by anti-FLAG M2 beads followed by immunoblotting using antibody against HA. As shown in Fig. 3C, whereas a minor amount of KLF4 ubiquitin conjugates was detected without expression of Cdh1, overexpression of Cdh1 dramatically enhanced KLF4 ubiquitylation, further suggesting that Cdh1/APC is the ligase that catalyzes KLF4 for ubiquitylation and degradation. Cdh1 has been shown to function as a substrate receptor, which recruits substrates for APC (51, 52). To test if Cdh1 physically interacts with KLF4, we determined biochemical interaction between Cdh1 and KLF4. FLAG-KLF4 and Myc-Cdh1 were co-transfected into HEK293T cells. The KLF4 complex was pulled down and Cdh1 expression in the complex was detected by immunoblotting. As shown in Fig. 3D, expression of Myc-Cdh1 was examined on the KLF4 complex suggesting that Cdh1 physically interacts with KLF4. To further confirm the interaction between endogenous KLF4 and Cdh1, we conducted a pull-down experiment in Mv1Lu cells after stimulation with TGF-β. Mv1Lu cells were treated with TGF-β and collected 4 h after the stimulation. The endogenous Cdh1 complex was pulled down by immunoprecipitation using antibody against Cdh1 following by immunoblotting using antibody against KLF4. As shown in Fig. 3E, a significant amount of KLF4 protein was detected on the Cdh1-IP complex but not in the control. All together, our results suggest that Cdh1/APC could be a putative E3 ligase that governs TGF-β-induced KLF4 degradation.
FIGURE 3.
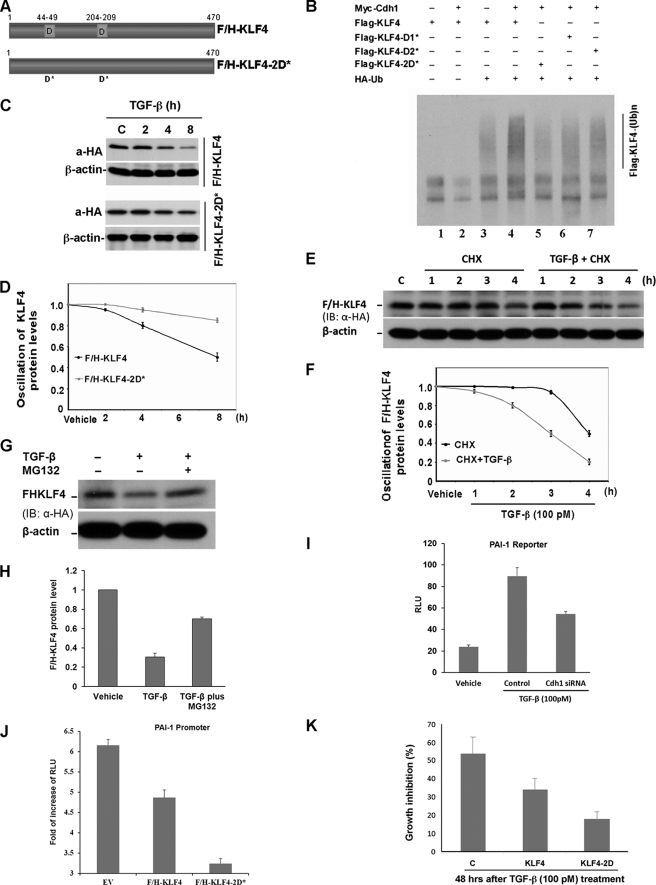
Cdh1/APC is a putative E3 ligase that facilitates TGF-β-induced KLF4 ubiquitylation. A, identification of destruction motifs on KLF4. Like most APC substrates, KLF4 bears a conserved destruction box as indicated by sequence alignment with Cyclin B and securin. The alignment was performed using the CLUSTAL W methods. Hs, human. B, elevation of Cdh1 results in degradation of KLF4. HEK293T cells were co-transfected with FLAG- and HA-tagged KLF4 (F/H-KLF4) and Myc-tagged Cdh1 (Myc-Cdh1). Cells were collected 40 h after the transfection. Expression levels of F/H-KLF4 and Myc-Cdh1 were measured by immunoblotting (IB) using the antibodies against FLAG and the Myc tag, respectively. β-Actin was used as loading control. KLF4 protein levels were dropped in cells with expression of Cdh1. C, overexpression of Cdh1 enhances KLF4 ubiquitylation. FLAG-tagged KLF4 (Flag-KLF4) was transfected into HEK293T cells together with Myc-Cdh1 and HA-tagged ubiquitin (HA-Ub). Transfected cells were treated with MG132 for 6 h. The accumulated ubiquitin-conjugated FLAG-KLF4 was examined by immunoprecipitation (IP) using anti-FLAG M2 affinity gel. KLF4 ubiquitin conjugates were then detected by immunoblotting using antibody against HA. The expression of FLAG-KLF4 and Myc-Cdh1 in the whole cell lysates (WCL) was estimated by immunoblotting. Overexpression of Cdh1 significantly enhances the formation of KLF4 ubiquitin conjugates. D, KLF4 interacts with Cdh1 in vivo. F/H-KLF4 and Myc-Cdh1 were co-transfected into HEK293T cells. Interaction between KLF4 and Cdh1 was measured by immunoprecipitation using anti-FLAG-M2 gel (Flag-KLF4) following by immunoblotting using anti-Myc (Myc-Cdh1). Expression of both FLAG-KLF4 and Myc-Cdh1 was estimated by immunoblotting using whole cell lysates. E, endogenous KLF4 interacts with Cdh1 in Mv1Lu cells. Mv1Lu cells were treated by TGF-β and applied to immunoprecipitation assay using anti-Cdh1 antibody and subsequent immunoblotting using anti-KLF4 antibody. Cdh1 was pulled down with KLF4 immunoprecipitation. The expression of Cdh1 in the whole cell lysates was measured by immunoblotting.
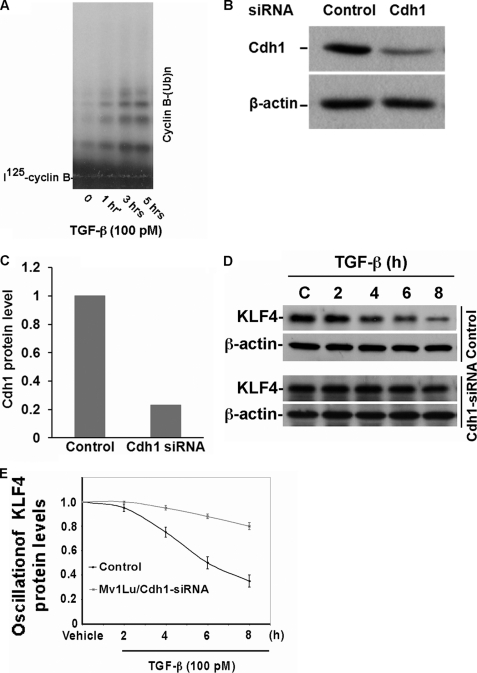
To further validate the hypothesis that Cdh1/APC is a putative E3 ligase governing KLF4 degradation in response to TGF-β signaling, we depleted Cdh1 by RNA interference and then tested the effect of Cdh1 depletion on TGF-β-induced KLF4 degradation. To date, we initially measured activation of APC by TGF-β signaling. Mv1Lu cells were treated with TGF-β and collected at different time points. Endogenous APC was purified by immunoprecipitation using antibody against Cdc27 (a hardcore subunit of APC). Iodine 125-radiolabeled Cyclin B was used as a test substrate of APC. As shown in Fig. 4A, upon stimulation with TGF-β, cyclin ubiquitylation as catalyzed by APC was significantly increased after stimulation with TGF-β. With Cdhl stably depleted in Mv1Lu cells (Fig. 4, B and C), we next tested the effect of Cdh1 depletion on TGF-β-induced KLF4 degradation. As shown in Fig. 4, D and E, depletion of Cdh1 significantly attenuated the TGF-β-induced KLF4 degradation, further confirming that Cdh1/APC is a putative E3 ligase that governs KLF4 degradation in response to TGF-β signaling.
FIGURE 4.
Depletion of Cdh1 attenuates TGF-β-induced KLF4 degradation. A, APC activity is enhanced in response to TGF-β signaling. Mv1Lu cells were treated with TGF-β and collected at the indicated time points. Endogenous APC was then purified by immunoprecipitation using antibody against Cdc27. Iodine 125-radiolabeled cyclin B recombinant protein was used as putative substrate for APC in vitro ubiquitylation assay. Upon stimulation with TGF-β, APC activity was profoundly enhanced as reflected by increased formation of cyclin B polyubiquitin conjugates. B, Cdh1 knockdown by RNAi in Mv1Lu cells. Mv1Lu cells were infected with Cdh1 shRNA expressing retrovirus and the stable cell line was generated. The knockdown effect of Cdh1 shRNA was evaluated by immunoblotting. The Cdh1 expression was normalized to the loading control β-actin. C, quantification of B. D, TGF-β-induced KLF4 degradation is attenuated in Cdh1-depleted Mv1Lu cells. Both Cdh1-depleted and control Mv1Lu cells were treated by TGF-β and harvested at different time points as indicated. KLF4 expression was measured by immunoblotting using antibody against KLF4. E, summary of D.
Mutation of Destruction Boxes on KLF4 Blocks TGF-β-induced KLF4 Degradation, Which Impairs TGF-β-mediated PAI-1 Transactivation
Mutation of destruction boxes should stabilize KLF4 and thus antagonize TGF-β response. To test this hypothesis, KLF4 mutants with deleted destruction boxes were generated as shown in Fig. 5A. Wild-type Klf4 or the Klf4 mutant were co-transfected with the Cdh1 construct into HEK293T cells and ubiquitylation of KLF4 was analyzed. As shown in Fig. 5B, basal KLF4 ubiquitylation was measured (lane 3). Although overexpression of Cdh1 significantly increased the ubiquitin signal for wild-type KLF4 (lane 4 compared with lane 3), disruption of the destruction box on KLF4 significantly attenuated the Cdh1-enhanced KLF4 ubiquitylation. In addition, mutation of the destruction box on KLF4 also stabilized KLF4 in response to TGF-β stimulation (Fig. 4, C and D). To confirm that ectopic KLF4, similar to endogenous KLF4 as described above, is also targeted by the ubiquitin-proteasome pathway for degradation in response to TGF-β signaling, we performed a protein chase experiment. As shown in Fig. 5, E and F, TGF-β stimulation greatly promoted turnover for the ectopically expressed KLF4. The TGF-β-induced turnover of ectopic KLF4 could also be blocked by MG132 (Fig. 5, G and G).
FIGURE 5.
Disruption of the molecular degron blocks TGF-β-induced KLF4 degradation, which then impairs TGF-β-mediated PAI-1 transactivation. A, diagram of the constructs expressing F/H-KLF4 and F/H-KLF4 with mutation of destruction boxes named F/H-KLF4–2D*. B, mutation of destruction boxes attenuates Cdh1-induced KLF4 ubiquitylation. HEK293T cells were co-transfected with Myc-Cdh1 and FLAG-KLF4 or various FLAG-tagged Klf4 mutants (FLAG-KLF4-D1*, FLAG-KLF4-D2*, and FLAG-KLF4–2D*), respectively. The transfected cells were treated with MG132 for 6 h. The accumulated ubiquitin conjugates of KLF4 were precipitated by anti-FLAG M2 gel and assayed by immunoblotting (IB) using antibody against HA. C, mutation of destruction boxes inhibits KLF4 degradation in response to TGF-β signaling. Mv1Lu cells were transfected with F/H-KLF4 or F/H-KLF4–2D* and subsequently stimulated with TGF-β. Cells were harvested at different time points as indicated. The expression of F/H-KLF4 and F/H-KLF4–2D* was analyzed by immunoblotting using antibody against HA. D, summary of C. E, ectopic KLF4 protein chase analysis in response to TGF-β signaling. Mv1Lu cells were transfected with F/H-KLF4 and treated with CHX in the absence and presence of TGF-β for different times as indicated. The dynamic of F/H-KLF4 expression was evaluated by immunoblotting using antibody against HA. F, summary of E. G, TGF-β-induced KLF4 degradation is blocked by MG132. Mv1Lu cells were transfected with F/H-KLF4 and treated with TGF-β in the absence and presence of MG132. The expression of F/H-KLF4 was analyzed by immunoblotting using antibody against HA. H, summary of G. I, Cdh1 depletion leads to inhibition of TGF-β-induced PAI-1 transactivation. Cdh1-depleted Mv1Lu cells were transfected with PAI-1 reporter plasmid together with a Renilla construct. The transfected cells were stimulated with TGF-β and harvested for luciferase activity assay. Relative light units (RLU) were plotted against various treatments. J, stabilization of KLF4 enhances its inhibition on TGF-β-induced PAI-1 transactivation. Mv1Lu cells were transfected with F/H-KLF4 or F/H-KLF4–2D* together with the PAI-1 reporter plasmid and a Renilla construct. The transfected cells were subjected to TGF-β treatment and harvested for luciferase activity assay. The induction of PAI-1 activity by TGF-β was presented as fold-increase of RLU and plotted against various treatments. K, Mv1Lu cells were infected with pLenti6-HA-KLF4 or pLenti6-HA-KLF4–2D. The GFP positive cells were sorted out and treated with TGF-β for 48 h. The effect of stabilization of KLF4 on TGF-β-induced growth inhibition was measured by the MTS assay.
To test the impact of Cdh1 depletion and KLF4 stabilization on TGF-β-mediated transcriptional activation, we measured their effect on TGF-β-mediated PAI-1 activation. The Cdh1 siRNA duplex was transfected into Mv1Lu cells. The transfected cells were stimulated with TGF-β and harvested for the luciferase activity assay. As shown in Fig. 5I, the PAI-1 reporter activity was induced 4-fold upon TGF-β treatment. Depletion of Cdh1 significantly inhibited TGF-β-activated PAI-1 reporter activity. As expected, stabilization of KLF4 by disruption of the destruction box on KLF4 significantly attenuated TGF-β-activated PAI-1 reporter activity (Fig. 5J). To further examine whether stabilization of KLF4 could affect TGF-β-induced growth inhibition, we engineered Mv1Lu cells that stably express non-degradable KLF4 using the lentiviral technique. As shown in Fig. 5K, expression of D-boxes mutated by KLF4 significantly attenuate the TGF-β-induced growth inhibition measured by MTS analysis. All together, the present results demonstrate that KLF4 is targeted for proteolysis in response to TGF-β signaling. Cdh1/APC is a putative E3 ligase for TGF-β-induced KLF4 degradation. Stabilization of KLF4 impairs the TGF-β-induced transcriptional activation that in turn antagonizes TGF-β-induced growth inhibition.
DISCUSSION
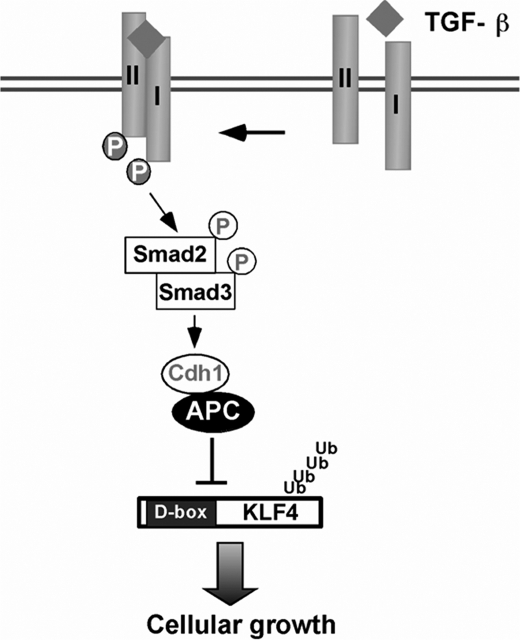
KLF4 is a critical transcriptional regulator that controls the switch from somatic cell to stem cell as well as orchestrates tumorigenesis (49). Although it has been demonstrated that KLF4 is regulated by various post-translational modifications, including phosphorylation, acetylation, and sumoylation, less is known if KLF4 is regulated by UPS (19, 27, 28, 53). The present study has demonstrated that KLF4 is regulated by the proteolysis depending on ubiquitin. Destruction of KLF4 by ubiquitin is necessary to ensure TGF-β-induced transcriptional activation. Our present identification that Cdh1/APC catalyzes KLF4 for ubiquitylation followed by degradation has elucidated the mechanism by which KLF4 is degraded in response to TGF-β signaling (Fig. 6). The findings from this study have filled a gap in our knowledge about KLF4 regulation and have further advanced our understanding of the TGF-β signaling pathway.
FIGURE 6.
Hypothetic model for TGF-β-regulated KLF4 proteolysis.
Post-translational modifications tightly regulate protein stability, subcellular localization, and functions. In addition to transcriptional regulation, KLF4 is also known to be subjected to multiple post-translational modifications, including acetylation (27), sumoylation (28, 54), and phosphorylation (53). Our findings have provided another layer to our understanding of the KLF4 regulatory circuitry, which in addition could enhance our understanding about the pivotal role of KLF4 in stem cell biology and carcinogenesis. KLF4 was found to be unstable in the human colon cancer cell line HCT116, where increased expression of KLF4 induces cell cycle arrest at the G1 phase through activation of p21Waf1 and/or repression of Cyclin D1 expression (14, 17, 55). After release from the serum starvation-induced G1 phase arrest, KLF4 is shown to be tagged by polyubiquitin and targeted for degradation. In our studies, we found the turnover of basal KLF4 in both Mv1Lu and HEK293T cells (Figs. 2, C and E, and 3C), suggesting that KLF4 turnover could be important in cell cycle control as well as non-cell cycle regulation. Identification of the precise role for KLF4 in regulating cell cycle progression and further elucidation of the mechanism by which KLF4 is ubiquitylated during the cell cycle are necessary to fill the current knowledge gap, bringing us closer to understanding the mystery of the double-blade function of KLF4 as both tumor suppressor and oncogene. Similarly to KLF4, TGF-β also has dual functions in its tumor suppressing role and malignant enhancing effect (29, 30, 33, 34). TGF-β stimulation inhibits cell cycle progression at the G1 phase through induction of p15INK4B and p21WAF1 (56, 57). Given that TGF-β and KLF4 are both involved in cell cycle regulation and KLF4 is negatively regulated by TGF-β in Mv1Lu cells, KLF4 could function as a positive regulator of the cell cycle in Mv1Lu cells. How KLF4 is regulated during cell cycle progression in Mv1Lu cells and whether the ubiquitin-proteasome pathway is involved in this regulation are both important aspects to understanding the relationship between KLF4 and TGF-β. Uncovering the connection between KLF4 and the TGF-β signaling pathway may bring some insights to the switch between their oncogenic role and tumor suppressing function. In addition to post-translational modifications, KLF4 is also targeted at the transcriptional level. For example, p53 and CDX2 can activate Klf4 transcription in different cellular settings (14, 58, 59). Interestingly, the transcriptional regulation of Klf4 by TGF-β signaling has been reported in other cell lines. This regulation is cell-context dependent. KLF4 is suppressed by TGF-β signaling in the macrophage (60), but induced in vascular smooth muscle cells (61). Our findings on the proteolytic regulation of KLF4 by TGF-β signaling further supports the notion that TGF-β-mediated KLF4 regulation is highly cell-context dependent.
TGF-β signaling regulates a large number of targeted genes through the activation of the downstream Smad pathway. Because the activated Smad complexes bind with weak affinity to Smad-binding elements located at the promoters of the targeted genes, the recruitment of Smad complexes to chromatin depends on their direct interaction with transcriptional factors that bind to DNA with higher affinity. Upon binding to DNA and to their transcriptional partners, Smads recruit co-activators, such as p300 and C/EBP-binding protein (CBP), or co-repressor, such as retinoblastoma-like 1 to activate or repress targeted genes (34). KLF4 is a zinc finger protein containing transcription factor. The regulation of KLF4 by TGF-β signaling is believed to occur at the transcriptional machinery. In our study, KLF4 was found to greatly antagonize TGF-β-mediated transcription (Fig. 1B). Likely, KLF4 may function as a co-repressor. Thus, the removal of KLF4 by TGF signaling will facilitate the activation of transcription by Smad. How KLF4 interplays with the Smad complex and its transcriptional co-factors to achieve the inhibitory effect will be interesting for further study. Two well characterized co-repressors, Ski (Sloan-Kettering Institute proto-oncogene) and SnoN (Ski-related novel gene N), are also targeted for degradation to facilitate the initiation of transcription. Several E3 ubiquitin ligases have been identified to govern the turnover of SnoN and Ski, including APC, SCF, RNF111, and Arkadia (62–65). In particular, we and others have demonstrated that Cdh1/APC can be activated by TGF-β signaling to degrade SnoN. Given that SnoN and KLF4 share the same E3 ubiquitin ligase, it is quite possible that KLF4 forms a complex with SnoN. Most recently, two studies have identified different mechanisms for the regulation of KLF4 on TGF-β-mediated transcription. In macrophages, KLF4 was found to compete with Smad 3 for the co-activator p300/cAMP-response element-binding protein and thus inhibit TGF-β-induced transcription (60). In contrast, in vascular smooth muscle cells, TGF-β induces KLF4 phosphorylation, allowing KLF4 to interact with Smad2. The KLF4-Smad2 complex in turn mediates the transcription of TβR1 (TGF-β type I receptor) (53). Taken together, like the regulation of TGF-β on KLF4 expression, the regulation of KLF4 on TGF-β signaling is also highly cell-text dependent.
TGF-β signaling is thought to be the common regulator of both epithelial cell cycle arrest and the EMT program, which mediates the cytostatic effect and tumor promoter functions. Similarly, KLF4 can suppress cell growth or inhibit apoptosis. Most recently, KLF4 is also found to be involved in the EMT program (66, 67). KLF4 can induce E-cadherin expression and thus inhibit EMT in mammary epithelial cells, which supports a suppressive role of metastasis by KLF4 in breast cancer (67). Interestingly, the induction of KLF4 on E-cadherin promotes mesenchymal-epithelial transition program, which is required for efficient reprogramming of fibroblast into iPS cells (66). Given that TGF-β signaling and KLF4 converge at E-cadherin expression in terms of EMT regulation, the negative regulation of KLF4 by TGF-β prompts us to further investigate the potential role of KLF4 in the TGF-β-induced EMT program in the future. In general, our finding that the proteolytic regulation of KLF4 by TGF-β signaling established a foundation for further study on the opposing role of KLF4 in carcinogenesis and paradoxical effect of TGF-β in both tumor suppression and oncogenic promotion.
Acknowledgments
We thank Drs. X. Liu, P Zhou, Z. Liu, and D. S. Peeper for cDNA clones. We are grateful to members of our laboratory for critical reading of the manuscript and related technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant CA115943.
- KLF4
- Krüppel-like factor 4
- UPS
- ubiquitin-proteasome system
- PAI-1
- plasminogen activator inhibitor-1
- APC
- anaphase promoting complex
- EMT
- epithelial-mesenchymal transition
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium.
REFERENCES
- 1. Rowland B. D., Peeper D. S. (2006) Nat. Rev. Cancer 6, 11–23 [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 3. Wei D., Kanai M., Huang S., Xie K. (2006) Carcinogenesis 27, 23–31 [DOI] [PubMed] [Google Scholar]
- 4. Wang N., Liu Z. H., Ding F., Wang X. Q., Zhou C. N., Wu M. (2002) World J. Gastroenterol. 8, 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tetreault M. P., Yang Y., Travis J., Yu Q. C., Klein-Szanto A., Tobias J. W., Katz J. P. (2010) Gastroenterology 139, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohnishi S., Ohnami S., Laub F., Aoki K., Suzuki K., Kanai Y., Haga K., Asaka M., Ramirez F., Yoshida T. (2003) Biochem. Biophys. Res. Commun. 308, 251–256 [DOI] [PubMed] [Google Scholar]
- 7. Hu W., Hofstetter W. L., Li H., Zhou Y., He Y., Pataer A., Wang L., Xie K., Swisher S. G., Fang B. (2009) Clin. Cancer Res. 15, 5688–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guan H., Xie L., Leithauser F., Flossbach L., Moller P., Wirth T., Ushmorov A. (2010) Blood 116, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 9. Foster K. W., Frost A. R., McKie-Bell P., Lin C. Y., Engler J. A., Grizzle W. E., Ruppert J. M. (2000) Cancer Res. 60, 6488–6495 [PubMed] [Google Scholar]
- 10. Pandya A. Y., Talley L. I., Frost A. R., Fitzgerald T. J., Trivedi V., Chakravarthy M., Chhieng D. C., Grizzle W. E., Engler J. A., Krontiras H., Bland K. I., LoBuglio A. F., Lobo-Ruppert S. M., Ruppert J. M. (2004) Clin. Cancer Res. 10, 2709–2719 [DOI] [PubMed] [Google Scholar]
- 11. Rowland B. D., Bernards R., Peeper D. S. (2005) Nat. Cell Biol. 7, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 12. Foster K. W., Ren S., Louro I. D., Lobo-Ruppert S. M., McKie-Bell P., Grizzle W., Hayes M. R., Broker T. R., Chow L. T., Ruppert J. M. (1999) Cell Growth Differ. 10, 423–434 [PubMed] [Google Scholar]
- 13. Foster K. W., Liu Z., Nail C. D., Li X., Fitzgerald T. J., Bailey S. K., Frost A. R., Louro I. D., Townes T. M., Paterson A. J., Kudlow J. E., Lobo-Ruppert S. M., Ruppert J. M. (2005) Oncogene 24, 1491–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang W., Geiman D. E., Shields J. M., Dang D. T., Mahatan C. S., Kaestner K. H., Biggs J. R., Kraft A. S., Yang V. W. (2000) J. Biol. Chem. 275, 18391–18398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei D., Kanai M., Jia Z., Le X., Xie K. (2008) Cancer Res. 68, 4631–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoon H. S., Yang V. W. (2004) J. Biol. Chem. 279, 5035–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shie J. L., Chen Z. Y., Fu M., Pestell R. G., Tseng C. C. (2000) Nucleic Acids Res. 28, 2969–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghaleb A. M., Katz J. P., Kaestner K. H., Du J. X., Yang V. W. (2007) Oncogene 26, 2365–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Z. Y., Wang X., Zhou Y., Offner G., Tseng C. C. (2005) Cancer Res. 65, 10394–10400 [DOI] [PubMed] [Google Scholar]
- 20. Shields J. M., Christy R. J., Yang V. W. (1996) J. Biol. Chem. 271, 20009–20017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cullingford T. E., Butler M. J., Marshall A. K., Tham el L., Sugden P. H., Clerk A. (2008) Biochim. Biophys. Acta 1783, 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shie J. L., Chen Z. Y., O'Brien M. J., Pestell R. G., Lee M. E., Tseng C. C. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279, G806–814 [DOI] [PubMed] [Google Scholar]
- 23. Liu S., Zhang H., Zhu L., Zhao L., Dong Y. (2008) Mol. Cancer Res. 6, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Z. Y., Shie J., Tseng C. (2000) FEBS Lett. 477, 67–72 [DOI] [PubMed] [Google Scholar]
- 25. Birsoy K., Chen Z., Friedman J. (2008) Cell Metab. 7, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou Q., Hong Y., Zhan Q., Shen Y., Liu Z. (2009) Cancer Res. 69, 8284–8292 [DOI] [PubMed] [Google Scholar]
- 27. Evans P. M., Zhang W., Chen X., Yang J., Bhakat K. K., Liu C. (2007) J. Biol. Chem. 282, 33994–34002 [DOI] [PubMed] [Google Scholar]
- 28. Du J. X., McConnell B. B., Yang V. W. (2010) J. Biol. Chem. 285, 28298–28308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siegel P. M., Massagué J. (2003) Nat. Rev. Cancer 3, 807–821 [DOI] [PubMed] [Google Scholar]
- 31. Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 32. Blyth K., Cameron E. R., Neil J. C. (2005) Nat. Rev. Cancer 5, 376–387 [DOI] [PubMed] [Google Scholar]
- 33. Pardali K., Moustakas A. (2007) Biochim. Biophys. Acta 1775, 21–62 [DOI] [PubMed] [Google Scholar]
- 34. Ikushima H., Miyazono K. (2010) Nat. Rev. Cancer 10, 415–424 [DOI] [PubMed] [Google Scholar]
- 35. Wrighton K. H., Feng X. H. (2008) Cell. Signal. 20, 1579–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Datto M., Wang X. F. (2005) Cell 121, 2–4 [DOI] [PubMed] [Google Scholar]
- 37. Chen Y. G., Wang X. F. (2009) Cell 139, 658–660 [DOI] [PubMed] [Google Scholar]
- 38. Lönn P., Morén A., Raja E., Dahl M., Moustakas A. (2009) Cell Res. 19, 21–35 [DOI] [PubMed] [Google Scholar]
- 39. Wrighton K. H., Lin X., Feng X. H. (2009) Cell Res. 19, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 41. Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 42. Stroschein S. L., Bonni S., Wrana J. L., Luo K. (2001) Genes Dev. 15, 2822–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wan Y., Liu X., Kirschner M. W. (2001) Mol. Cell 8, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 44. Bengoechea-Alonso M. T., Ericsson J. (2010) Oncogene 29, 5322–5328 [DOI] [PubMed] [Google Scholar]
- 45. Koinuma D., Shinozaki M., Komuro A., Goto K., Saitoh M., Hanyu A., Ebina M., Nukiwa T., Miyazawa K., Imamura T., Miyazono K. (2003) EMBO J. 22, 6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li L., Xin H., Xu X., Huang M., Zhang X., Chen Y., Zhang S., Fu X. Y., Chang Z. (2004) Mol. Cell. Biol. 24, 856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu W., Wu G., Li W., Lobur D., Wan Y. (2007) Mol. Cell. Biol. 27, 2967–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inoue Y., Imamura T. (2008) Cancer Sci. 99, 2107–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Evans P. M., Liu C. (2008) Acta Biochim. Biophys. Sin. 40, 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Attisano L., Wrana J. L., López-Casillas F., Massagué J. (1994) Biochim. Biophys. Acta 1222, 71–80 [DOI] [PubMed] [Google Scholar]
- 51. Pfleger C. M., Lee E., Kirschner M. W. (2001) Genes Dev. 15, 2396–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wan Y., Kirschner M. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13066–13071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li H. X., Han M., Bernier M., Zheng B., Sun S. G., Su M., Zhang R., Fu J. R., Wen J. K. (2010) J. Biol. Chem. 285, 17846–17856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawai-Kowase K., Ohshima T., Matsui H., Tanaka T., Shimizu T., Iso T., Arai M., Owens G. K., Kurabayashi M. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen Z. Y., Shie J. L., Tseng C. C. (2002) J. Biol. Chem. 277, 46831–46839 [DOI] [PubMed] [Google Scholar]
- 56. Datto M. B., Li Y., Panus J. F., Howe D. J., Xiong Y., Wang X. F. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5545–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hannon G. J., Beach D. (1994) Nature 371, 257–261 [DOI] [PubMed] [Google Scholar]
- 58. Dang D. T., Mahatan C. S., Dang L. H., Agboola I. A., Yang V. W. (2001) Oncogene 20, 4884–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoon H. S., Chen X., Yang V. W. (2003) J. Biol. Chem. 278, 2101–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Feinberg M. W., Cao Z., Wara A. K., Lebedeva M. A., Senbanerjee S., Jain M. K. (2005) J. Biol. Chem. 280, 38247–38258 [DOI] [PubMed] [Google Scholar]
- 61. King K. E., Iyemere V. P., Weissberg P. L., Shanahan C. M. (2003) J. Biol. Chem. 278, 11661–11669 [DOI] [PubMed] [Google Scholar]
- 62. Le Scolan E., Zhu Q., Wang L., Bandyopadhyay A., Javelaud D., Mauviel A., Sun L., Luo K. (2008) Cancer Res. 68, 3277–3285 [DOI] [PubMed] [Google Scholar]
- 63. Levy L., Howell M., Das D., Harkin S., Episkopou V., Hill C. S. (2007) Mol. Cell. Biol. 27, 6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nagano Y., Mavrakis K. J., Lee K. L., Fujii T., Koinuma D., Sase H., Yuki K., Isogaya K., Saitoh M., Imamura T., Episkopou V., Miyazono K., Miyazawa K. (2007) J. Biol. Chem. 282, 20492–20501 [DOI] [PubMed] [Google Scholar]
- 65. Sun Y., Liu X., Ng-Eaton E., Lodish H. F., Weinberg R. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., Qin D., Feng S., Song H., Yang D., Zhang B., Zeng L., Lai L., Esteban M. A., Pei D. (2010) Cell Stem Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 67. Yori J. L., Johnson E., Zhou G., Jain M. K., Keri R. A. (2010) J. Biol. Chem. 285, 16854–16863 [DOI] [PMC free article] [PubMed] [Google Scholar]