Abstract
The ribosomal S6 kinase 2 (RSK2) is a member of the p90 ribosomal S6 kinase (p90RSK) family of proteins and plays a critical role in proliferation, cell cycle, and cell transformation. Here, we report that RSK2 phosphorylates caspase-8, and Thr-263 was identified as a novel caspase-8 phosphorylation site. In addition, we showed that EGF induces caspase-8 ubiquitination and degradation through the proteasome pathway, and phosphorylation of Thr-263 is associated with caspase-8 stability. Finally, RSK2 blocks Fas-induced apoptosis through its phosphorylation of caspase-8. These data provide a direct link between RSK2 and caspase-8 and identify a novel molecular mechanism for caspase-8 modulation by RSK2.
Keywords: Caspase, Protein Phosphorylation, Protein Stability, Rsk, Signal Transduction
Introduction
Ribosomal S6 kinase 2 (RSK2)2 is a member of the p90 ribosomal S6 kinase (p90RSK) family of proteins, which are serine/threonine kinases activated downstream of the Ras mitogen-activated protein kinase (MAPK) pathway (1). Several downstream substrates of RSK2 have been identified, including death-associated protein kinase (DAPK) (2), eukaryotic translation initiation factor 4B (eIF4B) (3), myelin transcription factor 1 (Myt1) (4), cAMP-responsive element-binding protein (CREB) (5, 6), histone H3 (7, 8), nuclear factor of activated T-cells 3 (NFAT3) (9), activating transcription factor 4 (ATF4) (10), and p53 (11). Based on known RSK2 substrates, the function of this kinase in proliferation, cell cycle, and transformation is fairly well understood. In contrast, accumulating evidence suggests that RSK2 is also involved in cell survival and apoptosis (5, 12–14), but the mechanism is not fully understand.
The caspases, a family of cysteine aspartyl proteases, play an essential role in the process of apoptosis. Caspases are divided into two types referred to as the large prodomain and the small prodomain families (15). The large prodomain family comprises the death effector domain found in procaspase-8 and -10 and the caspase recruitment domain in procaspase-2 and -9. Death effector domain and caspase recruitment domain, which are also known as death domain family members, are involved in procaspase activation and downstream caspase cascade regulation mediated through protein/protein interactions and are referred to as initiators (16). The small prodomain family is found in caspase-3, -6, and -7, which are downstream effectors in the caspase cascade (15, 17).
Here, we found that RSK2 directly interacts with and phosphorylates caspase-8 at a novel phosphorylation site, Thr-263. We also show that RSK2 mediates EGF, which induces caspase-8 ubiquitination and degradation, processes that are affected by Thr-263 phosphorylation. These data provide new insight into the role of RSK2 in apoptosis.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Chemical reagents, including Tris, NaCl, and SDS for molecular biology and buffer preparation, were purchased from Sigma-Aldrich. Restriction enzymes and some modifying enzymes were from New England Biolabs (Beverly, MA). The Taq DNA polymerase was obtained from Qiagen, Inc. (Valencia, CA). The DNA ligation kit (v2.0) was purchased from TAKARA Bio, Inc. (Otsu, Shiga, Japan). The pET-46 Ek/LIC His fusion bacterial expression vector and Ek/LIC cloning kit were from Novagen (Darmstadt, Germany). The cDNA4/HisMaxA plasmid used for the construction of the expression vector was from Invitrogen. Cell culture medium and other supplements were purchased from Invitrogen, and antibodies for Western blot analysis were obtained from Cell Signaling Technology, Inc. (Beverly, MA), Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), or Upstate Biotechnology, Inc. (Charlottesville, VA). Active RSK2 was purchased from Upstate. The caspase-8 peptides were from Genscript Corp. (Piscataway, NJ).
Construction of Expression Vectors
The expression constructs, including RSK2 and truncated RSK2 (9), the pMT123, 8x-HA-ubiquitin (HA-Ub; kindly provided by Dr. Stephen R. Hann, Department of Cell Biology, Vanderbilt University School of Medicine), the HA-caspase-8 (b) deletion mutants (18), and the RSK2 (Y707A) mutant (19), were amplified and used for expression in human embryonic kidney (HEK293) and HeLa cells. The open reading frame of the Casp-8 (b) (a kind gift from Dr. L. Zheng, Laboratory of Immunology, NIAID, NIH, Bethesda, MD) was amplified by polymerase chain reaction (PCR) using the following primers: sense, 5′-CGGGATCCCGATGGACTTCAGCAGAAATCTTTATGAT-3′; and antisense-5′-GCTCTAGAGCCATCAGAAGGGAAGACAAGTTTTTTTCT-3′. Primers were introduced into the BamHI/XbaI sites of the pcDNA3.1-v5-neo mammalian expression vector (Invitrogen) and the pET-46 Ek/LIC His fusion bacterial expression vector (Novagen). The mutants, including caspase-8-S119A, caspase-8-S256A, caspase-8-T263A, and caspase-8-S119A/S256A/T263A, were constructed using a site-directed mutagenesis kit (Stratagene, La Jolla, CA), and the small interfering RNA (siRNA) RSK2 was provided by Dr. Y. Y. Cho (20).
Cell Culture and Transfection
HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM; Hyclone, San Diego, CA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), 100 units/ml penicillin, and 100 mg/ml streptomycin and then cultured at 37 °C in a humidified incubator with 5% CO2. HeLa (human cervix adenocarcinoma) cells were cultured in Eagle's minimum essential medium (MEM) supplemented with 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin and then cultured at 37 °C in a humidified incubator with 5% CO2. Jurkat A3 and Jurkat I9.2 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin and then cultured at 37 °C in a humidified incubator with 5% CO2. The cells were maintained by splitting at 90% confluence, and media were changed every 3 days. When cells reached 50–60% confluence, transfection of the expression vectors, including RSK2 full-length and truncated constructs, caspase-8 and mutants, and pMT123 (HA-Ub), was performed using JetPEI (Polyplus-transfection Inc., New York, NY) following the manufacturer's suggested protocol. The cells were cultured for 36–48 h, and then proteins were extracted for further analysis.
In Vitro Kinase Assay
Wild type His-caspase-8 or mutant caspase-8 proteins or caspase-8 peptides were used as substrates for an in vitro kinase assay with active RSK2 (Upstate Biotechnology, Inc.). Reactions were carried out at 30 °C for 30 min in a mixture containing 50 μm unlabeled ATP and 10 μCi of [γ-32P]ATP and then were stopped by adding 6× SDS sample buffer. Samples were boiled, then separated by 10% SDS-PAGE or 20% SDS-PAGE (peptide), and visualized by autoradiography, Western blotting, or Coomassie Blue staining.
Immunoblotting and Immunoprecipitation
For immunoblotting, 30 μg of each protein sample from cells disrupted with Nonidet P-40 cell lysis buffer (50 mm Tris-Cl, pH 8.0, 150 mm NaCl, 0.5% Nonidet P-40, and protease inhibitor mixture) were used for immunoblotting with the appropriate specific primary antibodies and an alkaline phosphatase-conjugated secondary antibody and visualized using an enhanced chemifluorescence kit (Amersham Biosciences). For immunoprecipitation, protein samples were incubated with the appropriate antibody and agarose A/G beads (50% slurry) by rocking at 4 °C overnight. The beads were washed, mixed with 6× SDS sample buffer, and boiled, and then proteins were resolved by 10% SDS-PAGE. The proteins were visualized with the appropriate specific primary antibodies, an alkaline phosphatase-conjugated secondary antibody, and an enhanced chemifluorescence kit. For immunoprecipitation under denaturing conditions, proteins were extracted using regular immunoprecipitation buffer plus 1% SDS and heated at 95 °C for 5 min. The samples were diluted 1:10 in regular immunoprecipitation buffer before immunoprecipitation. The beads were washed, mixed with 6× SDS sample buffer, boiled, and then resolved by 10% SDS-PAGE. The proteins were visualized by immunoblotting.
Cycloheximide Chase and Protein in Vivo Ubiquitination Assays
To examine caspase-8 protein stability, wild type or mutants of caspase-8 mammalian expression vectors were transfected into HeLa cells in the presence of EGF. The cells were treated with cycloheximide (CHX; 30 μg/ml; Sigma) by adding it to the media. Cells were harvested at the indicated time points for immunoblotting. To explore ubiquitination of caspase-8, we introduced various combinations of expression vectors as described above and then pretreated cells with MG132 (20 μm) for 4 h followed by treatment with EGF and harvesting at the indicated time points. The proteins were extracted using Nonidet P-40 cell lysis buffer, and immunoprecipitation was carried out with a caspase-8 antibody. Immunoblotting was performed using the appropriate antibody and alkaline phosphatase-conjugated secondary antibodies.
In Vitro Caspase-8 and Caspase-3 Activity Assays
To determine caspase-8 and caspase-3 activities, cells transfected with si-mock or si-RSK2 were treated with anti-Fas mAb (500 ng/ml) and cycloheximide (10 μg/ml) and harvested at various time points. The proteins were extracted, and 50 μg of protein were used to determine caspase-8 and -3 activities using the caspase-8 and caspase-3 colorimetric assay kit (Millipore, Billerica, MA) following the manufacturer's suggested protocol. The absorbance values (405 nm) were converted to mg of protein.
Annexin V Staining
The apoptosis assay was carried out using the annexin V-FITC apoptosis detection kit following the manufacturer's suggested protocols (MBL International Corp., Watertown, MA). Apoptosis was compared between si-mock and si-RSK2 stably transfected cells treated with anti-Fas mAb (500 ng/ml) and cycloheximide (10 μg/ml) and then harvested at various time points. The cells were washed with phosphate-buffered saline and incubated for 5 min at room temperature with annexin V-FITC plus propidium iodide. Cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences). Apoptosis was compared in cells transfected with GFP-caspase-8-WT or GFP-caspase-8-T263A after treatment with anti-Fas mAb (200 ng/ml) in Jurkat A3 or Jurkat I9.2 for 6 h.
RESULTS
RSK2 Interacts Directly with Caspase-8
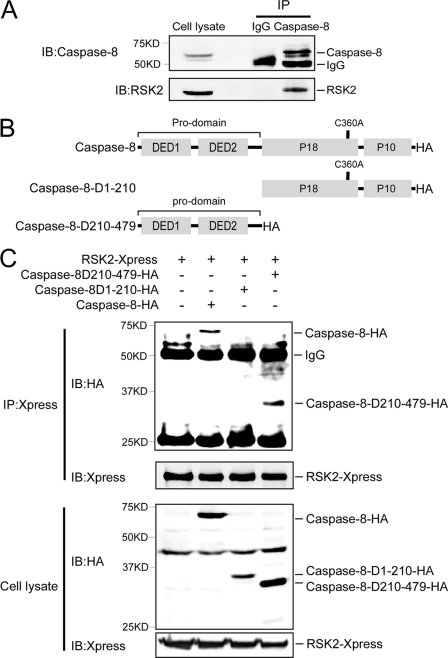
We found that caspase-8 is a novel interacting protein partner of RSK2. Endogenous RSK2 was detected in a HeLa cell extract immunoprecipitated with a caspase-8 antibody but not in the extract immunoprecipitated with the IgG control antibody (Fig. 1A). To identify the caspase-8 domain that is involved in the RSK2/caspase-8 interaction, full-length RSK2 combined with full-length caspase-8 or caspase-8 deletion mutants (Fig. 1B) was transfected into HEK293 cells. Because overexpression of caspase-8 is sufficient to induce cell death, all transfections were performed with caspase-8-C360A, which is a catalytically inactive point mutant. The results showed that full-length caspase-8 and caspase-8-D210–479, but not caspase-8-D1–210, co-precipitated with RSK2 (Fig. 1C, top). Expression of RSK2, caspase-8, and caspase-8 deletion mutants was detected in cell lysates (Fig. 1C, bottom). This result indicated that the death effector domain 1/2 is responsible for caspase-8 binding to RSK2.
FIGURE 1.
RSK2 binds to caspase-8 ex vivo. A, RSK2 interacts with endogenous caspase-8. A HeLa cell extract was used for immunoprecipitation (IP) with a caspase-8 antibody or control IgG overnight at 4 °C. Immunoprecipitated RSK2 was visualized by Western blot with an RSK2 antibody. B, structure and schematic diagrams of caspase-8 deletion mutant constructs. C, identification of the caspase-8 protein domain that interacts with RSK2. Various individual caspase-8-C360A-HA deletion mutants and RSK2 were co-transfected into HEK293 cells and cultured for 36 h. Extracted proteins (400 μg) were immunoprecipitated with anti-Xpress overnight at 4 °C. The immunoprecipitated caspase-8 was visualized with anti-HA by Western blotting, and the protein abundance of caspase-8 was also analyzed in cell lysates (20 μg) by Western blotting using anti-HA. IB, immunoblot.
RSK2 Phosphorylates Caspase-8 at Thr-263 in Vitro and ex Vivo
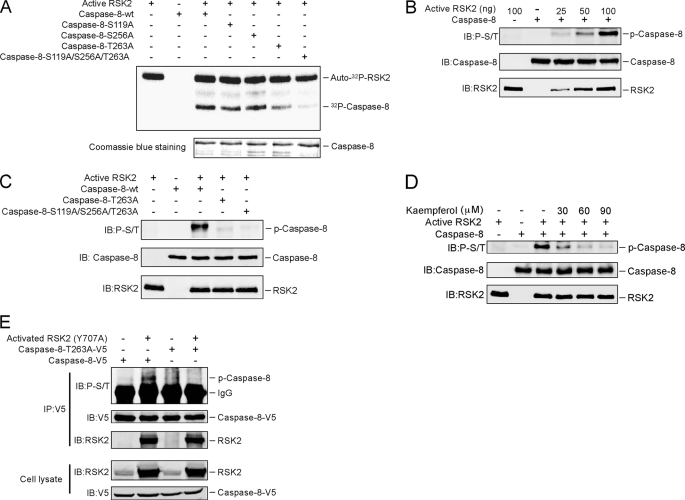
Based on the results above showing that RSK2 interacts with caspase-8, we hypothesized that caspase-8 could be a novel substrate for RSK2. To determine whether RSK2 can phosphorylate caspase-8, in vitro kinase reactions with active RSK2 and His-caspase-8 (as substrate) were performed using [γ-32P]ATP. Results indicated that RSK2 could phosphorylate caspase-8 (supplemental Fig. 1A). To determine the site(s) of caspase-8 that is potentially phosphorylated by RSK2, we first identified possible Ser/Thr sites using NetPhos 2.0 and then designed 15 peptides (supplemental Fig. 1B) that were based on the site(s) predicted by NetPhos 2.0. In vitro kinase assays were performed using the different caspase-8 peptides as substrates for RSK2. The results indicated that Ser-119, Ser-256, and Thr-263 on caspase-8 were potentially phosphorylated by RSK2 (supplemental Fig. 1C). We replaced each of the three potential phosphorylation sites of caspase-8 with alanine and also constructed a triple caspase-8 mutant encompassing all three sites. We then performed in vitro kinase reactions using active RSK2 and caspase-8-WT or individual caspase-8 mutants (caspase-8-S119A, caspase-8-S256A, caspase-8-T263A, and caspase-8-S119A/S256A/T263A) as substrates. The results indicated that the phosphorylation of the triple mutant of caspase-8 was almost gone (Fig. 2A). In addition, compared with the wild type caspase-8, phosphorylation was dramatically decreased for the caspase-8-T263A mutant and decreased slightly in the other two mutants (Fig. 2A). These results indicated that Thr-263 of caspase-8 is a major site for RSK2 phosphorylation.
FIGURE 2.
RSK2 phosphorylates caspase-8 (Thr-263). A, Thr-263 is a major phosphorylation site on caspase-8 for RSK2 in vitro. The indicated caspase-8 mutant proteins were purified and subjected to an in vitro kinase assay with 100 ng of active RSK2, 10 μCi of [γ-32P]ATP, and 50 μm unlabeled ATP. The 32P-labeled bands were visualized by autoradiography. B, the phospho-Ser/Thr (p-S/T) antibody recognizes the phosphorylation of caspase-8 in vitro. An in vitro kinase assay was performed with the caspase-8 protein together with increasing amounts of active RSK2. Phosphorylation of caspase-8 was detected by Western blot with a phospho-Ser/Thr antibody. C, confirmation of caspase-8 phosphorylation sites by Western blot. An in vitro kinase assay was performed with caspase-8-WT or caspase-8 mutants together with 100 ng of active RSK2 and 200 μm unlabeled ATP. The phosphorylated caspase-8 (P-Caspase-8) was visualized by Western blot with a phospho-Ser/Thr antibody. D, kaempferol inhibits RSK2-mediated caspase-8 phosphorylation in vitro. An in vitro kinase assay was conducted with 100 ng of active RSK2, 5 μg of caspase-8, 200 μm unlabeled ATP, and the indicated dose of kaempferol. Phosphorylated caspase-8 was visualized by Western blot with a phospho-Ser/Thr antibody. E, constitutively active RSK2 phosphorylates caspase-8 ex vivo. HEK293 cells were transfected with caspase-8-WT or the caspase-8-T263A mutant with or without constitutively active RSK2 (Y707A). Cells were cultured for 30 h, and then proteins were extracted. The caspase-8 proteins were immunoprecipitated (IP) with anti-V5, and phosphorylated caspase-8 was visualized by Western blot with a phospho-Ser/Thr antibody. IB, immunoblot.
The caspase-8 Thr-263 site is within a classical motif (RXRXX(S/T)) for substrates of RSK2, so we determined whether the phospho-Ser/Thr antibody, which recognizes the RXRXX(S/T) motif, could detect the phosphorylation of caspase-8 by RSK2. Results of an in vitro kinase assay indicated that the phosphorylation of caspase-8 increased in a dose-dependent manner with increasing amounts of active RSK2 (Fig. 2B). Next, we used the phospho-Ser/Thr antibody to detect RSK2 phosphorylation of caspase-8 mutants. Results indicated no phosphorylation in the caspase-8-T263A mutant or the triple mutant (Fig. 2C), but compared with caspase-8-WT, phosphorylation was not substantially decreased in mutant S119A or S256A (supplemental Fig. 1D).
Kaempferol has been shown to specifically inhibit RSK2 activity (20), and therefore, kaempferol should suppress the RSK2 phosphorylation of caspase-8. Indeed, the results showed that kaempferol attenuated RSK2 phosphorylation of caspase-8 in a dose-dependent manner (Fig. 2D). Collectively, these data indicated that RSK2 can phosphorylate caspase-8 at Thr-263.
To show that caspase-8 is phosphorylated by RSK2 at Thr-263 ex vivo, we co-transfected caspase-8 or the caspase-8-T263A mutant together with the constitutively active RSK2 construct (Y707A) (19). Then caspase-8 or its mutant was immunoprecipitated with anti-V5, and phosphorylation of caspase-8 was detected with the phospho-Ser/Thr (RXRXX(S/T)) antibody. We found that caspase-8 was efficiently phosphorylated by active RSK2, whereas the T263A mutant caspase-8 did not undergo phosphorylation despite its efficient expression (Fig. 2E). In addition, RSK2 was detected with either caspase-8 or mutant caspase-8-T263A, indicating that the T263A mutant had no effect on caspase-8 binding to RSK2 (Fig. 2E).
RSK2 Is Involved in EGF Induction of Caspase-8 Ubiquitination and Degradation
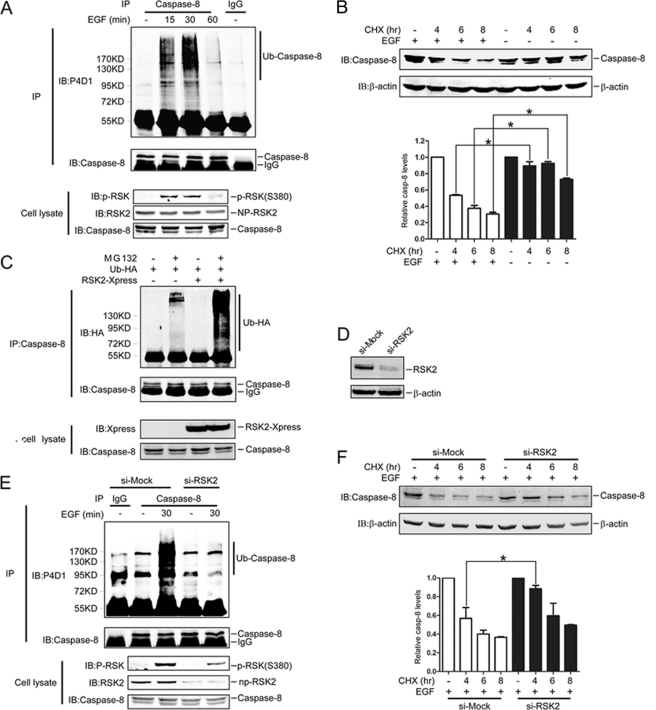
We found that EGF induces caspase-8 ubiquitination. HeLa cells were treated with MG132 prior to EGF (100 ng/ml) treatment. At 15 and 30 min after EGF treatment, caspase-8 ubiquitination was observed, and then ubiquitination was decreased at 60 min (Fig. 3A). Polyubiquitination is a death signal that targets proteins for degradation by the proteasome. Therefore, we tested whether EGF treatment would have an effect on caspase-8 stability, and as expected, the results showed that EGF treatment dramatically induced caspase-8 degradation (Fig. 3B).
FIGURE 3.
RSK2 is involved in EGF induction of caspase-8 ubiquitination and degradation. A, EGF induces endogenous caspase-8 ubiquitination. HeLa cells pretreated with MG132 (20 μm) for 4 h were immunoprecipitated (IP) under denaturing conditions with anti-caspase-8 after treatment with EGF (100 ng/ml) for different periods of time. Ubiquitination of caspase-8 was detected by Western blot with a P4D1antibody. B, EGF induces caspase-8 degradation. HeLa cells were treated with EGF (100 ng/ml) and CHX (30 μg/ml) for the indicated time, and proteins were extracted. The caspase-8 protein abundance was visualized by Western blot with a casapse-8 antibody. The graph shows data from triplicate experiments (mean ± S.D.). The asterisk (*) indicates a significant difference (p < 0.05, Student's t test). C, RSK2 enhances caspase-8 ubiquitination. HeLa cells were co-transfected with RSK2 and HA-Ub and cultured for 30 h. The cells were treated with MG132 (20 μm) for 4 h, and proteins were extracted. The caspase-8 proteins were immunoprecipitated with a caspase-8 antibody, and ubiquitinated caspase-8 proteins were visualized with anti-HA by Western blot. D, confirmation of RSK2 knockdown. HeLa cells were transfected with si-mock or si-RSK2 and selected in the presence of G418 (800 μg/ml) for 14 days, and then colonies were pooled. The proteins were extracted, and the RSK2 protein level was visualized by Western blot with an RSK2 antibody. Equal protein loading was confirmed by reprobing with a β-actin antibody. E, RSK2 attenuates EGF induction of caspase-8 ubiquitination. Knockdown RSK2 cells that were pretreated with MG132 (20 μm) for 4 h were immunoprecipitated with anti-caspase-8 under denaturing conditions after treatment with EGF (100 ng/ml) for 30 min. Endogenous ubiquitination of caspase-8 was detected with a P4D1antibody. F, RSK2 mediates EGF induction of caspase-8 degradation. Knockdown RSK2 cells were treated with EGF (100 ng/ml) plus CHX (30 μg/ml) for the indicated time, and proteins were extracted. The caspase-8 protein abundance was visualized by Western blot with a casapse-8 antibody. The graph shows data from triplicate experiments (mean ± S.D.). The asterisk (*) indicates a significant difference (p < 0.05, Student's t test). P-RSK, phosphorylated RSK; NP-RSK2, non-phosphorylated RSK2; IB, immunoblot; casp, caspase.
RSK2 is downstream of the Ras MAPK pathway and is activated by EGF. We found that overexpression of RSK2 can promote caspase-8 ubiquitination (Fig. 3C) and degradation (supplemental Fig. 2, B and C). Therefore, we generated knockdown RSK2 cells (Fig. 3D) and found that endogenous ubiquitination of caspase-8 was decreased in these cells after treatment with EGF (Fig. 3E). Furthermore, degradation of caspase-8 was lower in RSK2 knockdown cells (Fig. 3F). Overall, these results indicated that RSK2 contributes to EGF induction of ubiquitination and degradation of caspase-8.
Thr-263 Phosphorylation Is Associated with Caspase-8 Stability
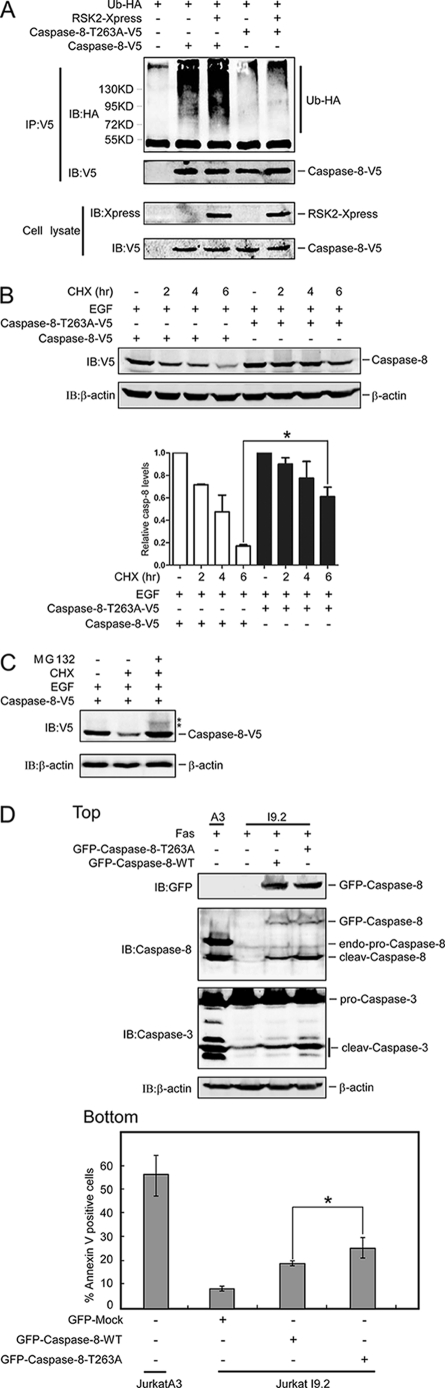
To explore the relationship between phosphorylation (Thr-263) and ubiquitination of caspase-8, caspase-8 or mutant caspase-8-T263A together with HA-Ub and RSK2 were transfected into cells. We found, compared with full-length caspase-8, that polyubiquitination of the T263A caspase-8 mutant was dramatically reduced in both the presence and absence of RSK2 transfection (Fig. 4A). Using our caspase-8 phosphorylation mutants, we also compared two other sites, Ser-119 and Ser-256, with Thr-263 for caspase-8 ubiquitination. The results showed that only Thr-263 is related to caspase-8 ubiquitination (supplemental Fig. 3). Next, the protein stability of the caspase-8-T263A mutant was compared with full-length caspase-8 following treatment of transfected HeLa cells with EGF plus CHX for the indicated time. The data showed that full-length caspase-8 was degraded more substantially compared with the caspase-8-T263A mutant, but this caspase-8 mutant could not totally block the degradation of caspase-8 (Fig. 4B). In addition, degradation of caspase-8 proteins in the presence of EGF plus CHX could be prevented by MG132 (Fig. 4C). We also detected ubiquitination of caspase-8 after MG132 treatment as indicated (Fig. 4C). Overall, these data indicated that the T263A caspase-8 mutant, which could not be phosphorylated by RSK2, is more stable.
FIGURE 4.
Phosphorylation of Thr-263 plays a critical role in stabilizing caspase-8. A, phosphorylation of caspase-8 at Thr-263 enhances ubiquitination of caspase-8. Caspase-8-WT and mutant caspase-8-T263A were each transfected with HA-Ub with or without RSK2 and cultured for 30 h. The cells were treated with MG132 for 4 h, and then proteins were extracted. The caspase-8 proteins were immunoprecipitated with anti-V5, and ubiquitinated caspase-8 proteins were visualized by Western blot with anti-HA. B, phosphorylation of Thr-263 plays an important role in caspase-8 stability. Caspase-8 or mutant caspase-8-T263A was transfected into HeLa cells. Cells were cultured for 30 h and then treated with EGF (100 ng/ml) plus CHX (30 μg/ml) for the indicated time, and then proteins were extracted. The caspase-8 protein abundance was visualized by Western blot with anti-V5 antibody. The graph shows data from triplicate experiments (mean ± S.D.). The asterisk (*) indicates a significant difference (p < 0.05, Student's t test). C, MG132 rescues caspase-8 expression. Caspase-8 was transfected into HeLa cells, and cells were cultured for 30 h. Cells were then pretreated with MG132 (20 μm) for 1 h followed by treatment with EGF (100 ng/ml) plus CHX (30 μg/ml) for 6 h. The proteins were extracted, and caspase-8 protein abundance was visualized by Western blot with anti-V5. D, Thr-263 is critical for Fas induction of apoptosis. GFP-caspase-8-WT or GFP-caspase-8-T263A was transfected into Jurkat I9.2 cells, and cells were cultured for 24 h. Cells were exposed to anti-Fas (200 ng/ml) for 6 h and then either subjected to immunoblotting to detect GFP, caspase-8, caspase-3, or β-actin (top), or apoptosis was analyzed by annexin V and propidium iodide staining using flow cytometry (bottom). Data are represented as means ± S.D. of the percentage of annexin V-positive cells determined from triplicate experiments. The asterisk (*) indicates a significant difference (p < 0.05, Student's t test; bottom). IB, immunoblot; cleav, cleaved; endo, endogenous.
FasL can induce substantial apoptosis in Jurkat A3 cells (supplemental Fig. 4). To determine the effect of the mutant caspase-8-T263A on apoptosis, we used the Jurkat I9.2 cell line, which is caspase-8 deficient and therefore is insensitive to Fas-induced apoptosis. However, reintroduction of caspase-8 into these cells should rescue sensitivity to Fas-induced apoptosis. GFP-caspase-8-WT or GFP-caspase-8-T263A was transfected into Jurkat I9.2 cells, and then cells were treated with Fas for 6 h. Jurkat A3 cells, which have normal caspase-8 expression and are sensitive to Fas-induced apoptosis, served as a positive control. As expected, the results indicated that Jurkat A3 cells exhibited dramatic apoptosis after treatment with Fas (Fig. 4D). For Jurkat I9.2 cells, reconstituting caspase-8-WT and caspase-8-T263A restored sensitivity to Fas-induced apoptosis to some extent (Fig. 4D). Compared with caspase-8-WT, Western blotting results showed that caspase-8-T263A exhibited more cleavage of caspase-8 or caspase-3 (Fig. 4D, top). In addition, more annexin V-positive cells were detected in cells transfected with caspase-8-T263A (Fig. 4D, bottom), suggesting that phosphorylation of Thr-263 is critical for caspase-8 function.
RSK2 Attenuates Fas-induced Apoptosis through Its Phosphorylation of Caspase-8
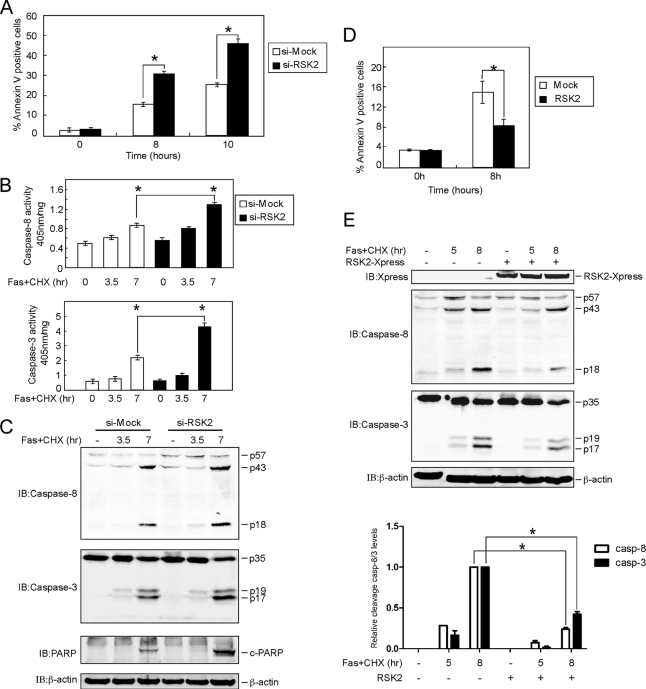
Fas-induced apoptosis is strictly dependent on caspase-8 activation (21). We found that knockdown of RSK2 sensitized HeLa cells to FasL-induced apoptosis (Fig. 5A). At the same time, an enzyme activity assay showed that caspase-8 and caspase-3 activities were enhanced in knockdown RSK2 cells (Fig. 5B). We also investigated the effect of knocking down RSK2 on FasL-induced cleavage of caspase-8, caspase-3, and poly(ADP-ribose) polymerase. The results indicated that cleavage of caspase-8, caspase-3, and poly(ADP-ribose) polymerase was increased in knockdown RSK2 cells treated with FasL (Fig. 5C). In contrast, overexpression of RSK2 blocked apoptosis induced by Fas treatment (Fig. 5D) and delayed the processing of pro-caspase-8 and procaspase-3 (Fig. 5E), which suggests that RSK2 protects cells from Fas-induced apoptosis.
FIGURE 5.
RSK2 blocks Fas-induced apoptosis mediated through caspase-8. A, knockdown of RSK2 induces apoptosis with FasL stimulation. Si-mock and si-RSK2 cells were treated with FasL (500 ng/ml) and CHX (10 μg/ml) and harvested at the indicated time points. Apoptosis was analyzed by annexin V and propidium iodide staining using flow cytometry. Data are represented as means ± S.D. of the percentage of annexin-positive cells as determined in triplicate experiments. The asterisk (*) indicates a significant difference (p < 0.05, Student's t test). B, RSK2 knockdown enhances caspase-8 and caspase-3 activities. HeLa cells stably expressing si-mock or si-RSK2 were treated with FasL (500 ng/ml) and CHX (10 μg/ml) and harvested at the indicated time points. The proteins were extracted, and 50 μg used to determine caspase-8 and -3 activities using the caspase-8 and caspase-3 colorimetric assay kit. Data are shown as means ± S.D. of percentage of activity (1 mg of protein) determined from triplicate experiments. The asterisk (*) indicates a significant difference (p < 0.05, Student's t test). C, RSK2 knockdown enhances FasL-induced cleavage of caspase-8, caspase-3, and poly(ADP-ribose) polymerase (PARP). HeLa cells stably expressing si-mock or si-RSK2 were treated with FasL (500 ng/ml) and CHX (10 μg/ml), and cells were harvested at the indicated time points. The proteins were extracted, and cleaved caspase-8, caspase-3, and poly(ADP-ribose) polymerase (c-PARP) were visualized by Western blot with specific antibodies. β-Actin was used to verify equal protein loading as an internal control. D, overexpression of RSK2 blocks apoptosis induced by FasL treatment. RSK2 was transfected into HeLa cells, and at 30 h after transfection, cells were treated with FasL (500 ng/ml) plus CHX (10 μg/ml) and harvested at the indicated time points. Apoptosis was analyzed by annexin V and propidium iodide staining using flow cytometry. Data are represented as means ± S.D. of the percentage of annexin-positive cells as determined in triplicate experiments. The asterisk (*) indicates a significant difference (p < 0.05, Student's t test). E, overexpression of RSK2 suppresses cleavage of caspase-8 and caspase-3 induced by FasL stimulation. RSK2 was transfected into HeLa cells, and cells were cultured for 30 h. The cells were treated with FasL (500 ng/ml) and CHX (10 μg/ml) for the indicated time, and proteins were then extracted. Cleaved caspase-8 and caspase-3 were visualized by Western blot with specific antibodies. β-Actin was used to verify equal protein loading as an internal control. The graph shows data from triplicate experiments (means ± SD). The asterisk (*) indicates a significant difference (p < 0.05, Student's t test). casp, caspase; IB, immunoblot.
DISCUSSION
The MAPK signaling pathways comprise families of proteins that include the extracellular signal-regulated kinases (ERKs), c-Jun N-terminal protein kinases (JNKs), and p38, which control cell proliferation, differentiation, and death. The MAPKs transmit and amplify signals that are initiated by a variety of extracellular stimuli ending in a specific cellular response. RSK2 is a member of the p90 ribosomal S6 kinase (RSK) family of proteins, which are serine/threonine kinases that are activated downstream of the ERK MAPK pathway.
We showed that RSK2 interacts with caspase-8 and that caspase-8 is a novel substrate of RSK2 with Thr-263 as a major phosphorylation site for RSK2. Unexpectedly, we observed that EGF induces caspase-8 ubiquitination and degradation (Fig. 3, A and B) that was attenuated by knockdown of RSK2 through the (Fig. 3, E and F). Moreover, a caspase-8 Thr-263 mutant was more stable and not easily polyubiquitinated (Fig. 4, A and B), suggesting that RSK2 regulates caspase-8 degradation and ubiquitination through its phosphorylation of caspase-8. However, because the caspase-8 Thr-263 mutant failed to totally block the degradation of caspase-8 (Fig. 4B), degradation of caspase-8 is very likely regulated by both phosphorylation-dependent and -independent mechanisms. In addition, compared with caspase-8-WT, mutant caspase-8-T263A induced more apoptosis in Jurkat I9.2 cells (Fig. 4D).
Evidence indicates that ubiquitination can regulate caspase protein stability and activity. Caspase-3 and -7 were reported to be substrates of xIAP2 in vitro (22), and the ubiquitin protein ligase activity of xIAP promotes caspase-3 ubiquitination and degradation in vitro and in vivo, which impairs FasL-induced apoptosis (23). The Skp1-cullin-F-box protein (SCF), another E3 ubiquitin ligase, was demonstrated to mediate caspase-3 ubiquitination and degradation (24). Recently, CUL3, a member of the cullin RING ligases, was identified as an E3 ligase for caspase-8. However, this ubiquitination of caspase-8 was not related to degradation but instead was related to its activity even though Lys-63- and Lys-48-linked ubiquitins were observed in caspase-8 (25). Lys-48-linked ubiquitin chains typically induce target proteins for proteasomal degradation, and Lys-63-linked ubiquitin chains play a role in DNA repair, act as protein adaptors, and influence protein kinase activity (26). Caspase-8 also is involved in the proteasome degradation system. Caspase-8- and caspase-10-associated RING proteins (CARPs), which have E3 ligase activity, were reported to promote caspase-8 degradation. However, evidence for ubiquitination of caspase-8 was not presented (27). Here, we showed that EGF induces caspase-8 ubiquitination and degradation, but whether CARPs acted as E3 ligases for caspase-8 in the presence of EGF treatment is not clear.
We found that knockdown of RSK2 accelerated procaspase-8 and procaspase-3 processing and the accumulation of their cleavage products. At the same time, the enzyme activity assay showed that caspase-8 and caspase-3 activities were enhanced by suppressing RSK2 activity. In contrast, overexpression of RSK2 blocked apoptosis induced by Fas treatment and delayed the procaspase-8 and procaspase-3 processes, suggesting that RSK2 protects cells from Fas-induced apoptosis. Taken together, these results suggest a direct link between RSK2 and caspase-8 and indicate that RSK2 is a new molecular target for sensitizing human carcinomas to drug-induced apoptosis.
Supplementary Material
This work was funded by The Hormel Foundation and by National Institutes of Health Grants CA077646, CA111536, CA120388, CA027502, and ES016548.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- RSK2
- ribosomal S6 kinase 2
- Ub
- ubiquitin
- CHX
- cycloheximide
- FasL
- Fas ligand
- xIAP
- X-linked inhibitor of apoptosis protein
- CARP
- caspase-8- and caspase-10-associated RING protein.
REFERENCES
- 1. Frödin M., Jensen C. J., Merienne K., Gammeltoft S. (2000) EMBO J. 19, 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anjum R., Roux P. P., Ballif B. A., Gygi S. P., Blenis J. (2005) Curr. Biol. 15, 1762–1767 [DOI] [PubMed] [Google Scholar]
- 3. Shahbazian D., Roux P. P., Mieulet V., Cohen M. S., Raught B., Taunton J., Hershey J. W., Blenis J., Pende M., Sonenberg N. (2006) EMBO J. 25, 2781–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palmer A., Gavin A. C., Nebreda A. R. (1998) EMBO J. 17, 5037–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonni A., Brunet A., West A. E., Datta S. R., Takasu M. A., Greenberg M. E. (1999) Science 286, 1358–1362 [DOI] [PubMed] [Google Scholar]
- 6. Xing J., Ginty D. D., Greenberg M. E. (1996) Science 273, 959–963 [DOI] [PubMed] [Google Scholar]
- 7. Sassone-Corsi P., Mizzen C. A., Cheung P., Crosio C., Monaco L., Jacquot S., Hanauer A., Allis C. D. (1999) Science 285, 886–891 [DOI] [PubMed] [Google Scholar]
- 8. Mahadevan L. C., Clayton A. L., Hazzalin C. A., Thomson S. (2004) Novartis Found. Symp. 259, 102–111; discussion 111–114, 163–169 [PubMed] [Google Scholar]
- 9. Cho Y. Y., Yao K., Bode A. M., Bergen H. R., 3rd, Madden B. J., Oh S. M., Ermakova S., Kang B. S., Choi H. S., Shim J. H., Dong Z. (2007) J. Biol. Chem. 282, 8380–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang X., Matsuda K., Bialek P., Jacquot S., Masuoka H. C., Schinke T., Li L., Brancorsini S., Sassone-Corsi P., Townes T. M., Hanauer A., Karsenty G. (2004) Cell 117, 387–398 [DOI] [PubMed] [Google Scholar]
- 11. Cho Y. Y., He Z., Zhang Y., Choi H. S., Zhu F., Choi B. Y., Kang B. S., Ma W. Y., Bode A. M., Dong Z. (2005) Cancer Res. 65, 3596–3603 [DOI] [PubMed] [Google Scholar]
- 12. Kang S., Dong S., Gu T. L., Guo A., Cohen M. S., Lonial S., Khoury H. J., Fabbro D., Gilliland D. G., Bergsagel P. L., Taunton J., Polakiewicz R. D., Chen J. (2007) Cancer Cell 12, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eisinger-Mathason T. S., Andrade J., Groehler A. L., Clark D. E., Muratore-Schroeder T. L., Pasic L., Smith J. A., Shabanowitz J., Hunt D. F., Macara I. G., Lannigan D. A. (2008) Mol. Cell 31, 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng C., Cho Y. Y., Zhu F., Xu Y. M., Wen W., Ma W. Y., Bode A. M., Dong Z. (2010) FASEB J. 24, 3490–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boatright K. M., Salvesen G. S. (2003) Curr. Opin. Cell Biol. 15, 725–731 [DOI] [PubMed] [Google Scholar]
- 16. Shi Y. (2004) Cell 117, 855–858 [DOI] [PubMed] [Google Scholar]
- 17. Slee E. A., Adrain C., Martin S. J. (2001) J. Biol. Chem. 276, 7320–7326 [DOI] [PubMed] [Google Scholar]
- 18. Finlay D., Vuori K. (2007) Cancer Res. 67, 11704–11711 [DOI] [PubMed] [Google Scholar]
- 19. Vaidyanathan H., Ramos J. W. (2003) J. Biol. Chem. 278, 32367–32372 [DOI] [PubMed] [Google Scholar]
- 20. Cho Y. Y., Yao K., Kim H. G., Kang B. S., Zheng D., Bode A. M., Dong Z. (2007) Cancer Res. 67, 8104–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Juo P., Kuo C. J., Yuan J., Blenis J. (1998) Curr. Biol. 8, 1001–1008 [DOI] [PubMed] [Google Scholar]
- 22. Huang H., Joazeiro C. A., Bonfoco E., Kamada S., Leverson J. D., Hunter T. (2000) J. Biol. Chem. 275, 26661–26664 [DOI] [PubMed] [Google Scholar]
- 23. Suzuki Y., Nakabayashi Y., Takahashi R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan M., Gallegos J. R., Gu Q., Huang Y., Li J., Jin Y., Lu H., Sun Y. (2006) Neoplasia 8, 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 26. Yang W. L., Wang J., Chan C. H., Lee S. W., Campos A. D., Lamothe B., Hur L., Grabiner B. C., Lin X., Darnay B. G., Lin H. K. (2009) Science 325, 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McDonald E. R., 3rd, El-Deiry W. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6170–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.