Abstract
Reactive carbonyls, especially α,β-unsaturated carbonyls produced through lipid peroxidation, damage biomolecules such as proteins and nucleotides; elimination of these carbonyls is therefore essential for maintaining cellular homeostasis. In this study, we focused on an NADPH-dependent detoxification of reactive carbonyls in plants and explored the enzyme system involved in this detoxification process. Using acrolein (CH2 = CHCHO) as a model α,β-unsaturated carbonyl, we purified a predominant NADPH-dependent acrolein-reducing enzyme from cucumber leaves, and we identified the enzyme as an alkenal/one oxidoreductase (AOR) catalyzing reduction of an α,β-unsaturated bond. Cloning of cDNA encoding AORs revealed that cucumber contains two distinct AORs, chloroplastic AOR and cytosolic AOR. Homologs of cucumber AORs were found among various plant species, including Arabidopsis, and we confirmed that a homolog of Arabidopsis (At1g23740) also had AOR activity. Phylogenetic analysis showed that these AORs belong to a novel class of AORs. They preferentially reduced α,β-unsaturated ketones rather than α,β-unsaturated aldehydes. Furthermore, we selected candidates of other classes of enzymes involved in NADPH-dependent reduction of carbonyls based on the bioinformatic information, and we found that an aldo-keto reductase (At2g37770) and aldehyde reductases (At1g54870 and At3g04000) were implicated in the reduction of an aldehyde group of saturated aldehydes and methylglyoxal as well as α,β-unsaturated aldehydes in chloroplasts. These results suggest that different classes of NADPH-dependent reductases cooperatively contribute to the detoxification of reactive carbonyls.
Keywords: Arabidopsis, Chloroplast, Enzyme Purification, Oxidative Stress, Reactive Oxygen Species, Reductase, Cucumber, Lipid Peroxidation, Reactive Carbonyls
Introduction
When plants are subjected to abiotic and/or biotic stresses, oxidative stress often results; this leads to the production of reactive oxygen species, which damage biomolecules such as proteins and lipids. More than half of the fatty acids found in the membranes of chloroplasts and mitochondria, two of the most highly oxidative organelles, are linoleic and linolenic acid. Because linoleic and linolenic acid are sources of many short chain carbonyls through their peroxidation (1), biomolecules in both types of organelles are challenged by the toxicity of reactive compounds, including α,β-unsaturated carbonyls, which are involved in the pathophysiological effects associated with oxidative stress in cells and tissues (2). In fact, α,β-unsaturated carbonyls from peroxidized polyunsaturated fatty acids cause loss of functions in mitochondria (3); chloroplasts have recently been shown to be a major production center of reactive aldehydes (4), and photosynthetic functions are highly sensitive to α,β-unsaturated carbonyls (5–7).
The high reactivity of α,β-unsaturated carbonyls is due to the ability of their α,β-unsaturated bonds to form Michael adducts with thiol and amino groups in biomolecules; in the case of α,β-unsaturated aldehydes, the aldehyde group also contributes to reactivity through the formation of Schiff base adducts with amino groups. Thus the targets in scavenging α,β-unsaturated aldehydes are saturation of the α,β-unsaturated bond and reduction/oxidation of the aldehyde group. In mammalian cells, several pathways are known to scavenge α,β-unsaturated aldehydes as follows: the glutathione (GSH) S-transferase (GST) pathway, which is fueled by the GSH or thioredoxin redox cycle, for example, conjugates aldehydes with GSH; NAD(P)H-dependent alkenal/one oxidoreductase (AOR)3 catalyzes the hydrogenation of α,β-unsaturated bonds (8); aldo-keto reductase (AKR) catalyzes the reduction of aldehyde to alcohol (9, 10); and aldehyde dehydrogenase catalyzes the oxidation of aldehyde to carboxylate (11). In plants, as described above, the scavenging of α,β-unsaturated carbonyl compounds is essential for maintaining their viability; nevertheless, knowledge of α,β-unsaturated carbonyl compound-scavenging enzymes, and especially of the NAD(P)H-dependent pathway, is very limited.
In this study, we used acrolein as a model α,β-unsaturated aldehyde. Acrolein (prop-2-enal, propenal) is the simplest compound having both an α,β-unsaturated bond and an aldehyde group, and it has the highest cytotoxicity among the α,β-unsaturated carbonyls (2, 7). It is potentially toxic to the process of photosynthesis because it strongly inhibits both thylakoid and stromal reactions (5, 6); moreover, acrolein levels in tobacco leaves increased under photoinhibitory illumination (12). Thus, the elimination of acrolein generated through lipid peroxidation is necessary to maintain photosynthetic activity under stressed conditions. As the first objective of this study, we purified a major scavenger of acrolein from cucumber leaves via an NAD(P)H-dependent pathway; this scavenger was subsequently identified as a novel NADPH-dependent AOR that catalyzes the hydrogenation of α,β-unsaturated bonds to form saturated aldehydes and ketones. This enzyme is found in various plant species, including Arabidopsis, and we confirmed that recombinant Arabidopsis AOR exhibited an activity similar to that of cucumber AOR.
Resultant aldehydes catalyzed by AOR are also cytotoxic, although not as strongly cytotoxic as the α,β-unsaturated aldehydes (5), and the AORs could not reduce longer (C ≧ 5) α,β-unsaturated aldehydes. Accordingly, we searched for an enzyme-reducing aldehyde group based on the heterogeneously expressed recombinant proteins predicted to be involved in the aldehyde reduction. The NAD(P)H-dependent reduction of the aldehyde group is known to be mediated by AKR and short-chain dehydrogenase/reductase (SDR) family proteins, including alcohol dehydrogenase and carbonyl reductase (CBR). Originally, AKR and CBR were thought to belong to the same family, based on their similar enzymological properties, but CBR was later shown to be a member of the SDR family of proteins, based on conserved amino acid residues within the SDR family (13). In this study, to identify which species were involved in the detoxification, we selected candidates from Arabidopsis AKR and SDR families based on their subcellular localization and expression profiles as determined using data base information, and we analyzed the catalytic activities of expressed recombinant AKRs and SDRs. Our results show that one AKR and two SDRs catalyze the NADPH-dependent reduction of aldehyde groups in both saturated and unsaturated aldehyde compounds in chloroplasts; the SDRs were identified as aldehyde reductases (ADRs) based on their catalyzing reaction. Taking all the evidence together, we propose an NADPH-dependent system for the detoxification of reactive carbonyls in chloroplasts, in which AOR, ADRs, and AKR cooperatively scavenge reactive carbonyls.
EXPERIMENTAL PROCEDURES
Chemicals
Acrolein, crotonaldehyde, 3-buten-2-one, (E)-2-pentenal, allyl alcohol, and 2-methyl-2-butenal were purchased from Tokyo Kasei Kogyo (Tokyo, Japan). 1-Penten-3-one, 3-penten-2-one, 3-methyl-2-butenal, 2-cyclohexen-1-one, and methylglyoxal were purchased from Sigma. Other reagents purchased from Wako Pure Chemical (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan) were of analytical grade.
Preparation of Concentrated Crude Extracts and Enzyme Assay
Field-grown cucumber (Cucumis sativus) or Arabidopsis (Arabidopsis thaliana, Columbia) leaves grown in a growth chamber (12 h of light (80 μmol photon m−2 s−1) at 25 °C) were homogenized with 5 volumes of 50 mm HEPES-NaOH, pH 7.0. After centrifugation, proteins in the supernatant were precipitated by 80% ammonium sulfate saturation. The proteins were then dissolved and dialyzed against 50 mm HEPES-NaOH, pH 7.0, and the dialysate was used as a concentrated crude extract. To measure carbonyl-reducing enzyme activity, a reaction mixture containing 50 mm HEPES-NaOH, pH 7.0, 0.1 mm NADPH, and 20 mm substrate was prepared in a cuvette, and the reaction was started by adding enzyme solution to a total volume of 1 ml. Oxidation of NADPH was observed in the form of a decrease at 340 nm (6.22 mm−1 cm−1) in the initial linear phase as seen on a spectrophotometer (U-2900, Hitachi, Tokyo, Japan). Enzymatic activity in the presence of other reductant or oxidants (0.1 mm NADH, 1 mm NADP, or 1 mm NAD) was measured by monitoring changes at A340 nm in a reaction mixture containing 20 mm substrate and 50 mm HEPES-NaOH, pH 7.0. Some hydrophobic substrates were dissolved in dimethyl sulfoxide because the final concentration (up to 5% (v/v)) of the solvent did not inhibit the activity. Km and kcat values were determined by fitting the experimental data to the Michaelis-Menten equation.
Products resulting from the carbonyl-reducing enzyme-catalyzed reaction were determined by 2,4-dinitrophenylhydrazine (DNPH) derivatization and subsequent HPLC analysis. A reaction mixture (total 100 μl) containing 50 mm HEPES-NaOH, pH 7.0, 20 mm NADPH, 20 mm substrate, and enzyme was incubated at 25 °C. After the reaction was stopped by the addition of 10 μl of DNPH solution (2.5 mg/ml in 6 m HCl), 200 μl of n-hexane was added, and tubes were vortexed well. The hexane layer was recovered and evaporated, and the residue was then dissolved in 200 μl of 60% acetonitrile. A filtrated sample was subjected to HPLC using an ODS-UG3 column (4.6 × 150 mm, Nomura Chemical Co. Ltd., Aichi, Japan) and 60% acetonitrile as a mobile phase. DNPH derivatives were detected at 370 nm.
Purification of Acrolein-reducing Enzyme
Cucumber leaves (100 g) were frozen with liquid nitrogen, and then proteins were extracted with 5 volumes of 50 mm HEPES-NaOH, pH 7.0, containing 1 mm dithiothreitol (DTT) and 0.1 mm EDTA. After centrifugation at 10,000 × g for 20 min, the supernatant was fractionated by the addition of solid ammonium sulfate. Proteins precipitating between 35 and 55% ammonium sulfate saturation were dissolved in a minimum volume of 50 mm HEPES-NaOH, pH 7.0, containing 1 mm DTT (buffer A) and then dialyzed against buffer A. The dialysate was applied to a DE52 column (3.5 × 27 cm, Whatman) equilibrated with buffer A. After washing with buffer A (500 ml), activity was eluted with buffer A containing 110 mm NaCl (1000 ml). Active fractions were combined and dialyzed against 10 mm sodium phosphate buffer (Na-PB, pH 6.8) containing 1 mm DTT. The dialysate was applied to a hydroxyapatite column (1.5 × 8 cm, Wako Pure Chemical) and eluted by a linear gradient of 0 to 0.2 m Na-PB (pH 6.8, total 200 ml). After an equal volume of 60% ammonium sulfate saturated in 50 mm K-PB, pH 7.0, containing 1 mm 2-mercaptoethanol was added to the active fraction, the solution was applied to a phenyl-Sepharose column (1.5 × 8 cm, GE Healthcare) equilibrated with 30% ammonium sulfate saturated in 50 mm K-PB, pH 7.0, containing 1 mm 2-mercaptoethanol and then eluted with a linear gradient of 30 to 0% ammonium sulfate (total 100 ml). Active fractions were concentrated with Microcon-30 (Millipore, Billerica, MA), then applied to a Superose 12 PC 3.2/30 column (0.32 × 30 cm, SMART system, GE Healthcare) equilibrated with buffer A, and eluted at a flow rate of 40 μl/min. All purification steps were done between 0 and 4 °C. Protein concentration was determined according to the method of Lowry et al. (14) with bovine serum albumin as the standard.
Electrophoresis
SDS-PAGE was performed according to the method of Laemmli (15) using 12% polyacrylamide gel under reducing conditions. Proteins were stained with Coomassie Brilliant Blue R-250.
Amino Acid Sequencing of Internal Peptide of Purified Enzyme
Partially purified enzyme was separated by 12% SDS-PAGE, and bands were excised with a razor blade. Enzyme protein was fragmented with trypsin (sequence grade, Roche Applied Science), and amino acid sequences were determined through de novo sequencing after LC/MS/MS analysis.
Determination of Nucleotide Sequence of Acrolein-reducing Enzyme
Total RNA was prepared using an RNeasy plant mini kit (Qiagen, Hilden, Germany), and cDNA was synthesized using a first strand synthesis kit (Roche Applied Science). The cDNA fragments of acrolein reductase genes were amplified by PCR using degenerated primers designed based on internal amino acid sequences shown in supplemental Table S1. Amplified cDNAs were cloned into pCRII vector (Invitrogen), and their sequences were determined using a Big Dye sequencing kit (Applied Biosystems). The full-length cDNA clone for cucumber acrolein reductase was obtained through RACE-PCR using 5′-RACE and 3′-RACE kits (Invitrogen).
Sequence Comparisons
Protein sequences were aligned using ClustalW, and the neighbor-joining method was used to construct a phylogenetic tree and to infer evolutionary history (16). Evolutionary distances were computed using the Poisson correction method (17). There were a total of 198 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (18).
Recombinant Protein Expression and Purification
Primer sequences used for the amplification of DNA fragments for expression of recombinant proteins are shown in supplemental Table S1. After PCR amplification using proofreading TaqDNA polymerase, the resulting PCR product was digested with restriction enzymes and ligated into the corresponding restriction sites of pGEX-4T-1 (GE Healthcare). The constructed expression plasmid was transformed into Escherichia coli BL21 cells. Transformed cells were grown at 37 °C in LB broth containing 100 μg ml−1 ampicillin until the A600 nm reached 0.8–1.2. After induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, the cultures were grown for 18–24 h at 15–18 °C. Cells were precipitated through centrifugation (10,000 × g for 10 min) and then resuspended in PBS containing 0.1% Triton X-100. After sonication and centrifugation, the recombinant protein in the resultant supernatant was purified by affinity chromatography using GSH-Sepharose (GE Healthcare). If necessary, recombinant proteins were further purified using hydroxyapatite and subsequent Mono Q column chromatographies. Purified protein was stored at −20 °C in the presence of 40% (v/v) glycerol.
Analysis of Subcellular Localization
The protein-coding region of tested gene was amplified by PCR using primers listed in supplemental Table S1 and ligated to pTH2, for transient expression plasmid generously provided by Dr. Niwa (University of Shizuoka, Shizuoka, Japan), to produce tested protein-GFP fusion proteins. The GFP was sGFP(S65T), a modified GFP (19), and the chimeric construct was cloned into competent cells. After structural confirmation of the construct, they were introduced into the dorsal side of epidermal cell layers peeled off from excised spiderwort leaves (Tradescantia reflexa) by particle bombardment using a PDS-1000/He device (Bio-Rad), according to the manufacturer's instructions. The autofluorescence (red) of chloroplasts and the GFP images were observed under fluorescence light microscopy (BX51, Olympus, Tokyo, Japan).
Accession Numbers
The full sequences of the C. sativus chloroplastic and cytosolic AORs have been deposited at the DDBJ data bank with accession numbers AB597302 and AB597303, respectively. Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: AtAOR, At1g23740; AtChlADR, At1g54870 and At3g04000; AtCytADR, At2g24190, and At3g61220; AtChlAKR, At2g37770; AtAER, At5g16970.
RESULTS
NADPH Is Predominantly Fueled as a Reductant for Reduction of Both Saturated and Unsaturated Carbonyls
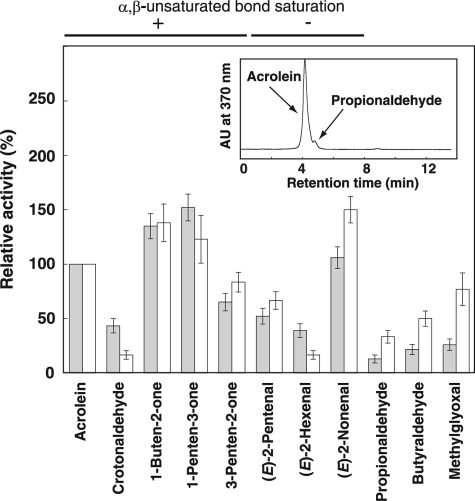
Using concentrated crude extracts prepared from cucumber and Arabidopsis leaves, we measured reducing and oxidizing activity against representative α,β-unsaturated and -saturated carbonyls found in plants using NADPH, NADH, NADP, or NAD, because known enzymes catalyzing reduction of the carbonyls need at least one of these biological reductants/oxidants. As a result, carbonyl-reducing activity using NADPH as a reductant was predominant in crude extracts of both cucumber and Arabidopsis (Fig. 1); the level of NADPH-dependent reduction was more than 10 times greater than that of NADH-dependent reduction (data not shown). Both α,β-unsaturated and saturated aldehydes were not oxidized in the presence of NAD or NADP (data not shown), suggesting that aldehyde dehydrogenase, which catalyzes the oxidation of aldehyde groups to carboxylic groups, is apparently not involved in the major detoxification pathway of α,β-unsaturated aldehydes. Neither reducing nor oxidizing activities against 2-butanone were detected. Results obtained by spectrophotometric analysis (Fig. 1) and decreases in peak areas of all substrates after enzymatic reaction observed by HPLC analysis (data not shown) indicated the presence of aldehyde group-reducing enzyme(s) in crude extracts. In addition, the presence of α,β-unsaturated bond-saturating activity was also indicated because a part of the acrolein, crotonaldehyde, methylvinyl ketone, ethylvinyl ketone, and 3-penten-2-one was reduced to their corresponding saturated carbonyl forms (a representative result is shown in the inset chromatograph in Fig. 1). These results indicate that the detoxification of reactive aldehydes is mediated by reduction of both the aldehyde group and α,β-unsaturated bonds with NADPH as a reductant.
FIGURE 1.
NADPH-dependent carbonyl-reducing activities in crude enzymes prepared from cucumber and Arabidopsis leaves. Activity levels shown in columns were calculated based on a decrease of absorbance at 340 nm of NADPH (6.22 mm−1 cm−1). Specific activity levels for acrolein are set to 100% activity (specific activity levels were 35 ± 6 (cucumber, gray bar) and 11.4 ± 3.4 (Arabidopsis, white bar) nmol/min·mg protein, respectively). Error bar indicates ±S.E. (n = 3). Relative activity for each substrate is shown in the graph. Whether the α,β-unsaturated bond was reduced (+) or not (−) was judged based on the appearance of the corresponding saturated carbonyl after enzymatic reaction by means of HPLC analysis (representative result is shown in inset chromatogram, in which acrolein was partly converted to propionaldehyde).
Purification of Acrolein-reducing Enzyme from Cucumber Leaves
To identify the acrolein-scavenging enzyme of plants, we used cucumber leaves as a biological source because they exhibit high levels of acrolein-reducing activity. This activity was completely eliminated by boiling, suggesting that a proteinaceous acrolein-reducing enzyme might be involved in the reduction of acrolein.
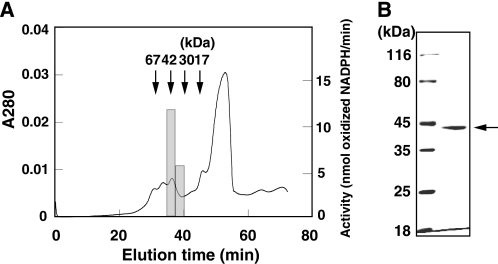
Purification of the acrolein-reducing enzyme is summarized in Table 1. The acrolein-reducing enzyme was purified through successive chromatographies using anion exchange chromatography (DE52), hydroxyapatite, phenyl-Sepharose, and gel filtration (Superose 12). Estimation of native molecular mass by Superose 12 column gel chromatography suggested that the molecular mass of the acrolein-reducing enzyme was about 42 kDa (Fig. 2A). SDS-PAGE of the purified enzyme showed a single band at 42 kDa (Fig. 2B). These data suggested that the acrolein-reducing enzyme was a monomeric protein of 42 kDa.
TABLE 1.
Summary of purification of acrolein-reducing enzyme (from 100 g of cucumber leaves)
| Protein | Total activity | Specific ctivity | Yield | Purification | |
|---|---|---|---|---|---|
| mg | μmol/min | μmol/min/mg | % | -fold | |
| Crude extract | 4160 | 58.0 | 0.014 | 100 | 1 |
| 35–55%(NH4)2SO4 | 485 | 49.6 | 0.102 | 85.5 | 7.29 |
| DE52 | 173 | 29.9 | 0.173 | 51.6 | 12.3 |
| Hydroxyapatite | 30 | 16.7 | 0.558 | 28.8 | 39.8 |
| Phenyl-Sepharose | 12 | 13.4 | 1.12 | 23.1 | 80.0 |
| Superose12 | 0.12 | 7.2 | 60.0 | 12.4 | 4280 |
FIGURE 2.
Molecular weight of the purified acrolein-reducing enzyme. A, final step in the purification of the acrolein-reducing enzyme by means of gel filtration. Horizontal bar indicates the peak level of acrolein-reducing activity. Arrows indicate elution time of marker proteins BSA (67 kDa), ovalbumin (42 kDa), trypsin (30 kDa), and lysozyme (17 kDa). B, SDS-PAGE profile of the purified enzyme (1 μg). Proteins were stained with Coomassie Brilliant Blue R-250.
Substrate Specificity, Kinetics, and Identification of Purified Enzyme
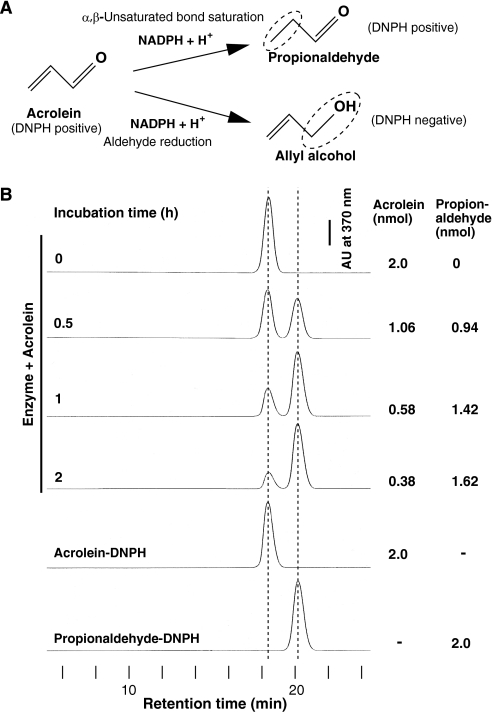
Acrolein has two potential sites to be reduced as follows: an α,β-unsaturated bond and an aldehyde group. Fig. 3A shows two theoretical reduction pathways of acrolein catalyzed by enzymes using NADPH as a reductant. One is an AOR-catalyzed pathway (8), and the other is an AKR (9) or alcohol dehydrogenase-catalyzed pathway. The former yields propionaldehyde as its product, whereas the latter yields allyl alcohol. To distinguish which site was reduced, the reduced product from acrolein was determined through HPLC after carbonyl-reactive DNPH derivatization. After incubation, the peak of acrolein-DNPH decreased accompanying the stoichiometric appearance of the other peak coinciding with propionaldehyde-DNPH at 20.3 min as incubation time increased (Fig. 3B). Furthermore, we confirmed through LC/MS in the negative ionization mode that the m/z of the product was 237, which corresponds to that of propionaldehyde-DNPH (data not shown). From these results, we concluded that the purified acrolein-reducing enzyme hydrogenated the α,β-unsaturated bond of acrolein and resulted in propionaldehyde as the product.
FIGURE 3.
Purified acrolein-reducing enzyme catalyzes the reduction of the α,β-unsaturated bond but not the aldehyde moiety of acrolein. A, two theoretical NADPH-dependent reduction pathways of acrolein. B, using NADPH as a reductant, acrolein was metabolized for the indicated length of time with the purified enzyme. After this reaction, the product was subjected to DNPH derivatization and subsequent HPLC analysis. DNPH reacts with the aldehyde group of acrolein and propionaldehyde but not the alcohol group of allyl alcohol.
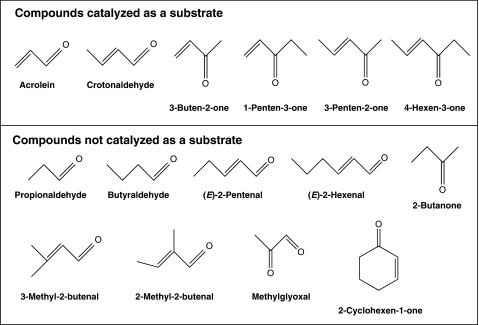
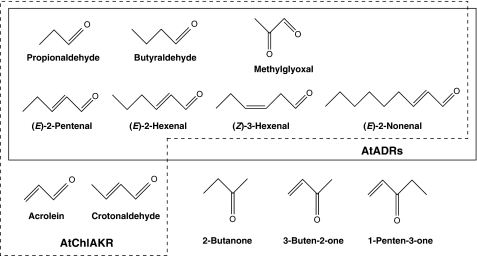
Using various related compounds, the substrate specificity of the purified enzyme was investigated. We found that the enzyme preferentially reduced 2-alkenones such as 3-buten-2-one, 1-penten-3-one, 3-penten-2-one, and 4-hexen-3-one (Fig. 4). Carbonyls without an α,β-unsaturated bond, such as propionaldehyde, butyraldehyde, and 2-butanone, did not serve as substrates. The purified enzyme reduced acrolein and crotonaldehyde but not 3-methyl-2-butenal and 2-methyl-2-butenal, suggesting that the presence of at least one hydrogen bound to both the α- and β-carbons is essential in a substrate. In addition, the enzyme did not reduce 2-cyclohexen-1-one, indicating that cyclic enone is not a substrate for the purified enzyme. Long carbon chain (C ≧ 5) α,β-unsaturated aldehydes such as (E)-2-pentenal and (E)-2-hexenal were not substrates for this enzyme.
FIGURE 4.
Structure of the compounds tested for substrate specificity analysis of purified CsAOR.
The kinetic parameters of the purified enzyme for α,β-unsaturated aldehydes and ketones are shown in Table 2. Its Km value against acrolein was 13.6 mm, whereas that against crotonaldehyde was 29.8 mm. Its Km value against α,β-unsaturated ketones was much lower than those against α,β-unsaturated aldehydes; as a result, the purified enzyme showed high specificity (kcat/Km) to α,β-unsaturated ketones. This substrate preference is a common characteristic among enzymes with α,β-unsaturated bond hydrogenation activity (8, 20). The acrolein-reducing enzyme operated 30 times faster in the presence of NADPH than in the presence of NADH (Table 2). Consequently, the purified enzyme was identified as an NADPH-dependent AOR using short chain α,β-unsaturated carbonyls as its substrates. Hereafter, we use the term C. sativus AOR (CsAOR) to denote the purified acrolein-reducing enzyme.
TABLE 2.
Kinetic parameters of the purified CsAOR
| Chain length | kcat | Km | kcat/Km | |
|---|---|---|---|---|
| s−1 | mm | s−1 mm−1 | ||
| Acrolein (propenal) | 3 | 63 ± 6 | 13.6 ± 1.4 | 4.63 ± 0.65 |
| Crotonaldehyde (butenal) | 4 | 42 ± 6.5 | 29.8 ± 3.0 | 1.41 ± 0.26 |
| 3-Buten-2-one | 4 | 74 ± 8.6 | 2.88 ± 0.29 | 25.7 ± 3.8 |
| 1-Penten-3-one | 5 | 61 ± 7.8 | 0.58 ± 0.06 | 105 ± 16 |
| 3-Penten-2-one | 5 | 22 ± 4.7 | 0.63 ± 0.06 | 34.9 ± 8.2 |
| 4-Hexen-3-one | 6 | 68 ± 8.2 | 0.023 ± 0.005 | 2960 ± 740 |
| NADPHa | 75 ± 8.7 | 0.185 ± 0.02 | 406 ± 64 | |
| NADHa | 75 ± 8 | 4.64 ± 0.5 | 16.2 ± 2.5 |
a Acrolein (20 mm) was used as a substrate.
Molecular Characteristics of CsAOR
The internal amino acid sequence of the purified enzyme was determined through de novo sequencing using LC/MS/MS after trypsin digestion. We designed degenerate primers from two obtained peptide sequences (Fig. 5) and determined full-length cDNAs encoding AOR protein using the RACE method. Cloning of the cucumber AOR resulted in the determination of two distinct sequences encoding AOR (Fig. 5). The isolated CsAOR cDNAs were 1219 bp encoding a polypeptide composed of 383 amino acids and 1066 bp encoding a polypeptide composed of 320 amino acids. To confirm that these polypeptides encoded functional AORs, we expressed recombinant GST-fused proteins in E. coli and found that both recombinant proteins had AOR activity (data not shown). A homology search suggested that homologous genes were found in various plant species (Fig. 5) and that they could be divided into two groups, one with variable N-terminal sequences and the other without. ChloroP analysis showed that these N-terminal sequences contained a chloroplast-targeting signal motif, indicating that chloroplastic and/or cytosolic AORs would be distributed widely in plant species. In these amino acid sequences, a glycine-rich sequence (GAGGVG), which contains the (GAGGXX) motif of the NADPH-binding Rossmann-like domain found in shikimate dehydrogenase (21), is strictly conserved.
FIGURE 5.
ClustalW sequence alignment of cucumber AORs with nearest sequences of various plants. The sequence is shown in single-letter amino acid code. Double arrows show peptide sequences used for designing degenerate primers according to the 5′- and 3′-RACE cloning method. Dotted arrow indicates a variable peptide sequence region. An NADPH-binding Rossmann-like domain is indicated by a box. The asterisks below the sequence indicate the positions of fully conserved residues. Accession numbers are BAJ23910 (cucumber chloroplastic), BAJ23911 (cucumber cytosolic), ACU19769 (soybean), AAK66565 (sunflower), BAF23605 (rice), ACG41966 (maize), and AAM16188 (Arabidopsis).
Subcellular Localization of CsAORs
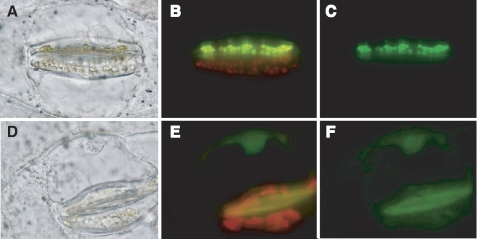
Because the CsAOR polypeptide composed of 383 amino acids contained a putative transit peptide for plastidial sorting at the N terminus as predicted by ChloroP, we constructed the GFP fusion proteins and transiently expressed them in spiderwort to determine the intracellular localization of these two CsAOR polypeptides. In the case of the CsAOR polypeptide with 383 amino acids, strong green fluorescence was distributed in the plastids; in the case of the CsAOR polypeptide with 320 amino acids, fluorescence was observed at the periphery and at the center of the cell. Autofluorescence of chlorophyll and GFP fluorescence was completely overlaid (Fig. 6, A–C), suggesting that the CsAOR with 383 amino acids was the chloroplastic protein. On the other hand, the GFP fluorescence of the CsAOR with 320 amino acids was distributed in the cytoplasm around vacuoles, along plasma membranes, and in the nucleoplasm of the nuclei (Fig. 6, D–F). Because GFP inherently tends to localize in nuclei (22), however, the localization of CsAOR in the nuclei cannot necessarily support a conclusion. Thus, we concluded that the CsAOR with 383 amino acids and that with 320 amino acids were the chloroplastic CsAOR (CsChlAOR) and the cytosolic CsAOR (CsCytAOR), respectively. The purified native CsAOR enzyme was thought to be a CsChlAOR because its native molecular mass (42 kDa) coincided with the theoretical molecular mass of CsChlAOR (40,527 Da) but not with that of CsCytAOR (34,423 Da).
FIGURE 6.
Intracellular localization of CsAOR-GFP fusion proteins in spiderwort cells. Optical microscopic images (A and D) and merged images of the same region of epidermal tissue showing GFP signals and red chlorophyll autofluorescence (B and E) or GFP fluorescence only (C and F) in the guard cell chloroplasts. Images represent the GFP-fused CsAOR of 383 amino acids (A–C) or 320 amino acids (D–F). The GFP construct was delivered only into the upper cell of the same guard cell pair that is shown in B and C.
Heterogeneous Expression of A. thaliana AOR (AtAOR)
Homology search of CsAOR revealed that a homologous gene was included in Arabidopsis genomic DNA. To confirm that this gene also encoded AOR, we cloned the cDNA of Arabidopsis AOR (AtAOR, At1g23740) and expressed the recombinant protein in E. coli. In constructing the expression plasmid, we removed about 80 residues of the N-terminal sequence because these regions are highly divergent among known sequences (Fig. 5), probably because of the plastid-targeted transit peptide. We observed that the GST-fused AtAOR protein was successively expressed in the soluble and functional fractions (supplemental Fig. S1A). The kinetic parameters of AtAOR were fairly similar to those of CsAORs (Table 3). At1g23740 protein has a predicted transit peptide in the N-terminal region and has been identified as a stromal protein in chloroplasts by two independent proteomic analyses (23, 24). Accordingly, we concluded that AtAOR was the chloroplast-localizing AOR.
TABLE 3.
Kinetic parameters of recombinant AtAOR
| Chain length | kcat | Km | kcat/Km | |
|---|---|---|---|---|
| s−1 | mm | s−1mm−1 | ||
| Acrolein (propenal) | 3 | 22 ± 4.7 | 3.5 ± 0.4 | 6.28 ± 1.5 |
| Crotonaldehyde (butenal) | 4 | 1.3 ± 0.2 | 1.8 ± 0.2 | 0.72 ± 0.14 |
| 3-Buten-2-one | 4 | 3.5 ± 0.4 | 0.13 ± 0.02 | 26.9 ± 5.2 |
| 1-Penten-3-one | 5 | 4.0 ± 0.4 | 0.29 ± 0.03 | 13.8 ± 2.0 |
| 4-Hexen-3-one | 6 | 3.3 ± 0.3 | 0.12 ± 0.02 | 27.5 ± 5.2 |
| NADPHa | 23 ± 4.8 | 0.01 ± 0.001 | 2300 ± 530 | |
| NADHa | 23 ± 4 | 0.053 ± 0.006 | 434 ± 90 |
a Acrolein (20 mm) was used as a substrate.
Aldehyde Reductases Belonging to SDR Superfamily Protein and AKR Are Also Involved in the α,β-Unsaturated Carbonyls Detoxification Pathway in Chloroplasts
The localization of AOR in chloroplasts suggests the importance of detoxification of α,β-unsaturated carbonyls for maintaining the photosynthetic process. However, as shown in Fig. 4, AOR could not reduce longer α,β-unsaturated aldehydes, suggesting that other enzyme(s) might be involved in the detoxification. The detoxification of α,β-unsaturated carbonyls can be achieved through the reduction or oxidation of their carbonyl groups. In crude extracts of cucumber and Arabidopsis, NADPH-dependent reducing activity predominantly occurred against both α,β-unsaturated and saturated aldehydes (Fig. 1), suggesting that SDR and/or AKR are involved in the reduction of these aldehyde groups. Therefore, to establish the detoxification pathway of α,β-unsaturated carbonyls in chloroplasts, we selected candidates involved in the detoxification of α,β-unsaturated carbonyls, i.e. the reduction of aldehyde group of both long α,β-unsaturated aldehydes and saturated aldehydes.
Selected candidate genes for SDR and AKR satisfied at least one of the following criteria: 1) co-expression with α,β-unsaturated bond-reducing enzymes, AtAOR (At1g23740) or Arabidopsis alkenal reductase (AtAER, At5g16970) as shown in the publicly available Arabidopsis co-expression data base (ATTED-II); 2) predicted chloroplastic protein status according to PSORT or ChloroP programs. Known genes having other enzymatic activity were excluded. The selected candidate genes were cloned into an expression vector, and encoded proteins were expressed in E. coli as GST fusion proteins (a list is shown in supplemental Table S1). Enzymes recovered in soluble form were purified through affinity chromatography using a GSH-Sepharose column (supplemental Fig. S1B), and the activity of recombinant proteins was measured. We successfully detected both α,β-unsaturated and -saturated aldehyde-reducing activity associated with two SDR family proteins (At1g54870 and At3g04000) and one AKR protein (At2g37770).
A summary of the substrate specificities and kinetic parameters of the proteins At1g54870 and At3g04000 are shown in Fig. 7 and Table 4, respectively. They reduced saturated aldehydes such as propionaldehyde and butyraldehyde in the presence of NADPH, but not NADH, as a reductant. This result does not contradict the results obtained using crude extracts, which suggested NADPH dependence (Fig. 1). These proteins also did not reduce 2-butanone and did not oxidize propanol, butanol, or ethanol in the presence of NADP or NAD (data not shown). They did reduce the aldehyde groups of α,β-unsaturated aldehydes such as (E)-2-pentenal, (E)-2-hexenal, and (E)-2-nonenal but not acrolein and crotonaldehyde. Aldehyde group reduction of α,β-unsaturated aldehydes was confirmed by the fact that saturated aldehydes were not detected as products after enzymatic reactions using α,β-unsaturated aldehydes as the substrates by HPLC analysis (data not shown). In addition, these proteins reduced methylglyoxal (MG), a representative α-dicarbonyl compound produced during glycolysis, and (Z)-3-hexenal, a major volatile involved in green leaf odor. The localizations of At1g54870 and At3g04000 were determined through a transient expression assay using GFP fused with the N-terminal regions of the polypeptides. Distribution of GFP fluorescence in both At1g54870 and At3g04000 completely corresponded to the autofluorescence of chloroplasts (supplemental Fig. S2, A–D), suggesting that the N-terminal regions of both proteins have chloroplast-targeting transit peptides. Based on these properties, we concluded that these proteins were NADPH-dependent chloroplastic aldehyde reductases (AtChlADRs) reducing saturated aldehyde and α,β-unsaturated aldehydes having more than 5 carbons (C ≧ 5) as substrates.
FIGURE 7.
Structure of the compounds tested for substrate specificity analysis of recombinant AtADRs. Substrates for AtADRs and AtChlAKR (At2g37770) are circumscribed by solid and dashed lines, respectively.
TABLE 4.
Kinetic parameters of AtChlADRs, AtCytADRs, and AtChlAKR
ND means no detectable activity. VH means very high, i.e. could not be fitted to Michaelis-Menten kinetics. NADPH (0.1 mm) was used as a reductant in all assays. In this table, only the average is shown.
| AtChlADR |
AtCytADR |
AtChlAKR, At2g37770 |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At1g54870 |
At3g04000 |
At2g24190 |
At3g61220 |
|||||||||||||||
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | ||||
| mm | s−1 | s−1mm−1 | mm | s−1 | s−1mm−1 | mm | s−1 | s−1mm−1 | mm | s−1 | s−1mm−1 | mm | s−1 | s−1mm−1 | ||||
| Saturated aldehyde | ||||||||||||||||||
| Propionaldehyde | VH | VH | VH | VH | 70 | 44 | 0.63 | |||||||||||
| Butyraldehyde | 10 | 4.3 | 0.43 | 7.8 | 29.6 | 3.8 | 40 | 9.1 | 0.23 | VH | 6.9 | 21 | 3.0 | |||||
| α,β-Unsaturated aldehydes | ||||||||||||||||||
| Acrolein | ND | ND | ND | ND | 2.6 | 6.6 | 2.54 | |||||||||||
| Crotonaldehyde | ND | ND | ND | ND | 1.63 | 18 | 11 | |||||||||||
| (E)-2-Pentenal | 0.09 | 1.7 | 18.9 | 0.6 | 6.4 | 10.7 | 0.2 | 0.4 | 2.0 | 0.07 | 0.27 | 3.9 | 0.63 | 9.5 | 15 | |||
| (E)-2-Hexenal | 4.4 | 1.1 | 0.25 | 2.5 | 1.8 | 0.72 | 1.7 | 0.6 | 0.35 | 0.53 | 0.4 | 0.75 | 0.72 | 7.3 | 10 | |||
| (E)-2-Nonenal | 70 | 7.3 | 0.10 | VH | VH | VH | 1.67 | 7.9 | 4.73 | |||||||||
| α-Dicarbonyl | ||||||||||||||||||
| Methylglyoxal | 4.7 | 2.4 | 0.51 | 5.0 | 28.8 | 5.76 | 69 | 24 | 0.35 | VH | 0.24 | 11.5 | 48 | |||||
AKR encoded by At2g37770 showed reducing activity of both saturated and unsaturated aldehydes (Fig. 7). As in the case of AtChlADRs, the activity of this AKR is completely dependent on NADPH. Analysis of subcellular localization indicated that this AKR is localized in chloroplasts (supplemental Fig. S2, E and F); accordingly, it was named AtChlAKR. The substrate specificity of AtChlAKR overlapped with those of the AtChlADRs, and AtChlAKR reduced acrolein and crotonaldehyde (Fig. 7). Ketones were not substrates for AtChlAKR. The kinetic parameters of AtChlAKR suggest that it reduces aldehydes with a higher efficiency than the AtChlADRs; AtChlADRs had higher Km and lower kcat values than AtChlAKR, except for butyraldehyde and (E)-2-pentenal (Table 4).
Cytosolic ADRs Also Reduce Aldehyde Groups of Saturated and Unsaturated Aldehydes
Two SDR family proteins (At2g24190 and At3g61220) are homologs of mammalian CBR. These are predicted to be chloroplastic proteins with low reliability. The distribution of GFP-fused chimeric proteins suggested that the subcellular localization of these proteins was cytosolic (supplemental Fig. S2, G and H). Recombinant protein studies showed that they exhibited NADPH-dependent reducing activity against saturated and unsaturated aldehydes, in a pattern of enzymatic properties similar to that of the AtChlADRs (Table 4). Based on these results, we concluded that these SDRs were NADPH-dependent cytosolic ADRs (AtCytADRs). CBRs found in bacteria, insects, and animals reduce carbonyl groups of various secondary carbonyls, and thus they are thought to be involved in the detoxification and inactivation of reactive carbonyls (13). Also in Arabidopsis, AtCytADRs probably act as detoxifying enzymes of reactive carbonyls in cytosol.
DISCUSSION
AOR, ADR, and AKR Cooperatively Scavenge Reactive Carbonyls in Chloroplasts of Arabidopsis
In this study, we exhibited that AOR, ADR, and AKR might cooperatively detoxify reactive carbonyls in chloroplasts where reactive carbonyls are abundantly formed. Substrate specificity showed that plant AOR preferentially reduced the α,β-unsaturated bonds of small carbonyls, especially α,β-unsaturated ketones. Major α,β-unsaturated aldehydes such as (E)-2-hexenal and (E)-2-pentenal are not substrates for plant AOR. Because Km values against α,β-unsaturated ketones were much lower than those against α,β-unsaturated aldehydes, a major physiological significance of plant AOR might be the reduction of α,β-unsaturated ketones. This conclusion is also supported by the observation that AtChlADRs and AtChlAKR could not reduce α,β-unsaturated ketones.
AtChlADRs catalyzed the reduction of aldehyde groups of long chain α,β-unsaturated aldehydes (C ≧ 5) and saturated aldehydes. AtChlADRs belong to the alcohol dehydrogenase family, whose other protein subfamilies include medium- and short-chain dehydrogenase/reductases, the proteins that metabolize saturated aldehydes in cucurbitaceae (25). The reaction catalyzed by these alcohol dehydrogenases is reversible, however, so that the balance of NAD(P)H and NAD(P) essentially affects the direction of the reaction. The only reaction catalyzed by AtChlADRs that is irreversible at physiological pH is the reduction of aldehyde groups; this suggests that one physiological significance of AtChlADRs is the elimination of aldehyde groups from saturated and unsaturated aldehydes, especially butyraldehyde and (E)-2-pentenal as suggested by their kinetic parameters (Table 4). In E. coli, the ADR YqhD is thought to be involved in the elimination of aldehydes produced during lipid peroxidation (26). Although the amino acid sequence of YqhD is not homologous to those of plant AtChlADRs, the physiological functions of these AtChlADRs are likely to be similar to those of bacterial ADRs.
AKR is also thought to be involved in the detoxification of reactive aldehydes. Oberschall et al. (27) have reported that overexpression of an alfalfa AKR contributed to improve plant tolerance to chemical and drought stresses through enhanced reduction of reactive aldehydes produced during lipid peroxidation. Interestingly, the alfalfa AKR is a homolog of AtChlAKR (At2g37770); the physiological significance of this class of AKR in plant species might therefore be the detoxification of reactive aldehydes. A comparison of the enzymological characteristics of AtChlADR and AtChlAKR (Table 4) indicated that AtChlAKR might reduce aldehydes more efficiently in vivo because of its higher kcat/Km value.
A co-expression analysis using ATTED-II showed that the expression patterns of AtChlADR (At3g04000), AKRs (At2g37770), and AtAER (At5g16970) are highly correlative, suggesting that these enzymes might function as a group to reduce aldehydes produced under conditions of stress such as high salinity and drought. AtAOR (At1g23740) and the AtCytADRs (At2g24190 and At3g61220), on the other hand, are likely to be involved in the general elimination of reactive carbonyls because they are constituently expressed.
CsAORs and AtAOR Belong to a Novel Plant AOR Family
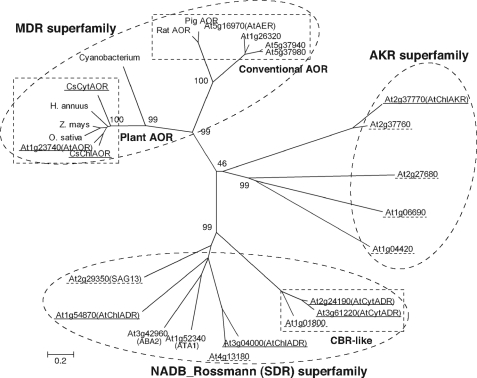
Although AtAOR and AtAER exhibit hydrogenation activity against the α,β-unsaturated bond of acrolein (20), phylogenetic analysis showed that the amino acid sequences of AOR and AtAER belong to different trees in the medium-chain dehydrogenase/reductase superfamily (Fig. 8). AtAER belongs to a different phylogenetic subgroup, which includes mammal AOR and deduced allyl alcohol dehydrogenases, whereas AtAOR and CsAORs belong to other classes, including deduced quinone-oxidoreductase family proteins. The enzymological properties of CsAORs and AtAORs, such as their optimum pH of around 6.0–6.5 (data not shown) and their kcat values (Tables 2 and 3), are similar to those of AtAER, but the CsAORs and AtAOR preferentially catalyze the hydrogenation of short chain α,β-unsaturated carbonyls, in contrast to AtAER, which preferentially catalyzes that of long carbon chain α,β-unsaturated aldehydes (20, 28). The subcellular localizations of AtAOR and AtAER are also quite different; specifically, AtAOR localizes in chloroplast, and AtAER localizes in the cytosol and nucleus (28). These results suggest that, although AtAOR and AtAER have similar enzymatic properties, the CsAORs and AtAOR belong to a novel plant-type AOR family that is responsible for scavenging α,β-unsaturated carbonyls. Homologs of plant AORs could be found in genetic information of cyanobacteria and Physcomitrella patens, implying that the presence and importance of AOR mediated detoxification in other photosynthetic organisms. However, it is necessary to confirm their enzymatic activities because their identities are below 50% compared with plant AORs.
FIGURE 8.
Phylogenetic analysis of amino acid sequences of enzymes involved in NADPH-dependent reduction of reactive carbonyls. Unrooted trees were constructed using MEGA version 4 software based on the ClustalW multiple alignment according to the neighbor-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Dashed circles and rectangles show superfamilies and subfamilies, respectively. Enzymes analyzed in this study are underlined by solid lines. Expressed proteins not showing carbonyl-reducing activity are underlined by dashed lines. Other sequences are representative example of ally alcohol dehydrogenase (At1g26320, At5g37940, and At5g37980) and plausible phytohormone-metabolizing SDRs (At1g52340 (ATA1) and At3g42960 (ABA2)). Accession numbers of plant AORs are BAJ23910 (CsChlAOR), BAJ23911 (CsCytAOR), AAK66565 (Helianthus annuus, sunflower), BAF23605 (Oryza sativa, rice), ACG41966 (Zea mays, maize), and AAM16188 (AtAOR). Homologous sequence of cyanobacterium is referred from accession number YP_722551. Accession numbers of rat and pig AORs are AAH89775 and BAA08381, respectively. MDR, medium-chain dehydrogenase/reductase.
ADRs and AKR Act as MG-scavenging Enzymes in Chloroplast and Cytosol
Another physiological significance of the AtADRs and AtChlAKR is the reduction of the aldehyde group of MG, because MG is also recognized as cytotoxic aldehydes. MG can react with biomolecules such as proteins and DNA and can potentially destroy their functional activity. It is produced as a nonenzymatic by-product of glycolysis (29) and lipid peroxidation (30) and also enzymatically either through stress-induced production from triose phosphates (31) or from dihydroxyacetone phosphate in a reaction catalyzed by MG synthase in bacteria (32). The mechanism(s) of MG production in plants is not yet understood; it is significant, however, that ascorbic acid is a potential precursor of MG (33). We have shown that glycation (protein modification by degraded products of sugars) occurs in the presence of 10 mm ascorbic acid (34), and the concentration of ascorbic acid in chloroplasts is estimated to be 12–25 mm (35). In addition, salt stress enhanced MG accumulation, whereas overexpression of glyoxalase I and II resulted in enhanced tolerance against salt stress (36), suggesting that MG is also a potential cytotoxic compound in plants. AtADRs and AtChlAKR might alleviate the toxicity of MG in chloroplasts and cytosol by mediating the NADPH-dependent reduction of the aldehyde groups of MG to acetol.
Detoxification Pathway of Reactive Carbonyls in Plants
In this study, we focused on the NADPH-dependent detoxification of reactive carbonyls. Eukaryotic cells also use other systems to cope with the harmful effects of reactive carbonyls; the GST pathway, for example, which is fueled by the GSH or thioredoxin redox cycle, conjugates aldehydes with GSH, supplying an important mechanism of detoxification of reactive carbonyls. In animals, the main detoxification pathway is GSH-dependent, and it has been shown that the alcohol/aldehyde dehydrogenase and GST pathways account for 10 and 55% of aldehyde elimination by hepatocytes, respectively (37). A GSH-dependent detoxification system is functional in plants also. The GST proteins constitute a large family; some of them are localized in the chloroplasts and mitochondria, and it is known that glyoxalase I and II cooperatively convert MG to lactate in a GSH-dependent manner (32). In the Arabidopsis genome, chloroplast-localizing glyoxalase I (At1g11840) and II (At1g06130) genes were predicted and found. In addition, GSH itself scavenges α,β-unsaturated carbonyls, because it can form adducts with α,β-unsaturated carbonyls (2). Because chloroplasts contain a millimolar order of GSH, the GSH-dependent detoxification system is thought to be substantially effective.
Aldehyde dehydrogenase activity, which catalyzes the oxidation of aldehyde groups to carboxylic groups, was not detected in our experiments. Sunkar et al. (38) have reported that stress-inducible aldehyde dehydrogenase contributed to tolerance against dehydration, NaCl, and heavy metal stresses. The accumulation of reactive aldehyde was suppressed in transgenic tobacco overexpressing aldehyde dehydrogenase, suggesting that stress-inducible, but probably not constitutive, aldehyde dehydrogenase might be involved in the detoxification pathway of reactive aldehydes.
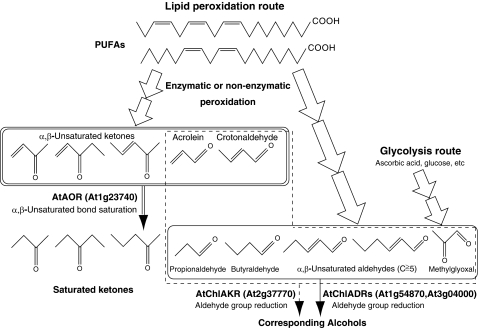
In this study, we revealed an NADPH-dependent system for the detoxification of reactive carbonyls in chloroplasts, in which AOR, ADRs, and AKR cooperatively scavenge reactive carbonyls as depicted in Fig. 9. In the model plant Arabidopsis, AtAOR reduces the α,β-unsaturated bonds of reactive α,β-unsaturated carbonyls derived from lipid peroxidation, forming saturated carbonyls. Importantly, the reduction of α,β-unsaturated ketones depends on AtAOR. AtChlAKR reduces the aldehyde groups of α,β-unsaturated and saturated aldehydes. AtChlADRs are also involved in the reduction of longer (C ≧ 5) α,β-unsaturated aldehydes and saturated aldehydes. MG derived from glycolysis and lipid peroxidation is reduced by AtChlAKR and AtChlADRs.
FIGURE 9.
AtAOR-, AtChlADR-, and AtChlAKR-related detoxification pathways of reactive carbonyls in Arabidopsis chloroplasts. Substrates for AOR, ADR, and AKR are circumscribed by double, solid, and dashed lines, respectively. α,β-Unsaturated carbonyls generated through multistep polyunsaturated fatty acids (PUFAs) peroxidation were reduced by AtAOR (At1g23740). The resulting saturated and α,β-unsaturated aldehydes (C ≧ 5) were reduced by AtChlADRs (At1g54870 and At3g04000). AtChlAKR (At2g37770) reduces saturated aldehydes and α,β-unsaturated aldehydes. MG is reduced by AtChlADRs and AtChlAKR.
Supplementary Material
Acknowledgment
We thank Dr. Shigeo Takumi of Kobe University for instructing us on subcellular localization analysis.
This work was supported by Grant-in-aid for Exploratory Research 21658112 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to Y. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- AOR
- alkenal/one oxidoreductase
- ADR
- aldehyde reductase
- AER
- alkenal reductase
- AKR
- aldo-keto reductase
- CBR
- carbonyl reductase
- DNPH
- 2,4-dinitrophenylhydrazine
- MG
- methylglyoxal
- SDR
- short-chain dehydrogenase/reductase
- RACE
- rapid amplification of cDNA ends.
REFERENCES
- 1. Dennis K. J., Shibamoto T. (1990) Lipids 25, 460–464 [DOI] [PubMed] [Google Scholar]
- 2. Esterbauer H., Schaur R. J., Zollner H. (1991) Free Radic. Biol. Med. 11, 81–128 [DOI] [PubMed] [Google Scholar]
- 3. Millar A. H., Leaver C. J. (2000) FEBS Lett. 481, 117–121 [DOI] [PubMed] [Google Scholar]
- 4. Yamauchi Y., Furutera A., Seki K., Toyoda Y., Tanaka K., Sugimoto Y. (2008) Plant Physiol. Biochem. 46, 786–793 [DOI] [PubMed] [Google Scholar]
- 5. Mano J., Miyatake F., Hiraoka E., Tamoi M. (2009) Planta 230, 639–648 [DOI] [PubMed] [Google Scholar]
- 6. Yamauchi Y., Sugimoto Y. (2010) Planta 231, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 7. Alméras E., Stolz S., Vollenweider S., Reymond P., Mène-Saffrané L., Farmer E. E. (2003) Plant J. 34, 205–216 [DOI] [PubMed] [Google Scholar]
- 8. Dick R. A., Kwak M. K., Sutter T. R., Kensler T. W. (2001) J. Biol. Chem. 276, 40803–40810 [DOI] [PubMed] [Google Scholar]
- 9. Kolb N. S., Hunsaker L. A., Vander Jagt D. L. (1994) Mol. Pharmacol. 45, 797–801 [PubMed] [Google Scholar]
- 10. Burczynski M. E., Sridhar G. R., Palackal N. T., Penning T. M. (2001) J. Biol. Chem. 276, 2890–2897 [DOI] [PubMed] [Google Scholar]
- 11. Sellin S., Holmquist B., Mannervik B., Vallee B. L. (1991) Biochemistry 30, 2514–2518 [DOI] [PubMed] [Google Scholar]
- 12. Mano J., Tokushige K., Mizoguchi H., Fujii H., Khorobrykh S. (2010) Plant Biotechnol. 27, 193–197 [Google Scholar]
- 13. Forrest G. L., Gonzalez B., Tseng W., Li X., Mann J. (2000) Cancer Res. 60, 5158–5164 [PubMed] [Google Scholar]
- 14. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 15. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 16. Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 17. Zuckerkandl E., Pauling L. (1965) in Evolutionary Divergence and Convergence in Proteins (Bryson V., Vogel H. H. eds) pp. 97–166, Academic Press, New York [Google Scholar]
- 18. Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 19. Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996) Curr. Biol. 6, 325–330 [DOI] [PubMed] [Google Scholar]
- 20. Mano J., Torii Y., Hayashi S., Takimoto K., Matsui K., Nakamura K., Inzé D., Babiychuk E., Kushnir S., Asada K. (2002) Plant Cell Physiol. 43, 1445–1455 [DOI] [PubMed] [Google Scholar]
- 21. Ye S., Von Delft F., Brooun A., Knuth M. W., Swanson R. V., McRee D. E. (2003) J. Bacteriol. 185, 4144–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanson M. R., Köhler R. H. (2001) J Exp. Bot. 52, 529–539 [PubMed] [Google Scholar]
- 23. Rutschow H., Ytterberg A. J., Friso G., Nilsson R., van Wijk K. J. (2008) Plant Physiol. 148, 156–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goulas E., Schubert M., Kieselbach T., Kleczkowski L. A., Gardeström P., Schröder W., Hurry V. (2006) Plant J. 47, 720–734 [DOI] [PubMed] [Google Scholar]
- 25. Manríquez D., El-Sharkawy I., Flores F. B., El-Yahyaoui F., Regad F., Bouzayen M., Latché A., Pech J. C. (2006) Plant Mol. Biol. 61, 675–685 [DOI] [PubMed] [Google Scholar]
- 26. Pérez J. M., Arenas F. A., Pradenas G. A., Sandoval J. M., Vásquez C. C. (2008) J. Biol. Chem. 283, 7346–7353 [DOI] [PubMed] [Google Scholar]
- 27. Oberschall A., Deák M., Török K., Sass L., Vass I., Kovács I., Fehér A., Dudits D., Horváth G. V. (2000) Plant J. 24, 437–446 [DOI] [PubMed] [Google Scholar]
- 28. Mano J., Belles-Boix E., Babiychuk E., Inzé D., Torii Y., Hiraoka E., Takimoto K., Slooten L., Asada K., Kushnir S. (2005) Plant Physiol. 139, 1773–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richard J. P. (1984) J. Am. Chem. Soc. 106, 4926–4936 [Google Scholar]
- 30. Niyati-Shirkhodaee F., Shibamoto T. (1993) J. Agric. Food Chem. 41, 227–230 [Google Scholar]
- 31. Sommer A., Fischer P., Krause K., Boettcher K., Brophy P. M., Walter R. D., Liebau E. (2001) Biochem. J. 353, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yadav S. K., Singla-Pareek S. L., Ray M., Reddy M. K., Sopory S. K. (2005) Biochem. Biophys. Res. Commun. 337, 61–67 [DOI] [PubMed] [Google Scholar]
- 33. Shipanova I. N., Glomb M. A., Nagaraj R. H. (1997) Arch. Biochem. Biophys. 344, 29–36 [DOI] [PubMed] [Google Scholar]
- 34. Yamauchi Y., Ejiri Y., Tanaka K. (2002) Plant Cell Physiol. 43, 1334–1341 [DOI] [PubMed] [Google Scholar]
- 35. Foyer C., Rowell J., Walker D. (1983) Planta 157, 239–244 [DOI] [PubMed] [Google Scholar]
- 36. Yadav S. K., Singla-Pareek S. L., Reddy M. K., Sopory S. K. (2005) FEBS Lett. 579, 6265–6271 [DOI] [PubMed] [Google Scholar]
- 37. Hartley D. P., Ruth J. A., Petersen D. R. (1995) Arch. Biochem. Biophys. 316, 197–205 [DOI] [PubMed] [Google Scholar]
- 38. Sunkar R., Bartels D., Kirch H. H. (2003) Plant J. 35, 452–464 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.