Abstract
Transcription factors of the nuclear factor 1 (NFI) family regulate normal brain development in vertebrates. However, multiple splice variants of four NFI isoforms exist, and their biological functions have yet to be elucidated. Here, we cloned and analyzed human NFI-X3, a novel splice variant of the nfix gene, which contains a unique transcriptional activation (TA) domain completely conserved in primates. In contrast to previously cloned NFI-X1, overexpression of NFI-X3 potently activates NFI reporters, including glial fibrillary acidic protein (GFAP) reporter, in astrocytes and glioma cells. The GAL4 fusion protein containing the TA domain of NFI-X3 strongly activates the GAL4 reporter, whereas the TA domain of NFI-X1 is ineffective. The expression of NFI-X3 is dramatically up-regulated during the differentiation of neural progenitors to astrocytes and precedes the expression of astrocyte markers, such as GFAP and SPARCL1 (Secreted Protein, Acidic and Rich in Cysteines-like 1). Overexpression of NFI-X3 dramatically up-regulates GFAP and SPARCL1 expression in glioma cells, whereas the knockdown of NFI-X3 diminishes the expression of both GFAP and SPARCL1 in astrocytes. Although activation of astrocyte-specific genes involves DNA demethylation and subsequent increase of histone acetylation, NFI-X3 activates GFAP expression, in part, by inducing alterations in the nucleosome architecture that lead to the increased recruitment of RNA polymerase II.
Keywords: Chromatin Structure, Gene Expression, Gene Regulation, Transcription Factors, Transcription Regulation, Astrocytes
Introduction
Astrocytes are critical for the normal functions of the brain but also play a destructive role in many diseases of the central nervous system. Nevertheless, mechanisms controlling their differentiation and astrocyte-specific gene expression are only partially understood. Astrocyte differentiation is promoted by the activation of several signaling pathways, including the JAK-STAT pathway (1–3), the activation of SMADs (Similar to Drosophila Mothers Against Decapentaplegic) (3, 4), the activation of Notch signaling (5, 6), and the activation of genes encoding the nuclear factor-1 (NFI)4 family of transcription factors (7–13). During the vertebrate embryonic development, neurons are generated first, followed by glia. This neurogenic-to-gliogenic switch is induced by the activation of the JAK-STAT signaling pathway in neural precursors by neuron-derived cardiotrophin-1 (2). Specifically, STAT3 induces production of BMP-2, which subsequently activates SMAD1 (3). In turn, SMAD1 forms a complex with STAT3 and induces astrogliogenesis (14). In addition to JAK-STAT and BMP-SMAD1 pathways, Notch signaling also affects gliogenesis by activation of RBP-Jκ transcriptional activity (6, 15), induction of NFI expression (13), and concomitant demethylation of the astrocyte-specific regulatory elements.
The NFI family transcription factors have recently emerged as important regulators of gliogenesis (13, 16, 17). These proteins, encoded by four genes highly conserved from chickens to humans (Nfia, Nfib, Nfic, and Nfix), regulate the transcription of various cellular and viral genes as well as viral DNA replication (16, 18). In vertebrates, products of these genes (NFI-A, -B, -C, and -X) share conserved N-terminal DNA binding and dimerization domains, followed by a subtype-specific domain and a variable C-terminal transactivation domain (19).
In mammals, the NFI genes are expressed in overlapping patterns during embryogenesis, with high levels of expression of NFI-A, -B, and -X in the developing neocortex (20). The Nfia and Nfib knock-out mice are characterized by neuroanatomical defects, including agenesis of the corpus callosum, loss of specific midline glial populations, and a 5–10-fold decrease in the expression of glial fibrillary acidic protein (GFAP), which is an astrocyte marker (11, 12, 21). In addition, Nfib-deficient mice present aberrant hippocampus and pons formation and die due to the defects in lung development (11). The immature nestin-positive glia populate the hippocampus of Nfib-deficient mice but fail to mature into GFAP-positive astrocytes (22). In contrast, disruption of the Nfic gene results in early postnatal defects in tooth formation, including the loss of molar roots and aberrant incisor development (23). The Nfix knock-out mice have recently been generated by two independent groups (24, 25), which reported multiple effects (26). The Nfix gene knock-out causes postnatal lethality in most of the animals and leads to hydrocephalus and partial agenesis of the corpus callosum (24). These mice also develop a deformation of the spine with kyphosis, due to a delay in ossification of vertebral bodies and a progressive degeneration of intervertebral disks. However, Nfix knock-out mice survive on a soft chow diet but are characterized by increased brain weight, expansion of the brain along the dorsal ventrical axis, and aberrant formation of the hippocampus (25). In summary, the knock-out phenotypes suggest that NFI-A, -B, and -X are important for normal brain development; however, the identity of the affected cell type(s) is not clear. Recently, both NFI-A and -B were shown to regulate gliogenesis in the chick embryo, with NFI-A also controlling the maintenance of neural precursors (27). More recently, NFI-C and -X were shown to regulate the expression of late astrocyte markers during the differentiation of neural precursors into astrocytes in vitro (28).
Alternative splicing is a common mechanism of generating transcription factors with diverse functions in the brain (29–32). Accordingly, transcripts of all four NFI genes are alternatively spliced, yielding many different proteins from a single gene (33, 34) with several different splice variants of NFI-X identified in human cells (35). Here, we cloned and characterized a novel human NFI-X splice variant X3 (NFI-X3), which regulates gene expression in primary human astrocytes. This splice variant contains a unique transcriptional activation (TA) domain that is remarkably conserved in mammals, including mice, rats, dogs, macaques, and humans. Mechanistically, the TA domain of NFI-X3 activates GFAP expression, in part, by inducing alterations in the nucleosome architecture that lead to the increased recruitment of RNA polymerase II.
EXPERIMENTAL PROCEDURES
Cell Culture
Human BG01V embryonic stem cells (ATCC, Manassas, VA) were cultured on a mitomycin C-inactivated mouse embryonic fibroblast layer. Cells were cultured in DMEM/F-12 medium, supplemented with 20% knock-out serum replacement (Invitrogen), 1 mm l-glutamine, 0.1 mm nonessential amino acids, 50 units/ml penicillin, 50 μg/ml streptomycin, 4 ng/ml basic fibroblast growth factor (PeproTech, Rocky Hill, NJ), and 0.1 mm β-mercaptoethanol. Cells were propagated in 4-day cycles and enzymatically passaged with collagenase and trypsin. Human cortical astrocyte cultures were established using dissociated human cerebral tissue as described previously (36). Cortical tissue was provided by Advanced Bioscience Resources (Alameda, CA), and the protocol for obtaining postmortem fetal neural tissue complied with the federal guidelines for fetal research and with the Uniformed Anatomical Gift Act. Human glioblastoma U373-MG cells and human embryonic kidney HEK293 cells were obtained from the ATCC, whereas human glioma U87, U251, D54, and T98G cells were obtained from Dr. Jaharul Haque (Cleveland Clinic Foundation, Cleveland, OH). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, antibiotics, sodium pyruvate, and non-essential amino acids.
Generation of Neural Progenitors
BG01V neural progenitors were generated exactly as described previously (28). Briefly, cells were grown in DMEM/F-12, supplemented with 15% defined FBS (Hyclone, Logan, UT), 5% knockout serum replacement, 2 mm l-glutamine, 0.1 mm nonessential amino acids, 50 units/ml penicillin, 50 μg/ml streptomycin, 4 ng/ml basic fibroblast growth factor, and 10 ng/ml leukemia inhibitory factor (Chemicon, Temecula, CA). Subsequently, cells were grown for 7 days in differentiation medium composed of DMEM/F-12, N2 supplement (Invitrogen), 2 mm l-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, and 4 ng/ml basic fibroblast growth factor. Subsequently, the mouse embryonic fibroblast layer was removed, and cells were dissociated by Dulbecco's PBS without calcium and magnesium, followed by collagenase and trypsin incubation. Cells were then plated on laminin- and poly-l-ornithin-coated dishes and propagated in NB27 medium consisting of neurobasal medium, B27 supplement (both from Invitrogen), 2 mm l-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, 20 ng/ml basic fibroblast growth factor, and 10 ng/ml leukemia inhibitory factor. Cells were maintained by passaging using trypsin.
Differentiation into Astrocytes
To generate astrocytes, neural progenitors were cultured on laminin- and poly-l-ornithin-coated dishes in DMEM, supplemented with 10% FBS, 2 mm l-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, and nonessential amino acids for 21–24 days.
Down-regulation of Target Genes
Expression of NFI-X mRNAs was down-regulated using SmartPool siRNAs (si-panNFI-X) from Dharmacon (Lafayette, CO). The NFI-X3-specific siRNAs (si-NFI-X3) was designed (software provided on the Dharmacon Web site) to specifically target mRNA encoded by exon 9 (5′-UCCUAUGCCUGAUUCCAAAUU-3′). siRNA was transfected into astrocytes using Dharmafect 1, according to the manufacturer's instructions.
RNA Isolation and Quantitative PCR
RNA was prepared by TRIzol (Invitrogen), following the manufacturer's protocol. Subsequently, 1 μg of total RNA was reverse-transcribed using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). GFAP, SPARCL1 (secreted protein acidic and rich in cysteine-like protein 1), NFI-X, and GAPDH mRNA levels were measured using premixed primer-probe sets, and TaqMan Universal PCR Master Mix according to the supplier's instructions (Applied Biosystems, Foster City, CA). The cDNAs were diluted 10-fold (for the target genes) or 100-fold (for GAPDH), and amplified using the ABI 7900HT cycler. Gene expression levels were normalized to GAPDH mRNA levels and presented as -fold induction with mean values ± S.D. Statistical analysis was performed by one-way analysis of variance. Differences were considered statistically significant when p values were <0.05. The expression of NFI-X3 splice isoform was analyzed using the Power SYBR Green PCR kit (Applied Biosystems, Foster City, CA) and the following primers: 5′-GTCGCTCGAGTCAG AGGAACCAGGACTG-3′ and 5′-GGTAGCGGCCAGGGCAAAG-3′ (the exon 9-specific primer).
Western Blotting
Cells were lysed in 10 mm Tris, pH 7.4, 150 mm sodium chloride, 1 mm EDTA, 0.5% Nonidet P-40, 1% Triton X-100, 1 mm sodium orthovanadate, 0.2 mm PMSF, and protease inhibitor mixture (Roche Applied Science). Samples were resolved using SDS-PAGE and electroblotted onto nitrocellulose membranes (Schleicher & Schuell). The anti-NFI, anti-GFAP, and anti-β-tubulin antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), whereas anti-SPARCL1 antibodies were purchased from R&D Systems (Minneapolis, MN). Antigen-antibody complexes were visualized by enhanced chemiluminescence using an Immobilon Western blotting kit (Millipore).
Synthetic Oligonucleotides
The following oligonucleotides were synthesized to amplify the NFI-X1 from the astrocyte cDNA: 5′-CGGAAGCTTCGCCATGTACTCCCCGTACTGCC-3′ and 5′-TGTGCTCGAGTCAGAAAGTTGCCGTCCCG-3′. The NFI-X3 was amplified in two steps to avoid the amplification of NFI-X1, which is the major splice isoform. The N-terminal part of NFI-X3 was amplified with the primers 5′-GTGAAGCTTCGCCATGGATGAGTTCCACCCGTTC-3′ and 5′-GGTGATGGAGGAGTCAAG-3′, whereas the C terminus was amplified with 5′-GGTGCGGCCAGGGCAAAG-3′ and 5′-GTCGCTCGAGTCAGAGGAACCAGGACTG-3′. Subsequently, both PCR products were combined, and the full-length NFI-X3 was amplified with the primers containing the HindIII and XhoI sites (underlined above). The NFI-X3Δ lacking the NFI-X3 transactivation domain was amplified using the N-terminal NFI-X3 and 5-TACCCTCGAGTCACTGTCCGGTGGCCTGGC-3′ primers. The transactivation domains of NFI-X1 and NFI-X3 were amplified using the following primers: 5′-AGGGGATCCTGAAG GAGTTTGTGCAGTTT-3′ and 5′-GTGTCTAGATCAGAAAGTTGCCGTC-3′ (X1TD) and 5′-AGGGGATCCTGAAGGAGTTTGTGCAGTTT-3′ and 5′-TCGTCTAGATCAGAGGAACCAGGACTGAG-3′ (X3TD). To generate plasmids expressing NFI-X1-FLAG and NFI-X3-FLAG, the following reverse primers were used: 5′-TACCAGATCTGAAAGTTGCCGTCCCGGGGTC-3′ and 5′-TACCAGATCTGAGGACCAGGACTGAGAC-3′.
Bisulfite Sequencing
250 ng of genomic DNA were converted using the EZ DNA methylation kit (Zymo Research, Orange, CA) according to the supplier's instructions. Subsequently, PCR products were amplified using a pair of primers: 5′-TGTAAGTAGATTTGGTAGTATTGGGTTGG-3′ and 5′-CCCTTTCCTAAACACAAACTAAATAAAACC-3′. Products were cloned into pCR2.1 (Invitrogen), and five clones were sequenced for each PCR product.
Plasmids
Reporter plasmids p10x(NFI)CAT and pGFAP(−1745)CAT were described previously (37). The p21(−2400)Luc reporter was provided by Dr. Bert Vogelstein (Baltimore, MD) (38). The p5xGAL4-Luc reporter and pCGal4 plasmid (pCDNA3.1 containing the Gal4 DNA-binding domain) were gifts of Dr. Joseph Fontes (Kansas City, KS) (39, 40). The PCR products containing full-length NFI-X1, NFI-X3, and NFI-X3 lacking its TA domain (NFI-X3Δ) were digested with HindIII and XhoI and cloned into the HindIII/XhoI sites of pcDNA5TO (Invitrogen). To generate vectors expressing full-length NFI-X1-FLAG and NFI-X3-FLAG, the PCR products were digested with HindIII and BglII and cloned into the HindIII/BamHI sites of pCMV5aFLAG (Sigma). Subsequently, HindIII/XhoI fragments were cloned into the HindIII/XhoI sites of pcDNA5TO (Invitrogen). The PCR products containing transactivation domains of NFI-X1 and NFI-X3 were digested with BamHI and XbaI and cloned into the BamHI/XbaI sites of pCGal4.
Transfections
Cells were transiently transfected in 12-well clusters using FuGENE6 transfection reagent (Roche Applied Science) according to the supplier's instructions. One day after transfection, the cells were harvested, protein extracts were prepared, and the protein concentration was determined by the BCA method (Sigma). Chloramphenicol acetyltransferase (CAT) and β-galactosidase assays were performed as described (41). Luciferase (Luc) assays were performed using a luciferase reporter assay kit (Promega Corp., Madison, WI). CAT and Luc activities were normalized to β-galactosidase activity and are means ± S.E. (5–7 determinations). Differences were considered statistically significant when p values were <0.05. To generate stable clones, 4 μg of NFI-X1, NFI-X1-FLAG, NFI-X3, or NFI-X3-FLAG expression plasmids were transfected into a 10-cm dish of U373 cells. Clones selected in DMEM containing 75 μg/ml hygromycin were subsequently pooled. Nucleofection was used to transiently transfect U373 cells. Briefly, the cells (0.5 × 106/well of a 6-well dish) were trypsinized, collected by centrifugation, and resuspended in 50 μl of T NucleofectorTM solution (Amaxa, Inc., Gaithersburg, MD). 0.8 μg of the respective plasmids were added to the solution, and transfection was performed using the Nucleofector device (Amaxa, Inc., Gaithersburg, MD) with the electrical setting of T-20. One ml of warm DMEM was added, and cells were incubated at 37 °C for 10 min and transferred to 6-well plates containing culture medium. Typical efficiencies were greater than 70% with cell viability close to 90%.
Chromatin Immunoprecipitation (ChIP) Assay
Cells were cross-linked with 1% formaldehyde for 10 min at 37 °C and then washed with ice-cold PBS containing 125 mm glycine and 1 mm PMSF. Chromatin was sonicated and immunoprecipitated using specific antibodies exactly as described in the chromatin immunoprecipitation protocol from Upstate Inc. (Charlottesville, VA). The following antibodies were used: anti-acetyl-histone H3 (06-599, Millipore), anti-trimethyl-histone H3 Lys4 (07-473, Upstate, Inc.), anti-Pol II (05-623, Millipore), and anti-FLAG (M2) (Sigma). The following primers were used in the qPCR: 5′-GCATCGCCAGTCTAGCCC-3′ and 5′-ATTCGAGCCAGGGAGAGGC-3′ for the GFAP promoter and 5′-TGCAAGCAGACCTGGCAGC-3′ and 5′-GCAAGCCCCCTGCTCAATG-3′ for the GFAP enhancer.
MNase Protection Assay
Three 10-cm dishes of cells were used for nucleosome mapping. Briefly, cells were trypsinized, washed with cold PBS, and resuspended in 2 ml of 60 mm KCl, 15 mm NaCl, 5 mm CaCl2, 10 mm Tris, pH 7.4, 300 mm sucrose, 0.1 mm EDTA, and 0.1% Nonidet P-40. Cells were lysed with a Dounce homogenizer, and nuclei were pelleted by centrifugation and subsequently washed with 2 ml of MNase digestion buffer (10 mm Tris, pH 7.4, 15 mm NaCl, 60 mm KCl, 0.15 mm spermine, 0.5 mm spermidine, and 1 mm CaCl2). Nuclei were resuspended in 150 μl of MNase digestion buffer (with 3 mm CaCl2), 10 units of MNase was added, and samples were incubated for 1 h at 37 °C. DNA was purified using Puregene DNA purification kit (Qiagen, Valencia, CA). DNA was analyzed by gel electrophoresis (>95% typically represented mononucleosomal DNA) and subsequently quantified by qPCR using the Power SYBR Green PCR kit (Applied Biosystems, Foster City, CA) and the following primers: F1, 5′-CAGTGGGGTGAGGGGAG-3′; F2, 5′-GCCTCTGGGCACAGTGAC-3′; F3, 5′-GGCGTGCCCAGGAAGCTC-3′; F4, 5′-CACCTGCCTCATGCAGGAG-3′; F5, 5′-ATG GTCCAACCAACCCTTTC-3′; R1, 5′-GCGAGGGCTTTATGAAGG-3′; R2, 5′-TGGGCTAGACTGGCGATG-3′; R5, 5′-GAGGCAGGTGGTACCTGG-3′; and −3kb, 5′-GGTCTGAGGGTTGAGAAGG-3′ and 5′-CCCTGTGTATGTGAGACAC-3′.
RESULTS
NFI-X3 Splice Variant Expressed in Astrocytes and Glioma Cells Is Highly Conserved in Mammals
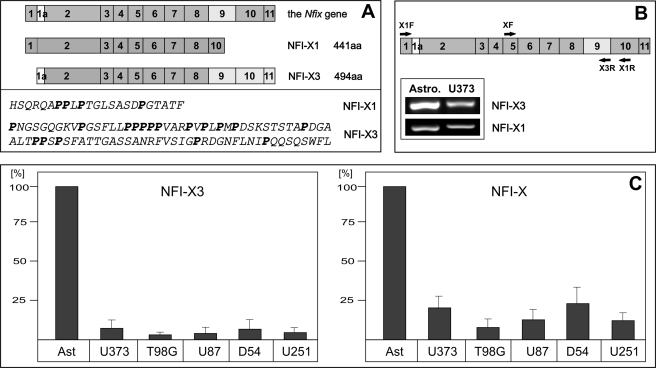
The structure of the Nfix gene is highly conserved in vertebrates, including Xenopus, rodents, canines, and primates. Moreover, the NFI-X mRNA is alternatively spliced in these species, producing several splice variants that have either been cloned or are predicted by the in silico analysis (reviewed in Ref. 35). In vertebrates, one of the splice variants (which we named NFI-X3 due to the homology of its C-terminal part to the previously cloned Xenopus NFI-X3) is generated by the inclusion of exon 9, which is absent in other splice variants, including NFI-X1 (see Fig. 2A). The incorporation of exon 9 in the NFI-X3 mRNA generates a frameshift placing the STOP codon of the alternative exon 11 in frame. As a result, the translated NFI-X3 protein possesses a long proline-rich C-terminal transcription activation (TA) domain, which is unique to this splice variant (Figs. 1 and 2A). On the amino acid level, the TA domain of NFI-X3 is 100% conserved in primates and dogs, whereas it differs by only 3 and 4 amino acids in mice and rats, respectively (Fig. 1). In contrast, the NFI-X3 TA domain of Xenopus differs by 36 amino acids, including a 3-amino acid-long insertion (Fig. 1). The functional properties of Xenopus NFI-X3 are not substantially different from the NFI-X1 and NFI-X2 splice variants (19). However, the absolute conservation of the TA domain of NFI-X3 in primates (and dogs) and over 95% identity in rodents, suggested that it may have acquired novel functions during the evolution of mammals. Because the Nfix gene is important for mouse brain development and regulates gene expression in human astrocytes, we analyzed whether NFI-X3 is expressed by these cells. In fact, NFI-X3 was expressed by primary human astrocytes and at much lower levels (5–10%) by glioma cell lines, which represent cells that escaped the astrocyte differentiation process (Fig. 2, B and C). Interestingly, glioma cells expressed low levels of all NFI-X transcripts, with the ratio of NFI-X3 to total NFI-X significantly reduced (by ∼60%), thus suggesting that NFI-X3 may regulate gene expression during the differentiation toward astrocytes, and therefore its expression in glioma cells is greatly diminished. In order to analyze the functions of NFI-X splice variants, we amplified NFI-X3 and NFI-X1 from primary human astrocytes. The C-terminal TA domains of astrocyte-derived NFI-X1 and NFI-X3 were identical to the software-predicted domains. However, in contrast to Xenopus NFI-X3 and human NFI-X1, the N terminus of human astrocyte-derived NFI-X3 is encoded by the alternative exon 1a, which results in a deletion of eight amino acids (Fig. 2A). Thus, the unique conserved NFI-X3 splice variant is expressed by astrocytes, and it may be important for the differentiation of these cells.
FIGURE 2.
NFI-X3 is expressed by astrocytes and glioma cells. A, a model of exon composition of the nfix gene and the NFI-X1 and NFI-X3 transcripts (top) and the amino acid (aa) sequences of the predicted TA domains (bottom). B and C, RNA was isolated from primary human astrocytes and the indicated glioma cell lines and reverse transcribed. B, NFI-X1 and NFI-X3 have been detected in both astrocytes and U373 glioma cells using the indicated isoform-specific primers (XF and X3R for NFI-X3, X1F and X1R for NFI-X1). C, the expression levels of NFI-X3 (left) (SYBR Green qPCR) and total NFI-X (right) (TaqMan qPCR) were analyzed as described under “Experimental Procedures.” Data were normalized to GAPDH mRNA and are presented as a percentage to human astrocytes set as 100%. Experiments were repeated twice in duplicate. Error bars, S.D.
FIGURE 1.
Alignment of the amino acid sequences of the TA domains of NFI-X3. Sequences were aligned with the ClustalW program. Dashes, gaps introduced to maximize the similarity; light gray boxes, identical sequences; dark gray boxes, sequences that are similar. The full-length sequence of human NFI-X3 was deposited into GenBankTM (FJ861276). The following sequences were aligned: Macaca fasicularis (AB172261), Macaca mulatta (XM_001110545), Canis familiaris (XM_542036), Xenopus laevis (52), Homo sapiens (AAD38241), Mus musculus (NM_001081981), and Rattus norvegicus (EDL92203).
The TA Domain of NFI-X3 Is Critical for the Regulation of Gene Expression in Human Astrocytes and Glioma Cells
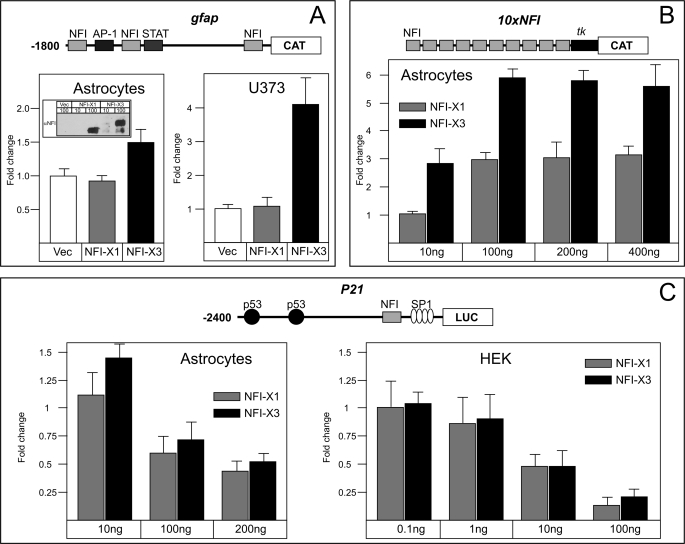
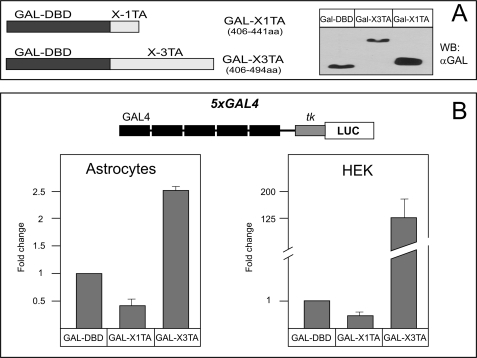
In order to analyze the transactivation potential of NFI-X3, we used several reporters that respond to NFI proteins (9, 37, 42). These reporters were analyzed in transient transfection experiments of primary human astrocytes, glioblastoma, and HEK293 cells (Fig. 3). In addition to NFI-X3, we also used NFI-X1 to compare the transactivation by these two NFI-X splice variants. Significantly, NFI-X3 but not NFI-X1 activated the GFAP reporter in both astrocytes and glioblastoma cells (Fig. 3A); however, this reporter was not functional in HEK293 cells (data not shown), indicating the requirement for the astrocyte/glioma-specific factors. Because expression levels of both NFI-X3 and NFI-X1 were comparable (Fig. 3A, inset), we conclude that NFI-X3 may be critical for the transcription of GFAP in glial cells. In fact, the artificial NFI reporter, containing 10 NFI binding sites linked to the minimal tk promoter (37), was similarly activated by 10-fold lower amounts of NFI-X3 than NFI-X1 in human astrocytes (Fig. 3B). In addition to the activator functions, NFIs also repress the expression of several genes, including p21(WAF1/CIP1) (42). Surprisingly, both NFI-X3 and NFI-X1 efficiently repressed the expression of the p21 reporter in astrocytes as well as HEK293 cells (Fig. 3C). These data suggest that the activation by NFI-X3 in glial cells may depend on its unique TA domain; however, the repression is probably mediated by a different part of the molecule, because the TA domains of NFI-X1 and NFI-X3 are very different (Fig. 2A). To test these TA domains, we fused them to the Gal4 DNA-binding domain (Fig. 4A) and co-transfected these constructs into astrocytes and HEK293 cells with the reporter plasmid p5xGAL4-luc, which contains five Gal4 binding sites linked to the firefly luciferase gene (40). The fusion protein containing the TA domain of NFI-X1 could not induce the Gal4 reporter either in astrocytes or HEK293 cells (Fig. 4B). In contrast, the fusion protein containing the TA domain of NFI-X3 activated the Gal4 reporter in both cell types (2.5- and 125-fold) (Fig. 4B). Thus, the TA domain of NFI-X3 substantially differs in function from the one present in NFI-X1 and may regulate the expression of target genes in astrocytes.
FIGURE 3.
Differential effects of NFI-X3 and NFI-X1 on various gene reporters. Primary human astrocytes and HEK293 cells were transiently transfected, whereas U373 cells were nucleofected as described under “Experimental Procedures.” Cells were transfected or nucleofected with 100 ng of the indicated reporter plasmids, 400 ng of NFI-X1 or NFI-X3 expression vectors (A), or the indicated amounts of the expression vectors (B and C) as well as 50 ng of β-galactosidase expression vector. One day after transfection, cells were harvested. CAT or Luc activities were normalized to β-galactosidase activities to account for transfection efficiency. The putative binding sites for NFI, AP-1, p53, and SP1 are depicted in the schematic diagrams representing the reporters. tk, a thymidine kinase minimal promoter. The inset shows the expression levels of NFI-X1 and NFI-X3. Experiments were performed three times (astrocytes) or two times (U373 and HEK cells) in duplicate. Error bars, S.D.
FIGURE 4.
Comparison of the activities of TA domains of NFI-X1 and NFI-X3. Primary human astrocytes and HEK cells were transiently transfected with 100 ng of p5xGAL4-Luc, 400 ng of the indicated expression plasmids, and 50 ng of β-galactosidase expression vector. One day after transfection, cells were harvested. (B) Luc activities were normalized to β-galactosidase activities to account for transfection efficiency. (A) The inset shows the expression levels of fusion proteins. Experiments were performed three times in duplicate. DBD, DNA-binding domain. Error bars, S.D.
Regulation of NFI-X3 Expression during Astrocyte Differentiation
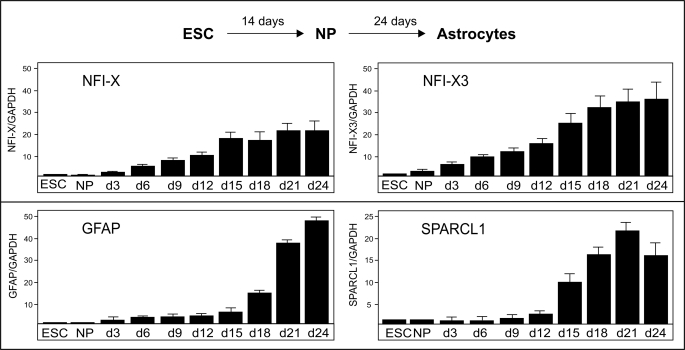
We have recently developed an in vitro astrocyte differentiation model that allows for the analysis of mechanisms regulating astrocyte-specific gene expression (28). We used BG01V embryonic stem cells and differentiated them into neural progenitors and thereafter to astrocytes (Fig. 5). Embryonic stem cells and neural progenitors expressed only minute amounts of NFI-X (all NFI-X transcripts), including NFI-X3 (Fig. 5, top right). However, differentiation of neural progenitors to astrocytes resulted in the significant activation of NFI-X expression (∼20-fold after 21 days), with even greater expression of the NFI-X3 (∼37-fold in 21 days). The expression of NFI-X3 preceded the expression of two markers of late astrocyte differentiation, GFAP and SPARCL1 (Fig. 5, bottom), therefore suggesting that NFI-X3 may regulate gene expression during the final stages of astrocyte differentiation.
FIGURE 5.
NFI-X3, GFAP, and SPARCL1 are coexpressed during the differentiation of astrocytes. Embryonic stem cells (ESC) were differentiated into neural progenitors (NP) and then into astrocytes for 24 days as described under “Experimental Procedures.” RNA was isolated during this process every 3 days and reverse-transcribed, and the expression of NFI-X3 (SYBR Green qPCR), total NFI-X, GFAP, and SPARCL1 (all TaqMan qPCR) was analyzed by qPCR. Target gene expression was normalized to GAPDH levels and is represented as -fold induction in reference to embryonic stem cells. Experiments were performed three times in triplicate. Error bars, S.D.
NFI-X3 Specifically Regulates the Expression of GFAP and SPARCL1 in Human Astrocytes and Glioma Cells
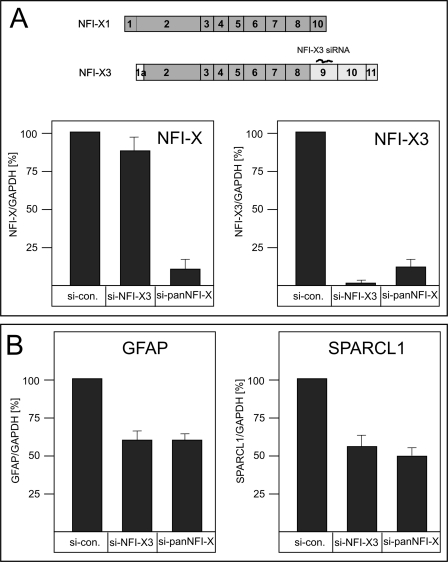
In order to evaluate whether NFI-X3 regulates the expression of GFAP and SPARCL1, we designed siRNA specifically targeting exon 9 of the nfix gene (Fig. 6A). For comparison, NFI-X SMARTPool siRNA was used to knock down the expression of all NFI-X splice variants. In primary human astrocytes, the siRNA targeting exon 9 of the nfix gene efficiently and specifically knocked down the expression of NFI-X3 (by 95%); however, this siRNA only diminished the expression of total NFI-X by 10% (Fig. 6A) and had limited effect on the expression of NFI-A, -B, and -C (supplemental Fig. 1). These results indicate that mRNA encoding NFI-X3 constitutes ∼10% of the total pool of the NFI-X mRNAs in astrocytes. More importantly, the NFI-X3 knockdown resulted in the inhibition of GFAP and SPARCL1 expression by 40 and 50%, respectively, which is almost identical to the repression found using siRNA targeting the total pool of NFI-X (Fig. 6B). Importantly, expression of NFI-X-independent genes (43), including CD44, vimentin, and S100β, was not significantly affected by these siRNAs (supplemental Fig. 1).
FIGURE 6.
NFI-X3 regulates the expression of GFAP and SPARCL1 in primary human astrocytes. Human astrocytes were transfected with 10 nm non-targeting siRNA (si-con), 10 nm siRNA targeting NFI-X3 (si-NFI-X3), or 10 nm siRNA targeting all NFI-X transcripts (si-panNFI-X). RNA was isolated after 48 h, and the expression of NFI-X3 (SYBR Green qPCR) and total NFI-X (A) and GFAP and SPARCL1 (B) (all TaqMan qPCR) was analyzed in triplicate (two experiments). Data were normalized to GAPDH mRNA and are presented as a -fold induction in reference to untreated astrocytes. Error bars, S.D.
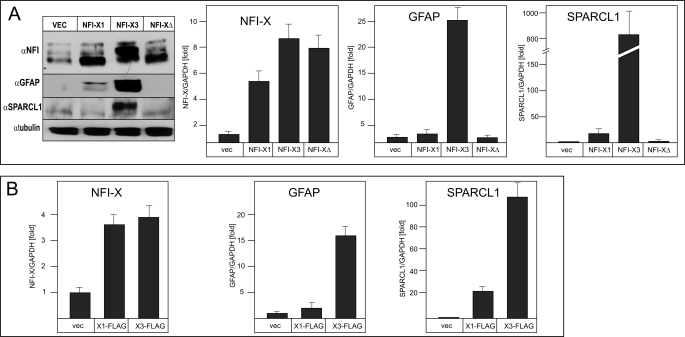
Next we generated NFI-X3 lacking its TA domain (NFI-X3Δ) and analyzed whether stable overexpression of NFI-X3, NFI-X3Δ, and NFI-X1 can induce the expression of GFAP and SPARCL1 in glioma cells. We have chosen U373 cells, which express relatively low levels of both of these markers, and generated pools of cells overexpressing NFI-X3, NFI-X3Δ, and NFI-X1 (Fig. 7A). Neither the morphology nor growth of U373 cells was affected by the overexpression of NFI-X1 or NFI-X3 (supplemental Fig. 2). However, cells overexpressing NFI-X1 and NFI-X3 up-regulated the expression of GFAP and SPARCL1 (Fig. 7A). Significantly, overexpression of NFI-X3 resulted in massive induction of both markers, whereas overexpression of NFI-X1 had a much lower effect (Fig. 7A). In contrast, NFI-X3Δ did not induce GFAP or SPARCL1 expression, proving that TA domains of NFI-X1 and NFI-X3 are indispensable for the activation. Significantly, overexpression of NFI-X1 or NFI-X3 did not affect expression of CD44, vimentin, and S100β (supplemental Fig. 2). The data of the knockdown and overexpression experiments suggest that NFI-X3, although accounting for only 10% of total NFI-X but possessing much greater transactivation potential than NFI-X1, regulates the expression of GFAP and SPARCL1 in both human astrocytes and glioma cells.
FIGURE 7.
Overexpression of NFI-X3 but not NFI-X3Δ dramatically induces the expression of GFAP and SPARCL1 in glioma cells. Pools of U373 stable clones transfected with plasmids expressing either NFI-X1, NFI-X3, NFI-X3Δ, or empty vector were cultured as described under “Experimental Procedures.” RNA and cell lysates were prepared from the duplicate cultures, and media were collected. A, the expression of NFIs, GFAP (cell lysates), and SPARCL1 (medium) was analyzed by Western blotting. RNA was reverse transcribed, and the expression of total NFI-X, GFAP, and SPARCL1 was analyzed in duplicate by qPCR (two experiments). B, pools of U373 stable clones transfected with plasmids expressing either NFI-X1-FLAG, NFI-X3-FLAG, or empty vector were cultured as described under “Experimental Procedures.” The expression of NFI-X, GFAP, and SPARCL1 was analyzed by qPCR as described above. Error bars, S.D.
Alterations in the Nucleosome Architecture at the GFAP Promoter Are Strongly Induced by the Unique TA Domain of NFI-X3
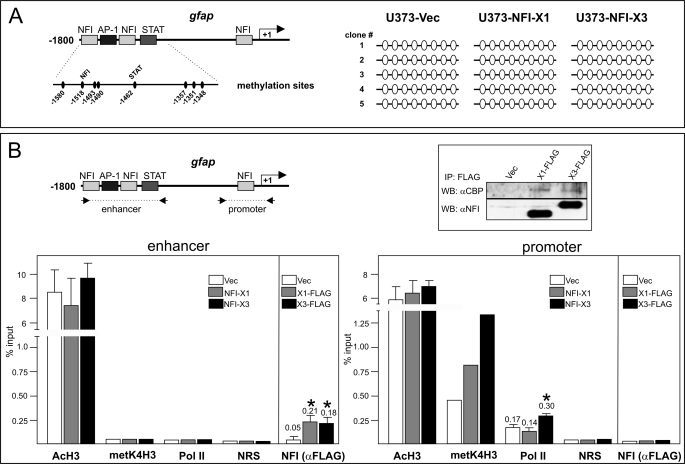
Recently, NFI-A has been shown to induce dissociation of DNA methyltransferase 1 from the gfap regulatory region and demethylation of the STAT3 binding site that is critical for the induction of GFAP expression (13). To examine whether NFI-X3 activates GFAP expression via a similar mechanism, we analyzed methylation of CpGs at the gfap gene in the pools of cells expressing either vector, NFI-X3 or NFI-X1 (Fig. 8A). However, all of the analyzed CpG, including the one located within the STAT3 binding element, were demethylated in vector, NFI-X3, and NFI-X1 clone pools. These data excluded DNA demethylation as a regulatory mechanism of NFI-X3-induced expression of the gfap gene. The recruitment of histone acetyltransferases by the STAT3-SMAD1 complex followed by acetylation of histones at the promoters of astrocyte-specific genes is another mechanism of their activation (14). However, despite drastically different activation of the gfap gene by NFI-X3 and NFI-X1 (Fig. 7), both of these splice isoforms similarly interacted with CBP (Fig. 8B, inset). Furthermore, histone acetylation at the gfap enhancer as well as the core promoter was comparable in vector, NFI-X3, and NFI-X1 clone pools as determined by ChIP analysis (Fig. 8B), excluding histone acetylation as a mechanism of activation. Nevertheless, the recruitment of RNA PolII to the gfap promoter was significantly increased in NFI-X3-expressing cells, which also correlated with the increased histone H3 Lys4 trimethylation, a conserved mark of actively transcribed chromatin (Fig. 8B). The increased recruitment of RNA PolII could be a result of stronger binding of NFI-X3 than NFI-X1 to the NFI binding elements. To examine the binding of both of these splice variants at the gfap regulatory regions, we generated U373 pools expressing FLAG-tagged NFI-X1 and NFI-X3. As expected, GFAP and SPARCL1 expression was strongly activated in NFI-X3-FLAG-expressing cells (Fig. 7B). Nevertheless, binding of both NFI-X1-FLAG and NFI-X3-FLAG to the gfap enhancer was similar (Fig. 8B), whereas they did not bind to the gfap promoter as previously reported (37).
FIGURE 8.
NFI-X3 induces the recruitment of RNA PolII to the gfap promoter. A, DNA was isolated from U373-Vec, U373-NFI-X1, and U373-NFI-X3 cells and bisulfite-converted, and methylation of suitable CpG was analyzed as described under “Experimental Procedures.” Empty circles indicate demethylated CpGs. B, lysates from U373-Vec, U373-NFI-X1-FLAG, and U373-NFI-X3-FLAG cells were immunoprecipitated with anti-FLAG antibodies, and precipitates were analyzed by Western blotting using anti-CBP and anti-NFI antibodies (inset). ChIP was performed using chromatin prepared from U373-Vec, U373-NFI-X1, and U373-NFI-X3 cells or from U373-Vec, U373-NFI-X1-FLAG, and U373-NFI-X3-FLAG cells as indicated. Both the gfap enhancer and core promoter were analyzed for the presence of acetylated histone H3, trimethylated histone H3 Lys4, RNA PolII, and NFI-X1 or NFI-X3 (anti-FLAG) using the antibodies described under “Experimental Procedures.” NRS, normal rabbit serum used for immunoprecipitation. Results are shown as a percentage of input. Experiments were performed three times. Error bars, S.D.
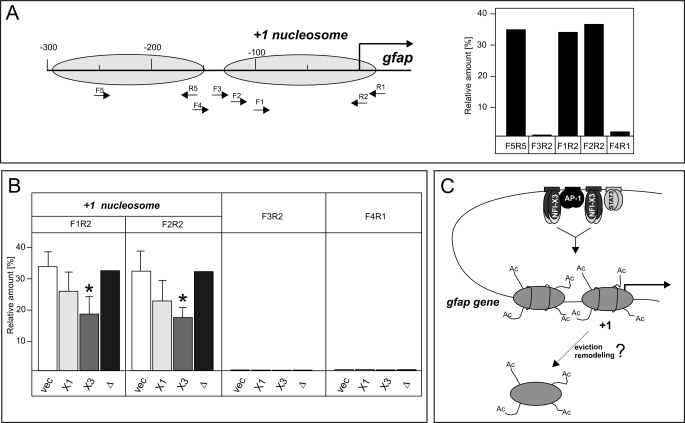
Nucleosomes are an impediment to transcription, and therefore they are evicted or remodeled during gene activation (44, 45). To understand the mechanism of NFI-X3-induced activation of GFAP expression, we analyzed nucleosome positioning and occupancy at the gfap promoter using MNase protection assay combined with qPCR (46). The +1 nucleosome was positioned at approximately −130 to +20 relative to the gfap transcription start site in vector, NFI-X3, NFI-X3Δ, and NFI-X1 clone pools (Fig. 9A) (data not shown). The upstream −1 nucleosome was also positioned at approximately −300 to −150 in these cells, indicating that both −1 and +1 nucleosomes are not repositioned in cells overexpressing NFI-X1, NFI-X3, or NFI-X3Δ. Subsequently, we analyzed the relative occupancy of the +1 nucleosome in all four cell types. Although the accessibility of DNA to MNase cleavage at the +1 nucleosome was increased in cells overexpressing NFI-X1 and NFI-X3, the accessibility was substantially greater in NFI-X3-overexpressing cells (Fig. 9B). Significantly, the accessibility of DNA to MNase cleavage of vector- and NFI-X3Δ-overexpressing cells was similar (Fig. 9B). Because NFI-X1 and NFI-X3 differ by their TA domains, we conclude that the unique TA domain of NFI-X3 induces stronger local changes in the promoter nucleosome architecture that allow for the increased RNA PolII recruitment (Fig. 8B) and subsequent activation of GFAP expression.
FIGURE 9.
NFI-X3 induces alterations in the nucleosome architecture at the gfap promoter. Nuclei were isolated from U373-Vec, U373-NFI-X1, U373-NFI-X3, and U373-NFI-X3Δ cells and digested with MNase as described under “Experimental Procedures.” A, the positions of mapped −1 and +1 nucleosomes are indicated, and locations of qPCR primers are shown. qPCRs were performed on both MNase-treated and total genomic DNA of U373-Vec cells, and the differences in Ct values are shown relative to the −3kb qPCR product (Ct values for −3kb were identical for genomic and MNase-treated DNA). B, qPCRs were performed on MNase-treated DNA of U373-Vec (vec), U373-NFI-X1 (X1), U373-NFI-X3 (X3), and U373-NFI-X3Δ (Δ) cells as indicated, and the differences in Ct values are shown relative to the −3kb qPCR product. Experiments were performed four times except for U373-NFI-X3Δ cells (one experiment). C, model of transcriptional activation of the gfap gene expression by NFI-X3. DNA is demethylated at the gfap enhancer, which allows for binding of the AP-1, NFI-X1/NFI-X3, and STAT3 in U373-NFI-X1/NFI-X3 cells. At the gfap promoter, histones are similarly acetylated in U373-NFI-X1 and U373-NFI-X3 cells. However, the unique TA domain of NFI-X3 induces stronger alteration of the nucleosome architecture (either remodeling or eviction) that allows for both the increased PolII occupancy and transcription of the gfap gene. Error bars, S.D.
DISCUSSION
It is apparent that NFIs are critical for the proper development of the brain; nevertheless, the vital target genes regulated by NFIs and the molecular mechanisms of their regulation remain elusive. To date, multiple NFI-responsive genes have been identified in the brain, including the genes encoding GFAP, ACT, SPARCL1, and S100B in astrocytes (28, 37, 47, 48); brain fatty acid binding protein in radial glial cells (7); myelin basic protein in oligodendrocytes (49); and midsized neurofilament in neurons (50). Because regulatory sequences of these genes can bind all NFIs, their transcriptional regulation in a given cell type, in part, depends on the availability of a particular NFI isoform, the presence of the specific splice variant, and probably the acquired post-transcriptional modifications of the NFIs. In fact, NFIs are expressed in complicated patterns during embryogenesis (20, 51), with NFI-A and NFI-B expressed during early gliogenesis (27), whereas NFI-X and NFI-C are expressed later in the differentiation of astrocytes and control the expression of late astrocyte markers (28). NFIs are also phosphorylated, and this modification affects their function (7).
To date, the functional importance of the multiple NFI splice variants as well as the mechanisms controlling their generation remain largely unknown. Significantly, the NFI splice variants are evolutionarily conserved in mammals, and their splicing mostly affects the C termini encoding their TA domains, thus suggesting that these variants probably fulfill very specific biological functions (35). In particular, the inclusion of the exon 9 found in the NFI-A, NFI-B, and NFI-C generates a subset of relatively active transcription factors (NFI-A1, NFI-B1, and CTF-1). In contrast to these isoforms, the importance of alternative splicing of the NFI-X transcript was controversial, because Xenopus and mouse splice variants containing exon 9 were reported not to be particularly active (19, 52). However, the TA domain of Xenopus NFI-X3 is drastically different from the TA of the X3 variants found in mammals (Fig. 1). Moreover, the repressive properties of the previously cloned mouse NFI-X3 can be attributed to the partial TA domain that lacks 14 C-terminal amino acids (52). In fact, the full-length TA domain of the NFI-X3 splice variant is remarkably conserved in mammals, with 100% identity in primates and dog, and contains 17 conserved proline residues (Fig. 1). These proline residues are probably responsible for the relatively strong activation of both the GFAP and NFI reporters by NFI-X3 in astrocytes and glioma cells (Fig. 3, A and B), activation of GFAP and SPARCL1 expression in differentiating astrocytes (Fig. 5), and activation of GFAP and SPARCL1 expression in cells overexpressing NFI-X3 (Fig. 7). In contrast, NFI-X3Δ, which lacks its proline-rich TA domain, did not activate GFAP and SPARCL1 expression when overexpressed (Fig. 7). The importance of the NFI-X3 TA domain is further supported by the findings that the Gal4-X3TA domain fusion protein efficiently activated Gal4 reporter (Fig. 4), whereas siRNA specific to exon 9 (present in NFI-X3) abolished the activation of GFAP and SPARCL1 expression (Fig. 6). In contrast, NFI-X1 did not activate the GFAP reporter in astrocytes and glioma cells (Fig. 3A), mildly activated the NFI reporter in astrocytes (with approximately 10 times lower efficiency than NFI-X3) (Fig. 3B), and relatively moderately activated SPARCL1 and GFAP expression when stably overexpressed in glioma cells (Fig. 7). Moreover, a fusion protein containing the NFI-X1 TA domain and the Gal4 DNA-binding domain did not activate the Gal4 reporter (Fig. 4). These data suggest that NFI-X3 serves as a strong transcriptional activator, whereas NFI-X1 is either a repressor or a relatively very weak activator. Surprisingly, both NFI-X1 and NFI-X3 repress some genes independently of their TA domains, as observed for the p21 reporter (Fig. 3C) and p21 mRNA in cells overexpressing NFI-X1 and NFI-X3 (data not shown); nevertheless, the mechanism of repression remains to be established. In contrast to the TA domain of NFI-X3, a deletion of 8 amino acids (exon 1A) did not affect NFI-X3 binding to the gfap enhancer (Fig. 8) or the repression of the p21 reporter (Fig. 3C).
The question remains as to whether NFI-X3 plays an important biological role. Our data argue that NFI-X3 is mainly responsible for the NFI-X-mediated regulation of GFAP and SPARCL1 expression in primary human astrocytes because the down-regulation of NFI-X3 expression has an identical effect as the down-regulation of expression of the total NFI-X pool (Fig. 6). Although NFI-X3 mRNA accounts for only ∼10% of the NFI-X mRNAs (Fig. 6A), the 10 times stronger activation potential (Fig. 3B) may explain its critical role in gene regulation. Moreover, the expression of NFI-X3 is strongly up-regulated during the differentiation of neural progenitors toward astrocytes and precedes the activation of GFAP and SPARCL1 expression (Fig. 5). Importantly, the ratio of NFI-X3 mRNA/total NFI-X mRNA increases ∼2-fold during the differentiation (Fig. 5). Conversely, this ratio is decreased in various glioma cells, already expressing low levels of all NFI-X transcripts (Fig. 2C), which may be viewed as cells that exited astrocyte differentiation. However, NFI-X3 expression is not restricted to glia because we were also able to detect NFI-X3 mRNA in other non-glial cells (data not shown), thus indicating that NFI-X3 probably regulates transcription in various cell types.
Although NFIs were discovered over 2 decades ago (53), the molecular mechanism of NFI-mediated gene activation still remains elusive. Interestingly, NFIs not only bind to their specific binding elements within the regulatory regions of target genes but also interact with other transcription factors (54) and histone H3 (55). Importantly, NFI-C (CTF-1) binding to histone H3, in part, depends on its TA domain (55). NFI-C can also be immunoprecipitated with remodeling complexes containing Brg1 (56), suggesting that chromatin remodeling may be the mechanism regulating NFI-dependent transcription. Recently, the induction of the c-fos gene by MAPK signaling has been shown to depend on the acetylation relay switch that induces a local change in the nucleosome architecture allowing for NFI binding (57). Nevertheless, how NFI recruitment increases c-fos expression is not clear. Our data allow us to postulate a novel model of gene activation that depends on the TA domain of NFI-X3. Our data suggest that binding of NFI-X3 (and to a lesser extent NFI-X1) to the gfap enhancer induces changes in the structure/occupancy of the nucleosomes positioned at the promoter of the gfap gene (Fig. 9B). This localized alteration is probably an eviction of the +1 nucleosome from the gfap promoter, which subsequently allows for the increased binding of the general transcription factors and the recruitment of PolII. In summary, our data suggest that NFI-X3 is a critical splice variant of the nfix gene, which regulates expression of late markers, such as GFAP, in astrocytes via a mechanism that depends, in part, on the induction of local alteration in chromatin architecture.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS044118 and R21NS063283 (to T. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) FJ861276.
- NFI
- nuclear factor 1
- CAT
- chloramphenicol acetyltransferase
- GFAP
- glial fibrillary acidic protein
- Luc
- luciferase
- TA
- transcriptional activation
- qPCR
- quantitative PCR
- MNase
- micrococcal nuclease
- PolII
- polymerase II.
REFERENCES
- 1. Miller F. D., Gauthier A. S. (2007) Neuron 54, 357–369 [DOI] [PubMed] [Google Scholar]
- 2. Barnabé-Heider F., Wasylnka J. A., Fernandes K. J., Porsche C., Sendtner M., Kaplan D. R., Miller F. D. (2005) Neuron 48, 253–265 [DOI] [PubMed] [Google Scholar]
- 3. Fukuda S., Abematsu M., Mori H., Yanagisawa M., Kagawa T., Nakashima K., Yoshimura A., Taga T. (2007) Mol. Cell Biol. 27, 4931–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomes W. A., Mehler M. F., Kessler J. A. (2003) Dev. Biol. 255, 164–177 [DOI] [PubMed] [Google Scholar]
- 5. Gaiano N., Nye J. S., Fishell G. (2000) Neuron 26, 395–404 [DOI] [PubMed] [Google Scholar]
- 6. Ge W., Martinowich K., Wu X., He F., Miyamoto A., Fan G., Weinmaster G., Sun Y. E. (2002) J. Neurosci. Res. 69, 848–860 [DOI] [PubMed] [Google Scholar]
- 7. Bisgrove D. A., Monckton E. A., Packer M., Godbout R. (2000) J. Biol. Chem. 275, 30668–30676 [DOI] [PubMed] [Google Scholar]
- 8. Cebolla B., Vallejo M. (2006) J. Neurochem. 97, 1057–1070 [DOI] [PubMed] [Google Scholar]
- 9. Krohn K., Rozovsky I., Wals P., Teter B., Anderson C. P., Finch C. E. (1999) J. Neurochem. 72, 1353–1361 [DOI] [PubMed] [Google Scholar]
- 10. Amemiya K., Traub R., Durham L., Major E. O. (1992) J. Biol. Chem. 267, 14204–14211 [PubMed] [Google Scholar]
- 11. Steele-Perkins G., Plachez C., Butz K. G., Yang G., Bachurski C. J., Kinsman S. L., Litwack E. D., Richards L. J., Gronostajski R. M. (2005) Mol. Cell Biol. 25, 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. das Neves L., Duchala C. S., Tolentino-Silva F., Haxhiu M. A., Colmenares C., Macklin W. B., Campbell C. E., Butz K. G., Gronostajski R. M., Godinho F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11946–11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T., Nakashima K. (2009) Dev. Cell 16, 245–255 [DOI] [PubMed] [Google Scholar]
- 14. Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. (1999) Science 284, 479–482 [DOI] [PubMed] [Google Scholar]
- 15. Hermanson O., Jepsen K., Rosenfeld M. G. (2002) Nature 419, 934–939 [DOI] [PubMed] [Google Scholar]
- 16. Gronostajski R. M. (2000) Gene 249, 31–45 [DOI] [PubMed] [Google Scholar]
- 17. Mason S., Piper M., Gronostajski R. M., Richards L. J. (2009) Mol. Neurobiol. 39, 10–23 [DOI] [PubMed] [Google Scholar]
- 18. Santoro C., Mermod N., Andrews P. C., Tjian R. (1988) Nature 334, 218–224 [DOI] [PubMed] [Google Scholar]
- 19. Roulet E., Armentero M. T., Krey G., Corthésy B., Dreyer C., Mermod N., Wahli W. (1995) Mol. Cell Biol. 15, 5552–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaudhry A. Z., Lyons G. E., Gronostajski R. M. (1997) Dev. Dyn. 208, 313–325 [DOI] [PubMed] [Google Scholar]
- 21. Shu T., Butz K. G., Plachez C., Gronostajski R. M., Richards L. J. (2003) J. Neurosci. 23, 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barry G., Piper M., Lindwall C., Moldrich R., Mason S., Little E., Sarkar A., Tole S., Gronostajski R. M., Richards L. J. (2008) J. Neurosci. 28, 12328–12340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steele-Perkins G., Butz K. G., Lyons G. E., Zeichner-David M., Kim H. J., Cho M. I., Gronostajski R. M. (2003) Mol. Cell Biol. 23, 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Driller K., Pagenstecher A., Uhl M., Omran H., Berlis A., Gründer A., Sippel A. E. (2007) Mol. Cell Biol. 27, 3855–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Campbell C. E., Piper M., Plachez C., Yeh Y. T., Baizer J. S., Osinski J. M., Litwack E. D., Richards L. J., Gronostajski R. M. (2008) BMC Dev. Biol. 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pekarik V., Belmonte J. C. (2008) J. Biol. 7, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deneen B., Ho R., Lukaszewicz A., Hochstim C. J., Gronostajski R. M., Anderson D. J. (2006) Neuron. 52, 953–968 [DOI] [PubMed] [Google Scholar]
- 28. Wilczynska K. M., Singh S. K., Adams B., Bryan L., Rao R. R., Valerie K., Wright S., Griswold-Prenner I., Kordula T. (2009) Stem Cells 27, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vihma H., Pruunsild P., Timmusk T. (2008) Genomics 92, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arteaga M. F., Coric T., Straub C., Canessa C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chu H. Y., Ohtoshi A. (2007) Mol. Cell Biol. 27, 3743–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Michelhaugh S. K., Vaitkevicius H., Wang J., Bouhamdan M., Krieg A. R., Walker J. L., Mendiratta V., Bannon M. J. (2005) J. Neurochem. 95, 1342–1350 [DOI] [PubMed] [Google Scholar]
- 33. Kruse U., Sippel A. E. (1994) J. Mol. Biol. 238, 860–865 [DOI] [PubMed] [Google Scholar]
- 34. Altmann H., Wendler W., Winnacker E. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3901–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gründer A., Qian F., Ebel T. T., Mincheva A., Lichter P., Kruse U., Sippel A. E. (2003) Gene 304, 171–181 [DOI] [PubMed] [Google Scholar]
- 36. Kordula T., Rydel R. E., Brigham E. F., Horn F., Heinrich P. C., Travis J. (1998) J. Biol. Chem. 273, 4112–4118 [DOI] [PubMed] [Google Scholar]
- 37. Gopalan S. M., Wilczynska K. M., Konik B. S., Bryan L., Kordula T. (2006) J. Biol. Chem. 281, 13126–13133 [DOI] [PubMed] [Google Scholar]
- 38. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 39. Mudhasani R., Fontes J. D. (2002) Mol. Cell Biol. 22, 5019–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Al-Kandari W., Koneni R., Navalgund V., Aleksandrova A., Jambunathan S., Fontes J. D. (2007) J. Mol. Biol. 369, 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Delegeane A. M., Ferland L. H., Mellon P. L. (1987) Mol. Cell Biol. 7, 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ouellet S., Vigneault F., Lessard M., Leclerc S., Drouin R., Guérin S. L. (2006) Nucleic Acids Res. 34, 6472–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilczynska K. M., Singh S. K., Adams B., Bryan L., Rao R. R., Valerie K., Wright S., Griswold-Prenner I., Kordula T. (2009) Stem Cells 27, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang C., Pugh B. F. (2009) Nat. Rev. Genet. 10, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cairns B. R. (2009) Nature 461, 193–198 [DOI] [PubMed] [Google Scholar]
- 46. Gévry N., Svotelis A., Larochelle M., Gaudreau L. (2009) Methods Mol. Biol. 543, 281–291 [DOI] [PubMed] [Google Scholar]
- 47. Besnard F., Brenner M., Nakatani Y., Chao R., Purohit H. J., Freese E. (1991) J. Biol. Chem. 266, 18877–18883 [PubMed] [Google Scholar]
- 48. Allore R. J., Friend W. C., O'Hanlon D., Neilson K. M., Baumal R., Dunn R. J., Marks A. (1990) J. Biol. Chem. 265, 15537–15543 [PubMed] [Google Scholar]
- 49. Tamura T., Miura M., Ikenaka K., Mikoshiba K. (1988) Nucleic Acids Res. 16, 11441–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwartz M. L., Hua Y., Cañete-Soler R., Schlaepfer W. W. (1998) Brain Res. Mol. Brain Res. 57, 21–30 [DOI] [PubMed] [Google Scholar]
- 51. Plachez C., Lindwall C., Sunn N., Piper M., Moldrich R. X., Campbell C. E., Osinski J. M., Gronostajski R. M., Richards L. J. (2008) J. Comp. Neurol. 508, 385–401 [DOI] [PubMed] [Google Scholar]
- 52. Nebl G., Cato A. C. (1995) Cell Mol. Biol. Res. 41, 85–95 [PubMed] [Google Scholar]
- 53. Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 6438–6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ravichandran V., Sabath B. F., Jensen P. N., Houff S. A., Major E. O. (2006) J. Virol. 80, 10506–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Müller K., Mermod N. (2000) J. Biol. Chem. 275, 1645–1650 [DOI] [PubMed] [Google Scholar]
- 56. Zhao L. H., Ba X. Q., Wang X. G., Zhu X. J., Wang L., Zeng X. L. (2005) Acta Biochim. Biophys. Sin. 37, 440–446 [DOI] [PubMed] [Google Scholar]
- 57. O'Donnell A., Yang S. H., Sharrocks A. D. (2008) Mol. Cell 29, 780–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.