Abstract
Serine-threonine kinase receptor-associated protein (STRAP) functions as a regulator of both TGF-β and p53 signaling. However, the regulatory mechanism of STRAP activity is not understood. In this study, we report that B-MYB is a new STRAP-interacting protein, and that an amino-terminal DNA-binding domain and an area (amino acids 373–468) between the acidic and conserved regions of B-MYB mediate the B-MYB·STRAP interaction. Functionally, B-MYB enhances STRAP-mediated inhibition of TGF-β signaling pathways, such as apoptosis and growth inhibition, by modulating complex formation between the TGF-β receptor and SMAD3 or SMAD7. Furthermore, coexpression of B-MYB results in a dose-dependent increase in STRAP-mediated stimulation of p53-induced apoptosis and cell cycle arrest via direct interaction. Confocal microscopy showed that B-MYB prevents the normal translocation of SMAD3 in response to TGF-β1 and stimulates p53 nuclear translocation. These results suggest that B-MYB acts as a positive regulator of STRAP.
Keywords: Apoptosis, p53, Protein-Protein Interactions, Transcription Factors, Transforming Growth Factor β (TGFβ), B-myb, Serine-Threonine Kinase Receptor-associated Protein (STRAP)
Introduction
Serine-threonine kinase receptor-associated protein (STRAP)2 is a transforming growth factor-β (TGF-β) receptor-interacting protein that inhibits TGF-β signaling by stabilizing the association between TGF-β receptors and SMAD7 (1). STRAP also contributes to tumor progression by blocking TGF-β-mediated signaling, especially in colon and lung carcinomas, indicating the possible involvement of STRAP in tumorigenesis (2, 3). Recently, STRAP has been found to act as a regulator of 3-phosphoinositide-dependent protein kinase-1 (PDK1) and apoptosis signal-regulating kinase 1 (ASK1) to stimulate cell growth (4, 5). In contrast to the findings described above, STRAP has also been shown to potentiate p53-induced apoptosis through direct interaction with p53 (6). These findings suggest that STRAP might have a bipartite role in the regulation of cell growth. However, the mechanism of STRAP regulation is not understood.
B-myb is a member of the Myb family of transcription factors, which is ubiquitously expressed and involved in controlling cell proliferation and differentiation (7–9). B-myb is phylogenetically the most divergent of the three myb proteins, A-myb, B-myb, and c-myb (10). Numerous reports have established a critical role of B-myb in the growth of normal and tumorigenic cell lines (7, 8, 11, 13). The most straightforward explanation for this effect is that B-myb could contribute to cell survival by inducing anti-apoptotic genes. For example, B-myb, like c-myb, stimulated transcription of Bcl2 and enhanced cell survival (15), suggesting that Bcl2 is a target gene for B-myb-mediated cell survival. In addition, anti-apoptotic ApoJ/Clusterin, which is highly induced in the presence of a variety of apoptotic stimuli, was also transactivated by B-MYB (16). Despite evidence supporting an anti-apoptotic function for B-myb, other studies have implicated B-myb in promoting cell death. Overexpression of B-myb accelerates apoptosis in TGF-β1-treated M1 cells without affecting the regulation of c-myb and c-myc expression (17). In addition, B-myb has been shown to induce neuronal apoptosis evoked by nerve growth factor deprivation and DNA damage (18). Thus, the function of B-myb in regulating cell growth is still unclear and awaits further evidence.
In this study, we found that B-MYB plays an important role in the regulation of STRAP-mediated TGF-β and p53 signaling by acting as a positive regulator of STRAP. Direct interaction between B-MYB and STRAP is essential for the positive regulation of STRAP-mediated TGF-β and p53 signaling.
MATERIALS AND METHODS
Antibodies and Plasmids
Anti-FLAG (M2), anti-GST, anti-PAI-1, anti-p21, anti-SMAD7, anti-CDK4, anti-cyclin D1, anti-B-MYB, anti-p53, anti-MDM2, anti-BAX, and anti-β-actin antibodies have been previously described (6, 19). Anti-phospho-SMAD3 (Ser-423/425) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Wild-type B-MYB and its deletion forms (partial B-MYB, R1, R2, CR, TA1, DBD, TA), p3TP-Lux and p53-Luc reporter plasmids, and pSuper and pSingle-tTS-shRNA vectors were also described previously (4, 6, 19–21).
Cell Culture and Stable Cell Lines
HEK293, HeLa, HCT116, Hep3B, HepG2, HaCaT, and MCF7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen) as previously described (19). HEK293 cells stably expressing pcDNA3.1-HisC-B-MYB plasmid (B-myb(OE)) were screened in the presence of 800 μg/ml of G418. HEK293 cells stably expressing B-MYB-specific shRNA (B-myb(KD)) were screened in the presence of 1.5 μg/ml of puromycin as described previously (19). To prepare B-myb(KD), the following double-stranded oligonucleotide was cloned into the pSuper vector: forward, 5′-GATCCCCGTTAAGAAGTATGGCACAATTCAAGAGATTGTGCCATACTTCTTAACTTTTTGGAAA-3′; and reverse, 5′-AGCTTTTCCAAAAAGTTAAGAAGTATGGCACAATCTCTTGAATTGTGCCATACTTCTTAACGGG-3′. The B-MYB sequence is underlined. HEK293 cells inducibly expressing B-MYB-specific shRNA (inducible B-MYB(KD)) were screened in the presence of 450 μg/ml of G418. For inducible knockdown of endogenous B-MYB expression, the following double-stranded oligonucleotide was cloned into the pSingle-tTS-shRNA vector as described previously (5): forward, 5′-TCGAGGGTTAAGAAGTATGGCACAATTCAAGAGATTGTGCCATACTTCTTAACCTTTTTTA-3′; and reverse, 5′-AGCTTAAAAAAGGTTAAGAAGTATGGCACAATCTCTTGAATTGTGCCATACTTCTTAACCC-3′. The B-MYB sequence is underlined. After treatment with 1 μg/ml of doxycycline (Sigma), a tetracycline analog, for 72 h, immunoblotting with an anti-B-MYB antibody was carried out to confirm endogenous B-MYB knockdown. For inducible overexpression of endogenous B-MYB, the full-length B-MYB was cloned into the pcDNA4TM/TO/myc-HisA vector (Invitrogen), and HEK293 cells stably expressing the pcDNA4/TO/myc-HisA-B-MYB plasmid (inducible B-myb(OE)) were screened in the presence of 250 μg/ml of zeocin (Invitrogen) and 5 μg/ml of blasticidin (Invitrogen).
Construction of B-MYB(373/468) Mutant
The B-MYB(373/468) deletion mutant was generated by PCR as described previously (19). PCR was performed using wild-type B-MYB as the template. The following primers were used: forward primer 5′-GCGAATTCCTGGATGGCCAC-3′ containing an EcoRI site (underlined); and reverse primer 5′-GCGGATCCCAGCTCCAATGT-3′ containing a BamHI site (underlined). The amplified PCR products were digested with EcoRI and BamHI and ligated into pBluescript KS (Stratagene). The ClaI/NotI fragment of the resulting plasmid was then cloned into pEBG vector (20) cut with ClaI and NotI, yielding GST-B-MYB(373/468).
Binding Assay, Immunoprecipitation, and Immunoblot
In vivo binding assays were performed as previously described (5, 19). Native PAGE to determine the in vitro interaction between B-MYB and STRAP was performed using in vitro translated 35S-labeled B-MYB, prepared with the TNT reticulocyte lysate system (Promega), and recombinant STRAP proteins (5). Immunoprecipitation and immunoblot analyses were performed as described (4, 22).
RNA Interference
B-MYB-specific siRNAs (number 1, 5′-GAUCUGGAUGAGCUGCACUTT-3′; number 2, 5′-GUUAAGAAGUAUGGCACAATT-3′) targeting two coding regions (number 1, amino acids 9–15; number 2, amino acids 98–104) of human B-MYB and p53-specific siRNA (5′-GACUCCAGUGGUAAUCUACTT-3′) targeting a coding region (amino acids 259–265) of human p53 were synthesized by Bioneer Corp. (Cheongwon, Korea). The control scrambled siRNA was described previously (23). WelFect-ExTM Plus (WelGENE, Daegu, Korea) was used to transfect cells with the indicated concentrations of siRNA oligonucleotides.
Reporter Assay
HepG2 or MCF7 cells were transfected with p3TP-Lux or p53-Luc reporter and the indicated expression vectors using WelFect-Ex Plus reagent. Luciferase activity was measured by a luciferase assay system (Promega) as described previously (5). Values were adjusted with respect to the expression level of a cotransfected β-galactosidase reporter control, and, for each experiment, at least three independent transfections were performed.
Apoptosis and Cell Cycle Analysis
Cell death experiments were performed as described previously using the green fluorescent protein (GFP) system in HaCaT and MCF7 cells transfected with the indicated expression vectors (4). The percentage of apoptotic cells was calculated as the number of GFP-positive cells with apoptotic nuclei divided by the total number of GFP-positive cells. The cells present in each phase of the cell cycle were analyzed in HaCaT and MCF7 cells by flow cytometry performed on a FACSCalibur-S system (BD Biosciences) (6, 22).
Statistical Analysis
All experiments were repeated at least three times, and results are expressed as the mean ± S.D. A p value relative to control of p < 0.05, as calculated by Student's t test, was considered statistically significant.
RESULTS
B-MYB Is an Interacting Partner of STRAP
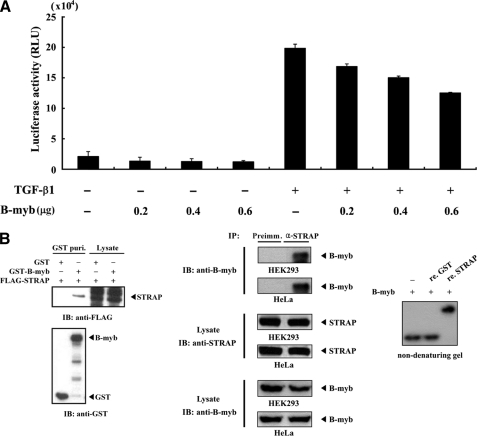
In an effort to identify proteins that interact with B-MYB, we previously performed a yeast two-hybrid test using the B-MYB library plasmid and found that B-MYB interacts with the TGF-β receptor-interacting protein STRAP (data not shown), in addition to several other baits tested, including ZPR9 (20), PTP-BAS (24), and B-MYB (self-association) (24). These results strongly indicated that B-MYB could interact with STRAP physically in mammalian cells. Because STRAP inhibits TGF-β signaling (1), we speculated that B-MYB might be involved in the regulation of STRAP-mediated TGF-β signaling. To test this, a luciferase reporter assay was performed using p3TP-Lux reporter plasmid containing elements from the PAI-1 promoter (25) to investigate the effect of B-MYB on TGF-β signaling. As shown in Fig. 1A, B-MYB decreased TGF-β-induced transcription in a dose-dependent manner, suggesting B-MYB as a potential regulator of TGF-β signaling.
FIGURE 1.
Physical interaction of B-MYB with STRAP. A, inhibition of TGF-β-induced transcription by B-MYB. HepG2 cells were transfected with 0.3 μg of p3TP-Lux reporter and increasing amounts of B-MYB, as indicated, in the presence or absence of TGF-β1 (100 pm). B, in vivo and in vitro association of B-MYB with STRAP. GST alone and GST-B-MYB were cotransfected with FLAG-STRAP into HEK293 cells. GST fusion proteins were purified on glutathione-Sepharose beads (GST Puri.), and the amounts of complex formation and FLAG-STRAP used for the in vivo binding assay were determined by anti-FLAG antibody immunoblot (left, upper panel). Cell lysates from HEK293 or HeLa cells were subjected to immunoprecipitation using either rabbit preimmune serum (Preimm.) or anti-STRAP antibody (α-STRAP), followed by immunoblot analysis using an anti-B-MYB antibody to determine the complex formation between endogenous B-MYB and STRAP (middle). For native PAGE of the B-MYB·STRAP complex, in vitro translated 35S-labeled B-MYB was prepared with the TnT reticulocyte lysate system. 35S-Labeled B-MYB was incubated with unlabeled recombinant GST, as a control, or STRAP (5 μg each) as indicated at room temperature for 1 h (right). IB, immunoblot; IP, immunoprecipitation; re., recombinant.
To examine the possible interaction of B-MYB with STRAP in cells, cotransfection experiments were first performed using GST-B-MYB and FLAG-STRAP. STRAP was detected in the coprecipitate when coexpressed with GST-B-MYB but not with control GST alone, demonstrating that B-MYB interacts with STRAP (Fig. 1B, left). To confirm the endogenous interaction between B-MYB and STRAP, coimmunoprecipitation experiments were performed in HEK293 and HeLa cells. Endogenous STRAP was immunoprecipitated by anti-STRAP antibody, and the presence of B-MYB was then analyzed by immunoblotting with an anti-B-MYB antibody. B-MYB was present in the STRAP, but not in the control, preimmune serum immunoprecipitates (Fig. 1B, middle). This endogenous interaction was also demonstrated by reciprocal coimmunoprecipitation experiments in which anti-B-MYB antibody, instead of anti-STRAP antibody, was used for immunoprecipitation (data not shown).
Non-denaturing polyacrylamide gel electrophoresis was then used to analyze the direct association of recombinant STRAP with B-MYB, which was translated in vitro. When 35S-labeled B-MYB was incubated with recombinant STRAP proteins, mobility clearly shifted relative to incubation with the control GST alone or in the absence of STRAP (Fig. 1B, right). These results demonstrated that a physical association between B-MYB and STRAP could occur both in vivo and in vitro.
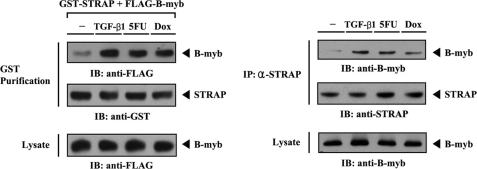
TGF-β and p53 Signals Stabilize a Complex Formation between B-MYB and STRAP
Because STRAP regulates TGF-β and p53 signaling (1, 6), the influence of 5-fluorouracil (5-FU), an inducer of p53-mediated apoptosis, and doxorubicin, an inducer of double-stranded DNA break-mediated cell cycle arrest, over B-MYB·STRAP complex formation in cells was next examined. Upon 5-FU and doxorubicin treatment, the association between B-MYB and STRAP was considerably increased compared with control untreated cells (Fig. 2, left, third and fourth lanes). This result was also confirmed by an immunoprecipitation experiment in which endogenous STRAP was immunoprecipitated by an anti-STRAP antibody, followed by immunoblotting with an anti-B-MYB antibody (Fig. 2, right). Similarly, exposure of the cells to TGF-β1 resulted in a remarkable increase in B-MYB·STRAP complex formation (Fig. 2, left and right, second lanes). These data indicate that the interaction between B-MYB and STRAP appears to be involved in the TGF-β and p53 signaling pathways.
FIGURE 2.
Modulation of B-MYB·STRAP interaction by TGF-β and p53 signals. HEK293 cells transfected with GST-STRAP and FLAG-B-MYB were incubated with or without the following stimuli: TGF-β1 (100 ng/ml, 20 h), 5-fluorouracil (0.38 mm, 30 h), or doxorubicin (Dox, 100 ng/ml, 24 h). Cell lysates were purified on glutathione-Sepharose beads (GST Purification), followed by immunoblotting with an anti-FLAG antibody to determine the association between B-MYB and STRAP (left). HEK293 cell lysates treated with or without the indicated stimuli were immunoprecipitated with an anti-STRAP antibody (IP: α-STRAP) and immunoblotted with an anti-B-MYB antibody to determine the endogenous association between B-MYB and STRAP (right).
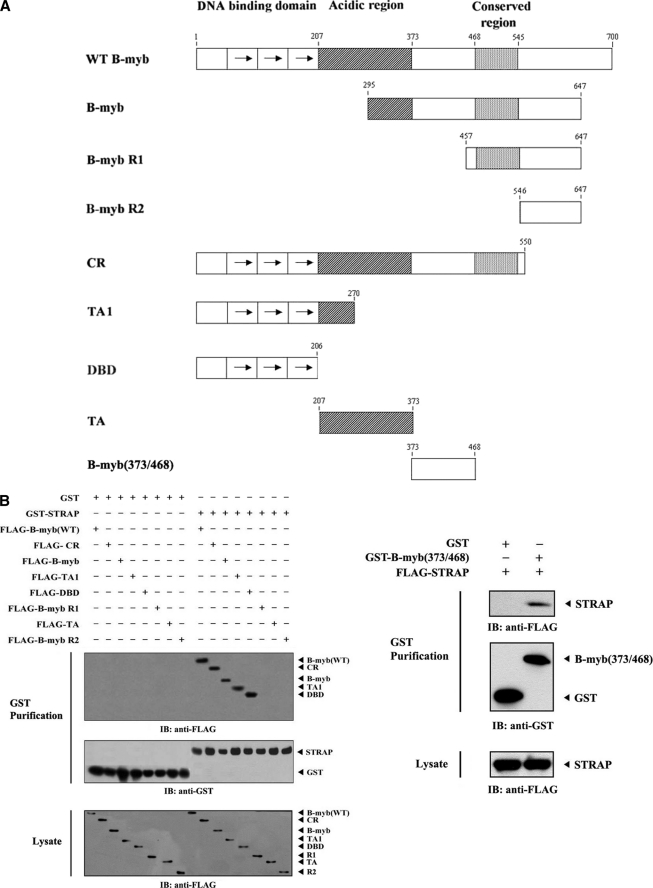
Identification of the STRAP-binding Domain of B-MYB
PDK1 was recently reported to bind to STRAP and inhibit TGF-β signaling (4). Thus, B-MYB, like PDK1, possibly modulates TGF-β signaling via direct interaction with STRAP. To determine the B-MYB regions required for STRAP binding, the binding properties of eight B-MYB deletion mutants fused to FLAG or GST were examined: FLAG-B-MYB (partial), FLAG-B-MYB R1, FLAG-B-MYB R2, FLAG-CR, FLAG-TA1, FLAG-DBD, FLAG-TA, and GST-B-MYB(373/468) (Fig. 3A). GST-STRAP binding was readily detectable with four deletion mutants, FLAG-B-MYB (partial), FLAG-CR, FLAG-TA1, and FLAG-DBD, as well as FLAG-wild-type B-MYB (Fig. 3B, left). However, no association was detected between STRAP and the B-MYB deletion mutants FLAG-B-MYB R1, FLAG-B-MYB R2, and FLAG-TA, implying that a region between the acidic and carboxyl-terminal conserved regions (amino acids 373–468) may also be involved in STRAP binding. To examine this possibility, another B-MYB deletion mutant, GST-B-MYB(373/468), was generated and used for an in vivo binding assay. Results showed that the region (amino acids 373–468) was also essential for STRAP binding (Fig. 3B, right). These results suggested that both the DNA-binding domain and a region (amino acids 373–468) between the acidic and conserved regions are responsible for STRAP binding in vivo.
FIGURE 3.
Domains of B-MYB involved in STRAP binding. A, the schematic structures of wild-type and deletion mutants of B-MYB are indicated. Numbers indicate the amino acid residues corresponding to the domain boundaries. B, HEK293 cells transfected with the indicated expression vectors were lysed, precipitated using glutathione-Sepharose beads (GST purification), and then immunoblotted with an anti-FLAG antibody to determine the level of B-MYB·STRAP complex formation (top panels). The same blots were re-probed with anti-GST antibody to confirm the expression of GST fusion proteins in the coprecipitates (middle panels).
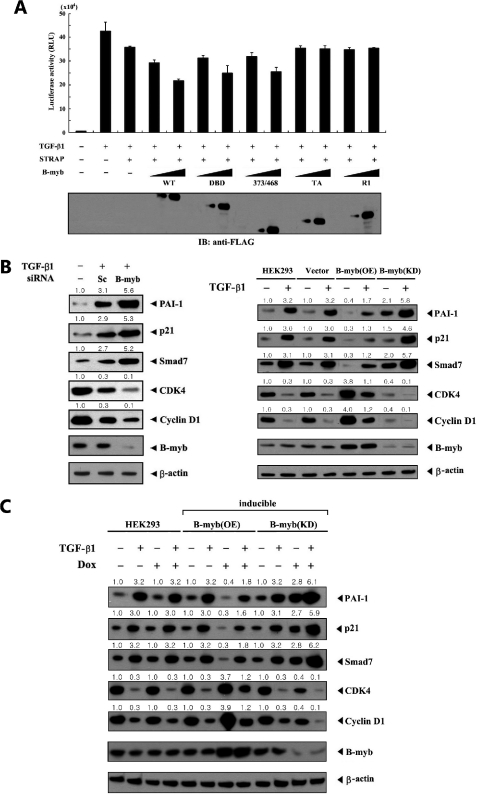
B-MYB Enhances STRAP-mediated Inhibition of TGF-β Signaling
STRAP has been previously shown to negatively regulate TGF-β-induced transcription dose-dependently (1). To investigate the role of the B-MYB·STRAP interaction, the effect of B-MYB on STRAP activity was examined. TGF-β-induced transcription was measured using the p3TP-Lux reporter in the presence of increasing amounts of B-MYB. Expression of wild-type B-MYB significantly enhanced the STRAP-mediated inhibition of TGF-β-induced transcription in a dose-dependent manner (Fig. 4A, third lane versus fourth and fifth lanes). Our next experiments aimed to determine whether the inhibition of TGF-β-induced transcription by B-MYB relates in a direct or indirect manner to the physical binding between B-MYB and STRAP. Results showed that the expression of two B-MYB deletion mutants (TA and R1) that are defective in STRAP binding (see Fig. 3B) did not affect TGF-β-induced transcription, whereas other B-MYB deletion mutants DBD and 373/468, like wild-type B-MYB, significantly enhanced STRAP-mediated inhibition of TGF-β-induced transcription in a dose-dependent manner (Fig. 4A, third lane versus sixth to ninth lanes). These results suggest that a direct interaction between B-MYB and STRAP plays a critical role in the modulation of TGF-β-induced transcription mediated by STRAP. To confirm the role of B-MYB in TGF-β-induced transcription, the effect of B-MYB on the expression of TGF-β target genes, which are normally up- (PAI-1, p21, and SMAD7) or down-regulated (CDK4 and cyclin D1) in response to TGF-β, was also examined using B-MYB-specific siRNA. Compared with the control scrambled siRNA, knockdown of B-MYB markedly increased the expression of positively regulated TGF-β target genes PAI-1, p21, and SMAD7, and decreased the expression of negatively regulated TGF-β target genes, CDK4 and cyclin D1 (Fig. 4B, left). This result indicates that B-MYB is a negative regulator of TGF-β signaling.
FIGURE 4.
Enhancement of STRAP-induced inhibition of TGF-β signaling by B-MYB. A, HepG2 cells were transiently transfected with 0.3 μg of p3TP-Lux reporter, 0.1 μg of expression plasmid for β-galactosidase as an internal control, and increasing amounts (0.3 and 0.6 μg) of wild-type B-MYB or its deletion mutants (DBD, 373/468, TA, and R1) in the presence of STRAP (0.6 μg), and subsequently stimulated by TGF-β1 (100 pm). Luciferase activity was measured 48 h after transfection and normalized to β-galactosidase activity. The expression level of FLAG-tagged wild-type B-MYB and its deletion mutants was analyzed by anti-FLAG antibody immunoblot. B, effect of B-MYB on the expression of TGF-β target genes. HepG2 cells were transiently transfected with 200 nm of control scrambled siRNA (Sc) or B-MYB-specific siRNA number 1 (B-myb) as indicated, and then treated with 100 pm TGF-β1 for 20 h (left). Cell lysates were subjected to immunoblot analysis using anti-PAI-1, anti-p21, anti-SMAD7, anti-CDK4, anti-cyclin D1, and anti-B-MYB antibodies. Parental HEK293 cells (HEK293), HEK293 cells stably expressing an empty vector (Vector), HEK293 cells stably overexpressing B-MYB (B-myb(OE)), and HEK293 cells stably expressing B-MYB-specific siRNA (B-myb(KD)) were lysed and subjected to immunoblot analysis using the indicated antibodies as described above (right). C, HEK293 cells harboring stably integrated pcDNA4/TO/myc-HisA vector containing wild-type B-MYB (inducible B-myb(OE)) or pSingle-tTS-shRNA vector containing B-MYB-specific shRNA (inducible B-myb(KD)) were cultured in the presence or absence of 1 μg/ml of doxycycline (Dox) for 72 h to determine the effect of B-MYB on the expression of TGF-β target genes. Inducible expression of endogenous B-MYB by doxycycline was assessed by immunoblotting with an anti-B-MYB antibody. Equal amounts of protein in each lane were confirmed by immunoblotting with an anti-β-actin antibody. The relative level of the expression of TGF-β target genes was quantified by densitometric analysis. The fold-increase relative to untreated HepG2 or HEK293 cells was calculated.
Our analysis was extended to investigate whether the stable expression of B-MYB (B-myb(OE)) could lead to the opposite effect to the above results obtained from siRNA experiments. Overexpression of B-MYB decreased the effect of TGF-β on the expression of its target genes (Fig. 4B, right, B-myb(OE)). Consistently, upon TGF-β1 treatment, HEK293 cells stably expressing B-MYB-specific siRNA (B-myb(KD)) showed a considerable increase in the expression of TGF-β target genes PAI-1, p21, and SMAD7, and decrease in the expression of TGF-β target genes CDK4 and cyclin D1 (Fig. 4B, right, B-myb(KD)). To further verify this in a physiological context, the effect of B-MYB on the expression of TGF-β target genes was also determined using an inducible B-MYB-specific shRNA system in HEK293 cells. Similar to the above results (see Fig. 4B), the knockdown of endogenous B-MYB increased the effect of TGF-β on the expression of its target genes (Fig. 4C, inducible B-myb(KD)). A similar result was also obtained in the analysis of the mRNAs of TGF-β targets using RT-PCR (supplemental Fig. S1A). These results indicate that B-MYB negatively regulates TGF-β signaling through direct interaction with STRAP.
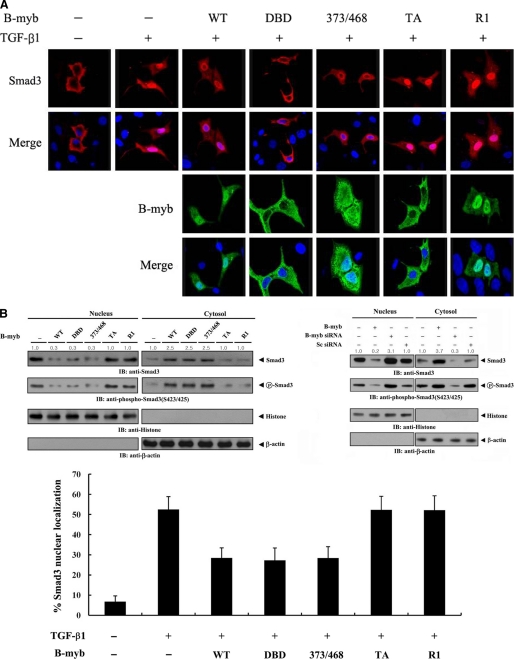
B-MYB Interferes with the Nuclear Translocation of SMAD3
Because B-MYB and STRAP coexpression resulted in a significant enhancement of STRAP-mediated inhibition of TGF-β-induced transcription (Fig. 4A), B-MYB could modify the intracellular localization of SMAD3, which is essential for TGF-β signaling. To address this possibility, Hep3B cells were transfected with SMAD3 alone or together with B-MYB or a deletion mutant (DBD, 373/468, TA, and R1) in the presence or absence of TGF-β1. Upon TGF-β1 treatment, SMAD3 predominantly exhibited a nuclear distribution (Fig. 5A, top, first lane versus second lanes). However, coexpression of wild-type B-MYB or deletion mutants DBD and 373/468 inhibited the nuclear translocation of SMAD3 in response to TGF-β1 (Fig. 5A, top, second lane versus third to fifth lanes). Analysis of B-MYB subcellular localization showed that the B-MYB-mediated inhibition of SMAD3 nuclear translocation was likely to occur in the cytoplasm (Fig. 5A, third lane). To further confirm whether B-MYB could alter the subcellular localization of SMAD3, Hep3B cells transfected with wild-type B-MYB or a deletion mutant, together with a nonspecific scrambled siRNA or a B-MYB-specific siRNA, were treated with TGF-β1 and separated into cytoplasmic and nuclear fractions. Each fraction was analyzed by immunoblot analysis. As expected, wild-type B-MYB and its deletion mutants DBD and 373/468, which are able to bind to STRAP (see Fig. 3B), decreased the nuclear localization of SMAD3 (Fig. 5B, upper left, top panels). This observation is in agreement with the above confocal microscopy data (Fig. 5A). Consistently, the accumulation of SMAD3 in the nuclear fraction was increased in B-MYB-knockdown cells compared with control cells expressing a nonspecific scrambled siRNA, whereas the cytoplasmic accumulation of SMAD3 was markedly decreased under the same conditions (Fig. 5B, upper right). Similar results were obtained in the analysis of SMAD3 phosphorylation at the C-terminal SXS motif, a key event in TGF-β signaling (Fig. 5B, upper, second panels), and the percentage of SMAD3 nuclear localization (Fig. 5B, lower). These data indicate that B-MYB inhibits TGF-β signaling by preventing the nuclear translocation of SMAD3 in response to TGF-β1.
FIGURE 5.
The effect of B-MYB on the subcellular localization of SMAD3. A, modulation of SMAD3 localization by B-MYB. Hep3B cells were transiently transfected with a plasmid vector expressing FLAG-SMAD3, together with GFP-tagged wild-type B-MYB or a deletion mutant (DBD, 373/468, TA, and R1), and incubated in the presence or absence of 100 pm TGF-β1. Cells were immunostained with anti-FLAG antibody, followed by Alexa Fluor-594 anti-mouse secondary antibody (red, for SMAD3), and analyzed by confocal microscopy. B-MYB subcellular localization was determined by immunostaining with anti-GFP and Alexa Fluor-488 anti-rabbit secondary antibodies (green, for B-MYB). Nuclei (blue) were stained with DAPI. B, cytoplasmic and nuclear fractions from the extracts of Hep3B cells transfected with the indicated plasmid vectors or siRNA duplexes were separated from each other, incubated with TGF-β1, and used for immunoblot analysis with the indicated antibodies. Nucleus and Cytosol indicate the nuclear and cytoplasmic fractions of cell extracts, respectively. The relative level of SMAD3 expression was quantified by densitometric analysis. The fold-increase relative to untransfected samples in parental Hep3B cells was calculated. The percentage of SMAD3 nuclear localization was calculated as the number of SMAD3-positive cells with nuclear localization divided by the total number of SMAD3-positive cells (lower panel). About 280 cells were analyzed per sample using a confocal microscopy.
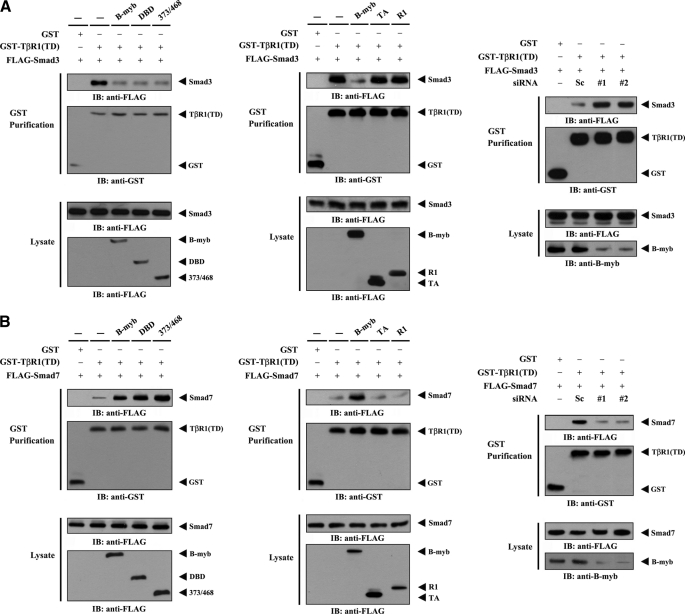
B-MYB Inhibits TGF-β Signaling by Modulating the Association of Activated Type 1 TGF-β Receptor with SMAD3 and SMAD7
To investigate the mechanism of B-MYB-mediated inhibition of TGF-β signaling, the contribution of B-MYB to modulating the interaction between activated type 1 TGF-β receptor (TβR1(TD)) and SMAD3 or SMAD7 was examined. HEK293 cells transfected with FLAG-SMAD3 and GST-TβR1(TD) in the presence or absence of wild-type B-MYB or a deletion mutant (DBD, 373/468, TA, and R1) were purified on glutathione-Sepharose beads (GST purification), followed by immunoblot analysis with an anti-FLAG antibody. Considerable decreases in complex formation between TβR1(TD) and SMAD3 (Fig. 6A, left), and increases in the association between TβR1(TD) and SMAD7 (Fig. 6B, left), were observed in cells expressing wild-type B-MYB or its deletion mutants DBD and 373/468, which are able to directly interact with STRAP, compared with the control in the absence of B-MYB. However, B-MYB deletion mutants TA and R1, which are defective in direct binding with STRAP, had no effect on the association (Fig. 6, A and B, middle). To further confirm whether the association between TβR1(TD) and SMAD3 and -7 is dependent on the expression of endogenous B-MYB, TβR1(TD)·SMAD protein complex formation was determined in HEK293 cells transfected with B-MYB-specific siRNAs. Opposite trends in the association between TβR1(TD) and SMAD3 and -7 were clearly observed in the B-MYB-knockdown cells (Fig. 6, A and B, right). These data indicate that the direct interaction between B-MYB and STRAP plays an important role in the B-MYB-mediated regulation of TGF-β signaling.
FIGURE 6.
Modulation of the activated type I TGF-β receptor·SMAD complex by B-MYB. HEK293 cells were transfected with the indicated combinations of plasmid vectors expressing GST-tagged TβR1(TD), an activated type I TGF-β receptor, FLAG-SMAD3, FLAG-SMAD7, and FLAG-tagged wild-type B-MYB or its deletion mutants (DBD, 373/468, TA, and R1), and the cell lysates were subjected to precipitation with glutathione-Sepharose beads (GST purification). Complex formation between TβR1(TD) and SMAD3 (A, left and middle, top panels) or TβR1(TD) and SMAD7 (B, left and middle, top panels) was determined by anti-FLAG antibody immunoblot. The expression level of GST-TβR1(TD) in GST precipitates was determined by anti-GST antibody immunoblot (A and B, left and middle, second panels). HEK293 cells were transfected with the indicated siRNA duplexes (B-MYB siRNAs (#1 and #2) or control scrambled siRNA (Sc)), together with plasmid vectors expressing GST-TβR1(TD), FLAG-SMAD3, or FLAG-SMAD7, and complex formation between TβR1(TD) and SMAD3 (A, right) or TβR1(TD) and SMAD7 (B, right) was determined by anti-FLAG antibody immunoblot (IB). The knockdown of endogenous B-MYB was determined by anti-B-MYB immunoblotting (A and B, right, bottom panels).
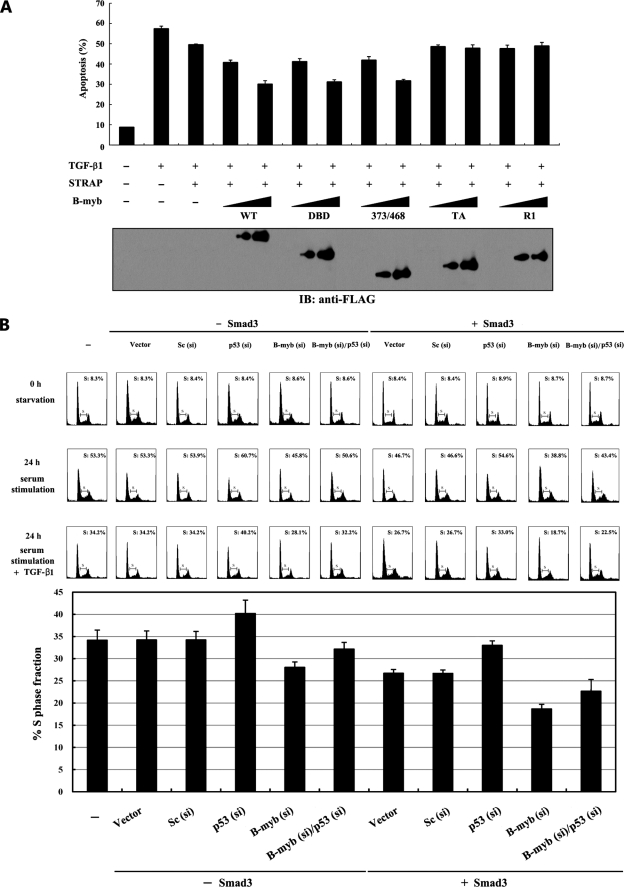
B-MYB Enhances STRAP-mediated Inhibition of TGF-β-induced Apoptosis and Growth Arrest
To explore the functional significance of the B-MYB·STRAP interaction, the effect of B-MYB on TGF-β-induced apoptosis was first determined using a GFP assay system (4). HaCaT cells were transfected with expression plasmids encoding GFP and STRAP, together with B-MYB or a deletion mutant (DBD, 373/468, TA, and R1), and incubated in the presence or absence of TGF-β1. Approximately 57% of HaCaT cells were apoptotic following TGF-β1 treatment (Fig. 7A, second lane). Cells transfected with wild-type B-MYB showed lower apoptosis rates than cells in the absence of B-MYB, and the effect was dose-dependent (Fig. 7A, third lane versus fourth and fifth lanes). Our next experiment aimed to determine whether the direct interaction of B-MYB with STRAP is crucial for the regulation of STRAP-mediated inhibition of TGF-β signaling. Results showed that wild-type B-MYB and its deletion mutants DBD and 373/468 decreased TGF-β-induced apoptosis to a similar extent in a dose-dependent manner, whereas B-MYB deletion mutants TA and R1, which are defective in STRAP binding, had no effect (Fig. 7A, third lane versus 10th–13th lanes), consistent with the results obtained from both luciferase assays (see Fig. 4A) and confocal microscopy analysis (see Fig. 5).
FIGURE 7.
Suppression of TGF-β-induced apoptosis and growth inhibition by B-MYB. A, B-MYB inhibits TGF-β-induced apoptosis in a dose-dependent manner. HaCaT cells were transiently transfected with increasing amounts (1.5 and 3 μg) of wild-type B-MYB or its deletion mutants (DBD, 373/468, TA, and R1) as indicated, together with an expression vector encoding GFP (2 μg). Transfected cells were treated with TGF-β1 (2 ng/ml) for 20 h to induce apoptosis. Apoptotic cell death was determined using the GFP expression system as described previously (4). The expression level of FLAG-tagged wild-type B-MYB and its deletion mutants was analyzed by anti-FLAG antibody immunoblot. B, effect of B-MYB on TGF-β-induced cell cycle arrest. HepG2 cells (∼2 × 105/dish) transfected with the indicated siRNA duplexes (200 nm B-MYB siRNA #1, B-myb (si), 100 nm p53 siRNA, p53 (si), or 200 nm control scrambled siRNA, Sc (si)) in the presence or absence of SMAD3 (3 μg) were synchronized in G0/G1 by hydroxyurea (2 mm) treatment for 20 h. Cells were collected before (0 h starvation) or after 10% serum treatment for 24 h in the absence (24 h serum stimulation) or presence (24 h serum stimulation + TGF-β1) of TGF-β1 (100 pm), and the percentage of cells in the G1, S, or G2/M phases was analyzed by flow cytometry. The bar graphs (lower panel) represent the fraction of cells in S phase (24 h serum stimulation + TGF-β1). si, siRNA.
Next, to determine whether TGF-β-induced cell cycle arrest was also affected by B-MYB, flow cytometry analysis was performed using HepG2 cells expressing a vector control, control scrambled siRNA (Sc (si)), p53-specific siRNA (p53 (si)), B-MYB-specific siRNA (B-myb (si)), B-myb (si)/p53 (si), SMAD3, SMAD3/Sc (si), SMAD3/p53 (si), SMAD3/B-myb (si), or SMAD3/B-myb (si)/p53 (si). As shown in Fig. 7B, ∼27% of cells expressing SMAD3 alone or SMAD3/Sc (si) accumulated in S phase after 24 h serum stimulation in the presence of TGF-β1. However, a lower number of cells (∼19%) were found to be in S phase in the presence of B-MYB siRNA compared with the control in the presence of scrambled siRNA (∼27%). A higher number of cells in S phase were detected in the presence of p53 siRNA when the effect of B-MYB siRNA on TGF-β-induced cell cycle arrest was analyzed (∼19 versus ∼23%). These results suggest that the physical interaction of B-MYB with STRAP is critical for the regulation of STRAP-mediated inhibition of TGF-β signaling pathways, such as apoptosis and cell cycle arrest, and that this effect is dependent on p53 signaling pathway.
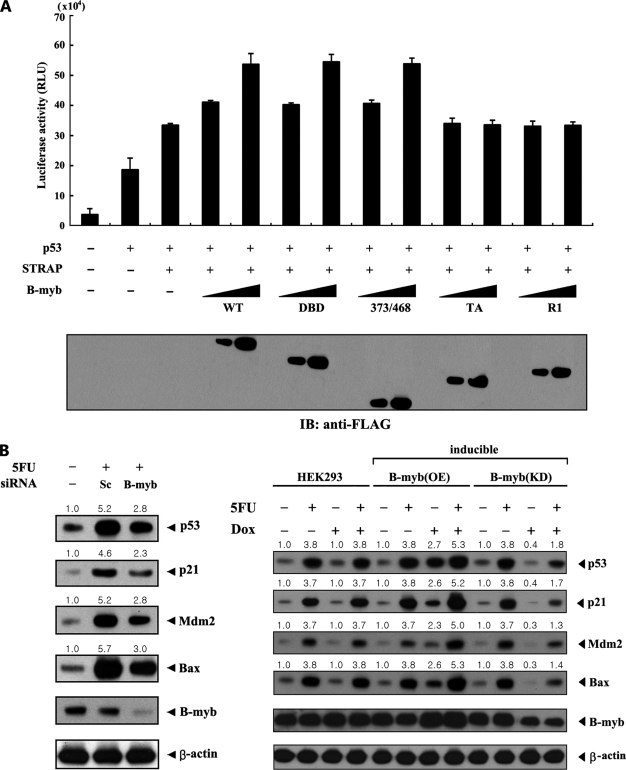
B-MYB Potentiates STRAP-mediated p53 Signaling
Because STRAP stimulates p53 signaling (6), the influence of B-MYB over STRAP-mediated p53 transcriptional activity was next examined. Coexpression of B-MYB resulted in a significant increase in STRAP-mediated p53 transcriptional activity in a dose-dependent manner (Fig. 8A, third lane versus fourth and fifth lanes). B-MYB deletion mutants (DBD, 373/468, TA, and R1) were then used to determine whether the direct interaction of B-MYB with STRAP is required for regulation of STRAP-mediated p53 transactivation. In contrast to B-MYB deletion mutants DBD and 373/468, no difference in STRAP-mediated p53 transactivation was observed in the presence of B-MYB deletion mutants TA and R1, which are unable to bind to STRAP (Fig. 8A, third lane versus 10th–13th lanes). These data indicate an essential role for the direct interaction between B-MYB and STRAP in the regulation of STRAP-mediated p53 transactivation.
FIGURE 8.
Enhancement of STRAP-mediated p53 signaling by B-MYB. A, effect of B-MYB on STRAP-mediated p53 transactivation. MCF7 cells were transfected with increasing amounts (0.2 and 0.4 μg) of wild-type (WT) or deletion mutants (DBD, 373/468, TA, and R1) of B-MYB as indicated, together with 0.2 μg of p53-Luc reporter plasmid and 0.1 μg of β-galactosidase internal control, in the presence or absence of p53 (0.5 μg). The expression level of FLAG-tagged wild-type B-MYB and its deletion mutants was analyzed by anti-FLAG antibody immunoblot (IB). B, effect of B-MYB on the expression of p53 target genes. MCF7 cells transiently transfected with 200 nm control scrambled siRNA (Sc) or B-MYB-specific siRNA #1 (B-myb), as indicated, were lysed and subjected to immunoblot analysis using anti-p53, anti-p21, anti-MDM2, anti-BAX, and anti-B-MYB antibodies (left). HEK293 cells harboring stably integrated pcDNA4/TO/myc-HisA vector containing wild-type B-MYB (inducible B-myb (OE)) or pSingle-tTS-shRNA vector containing B-MYB-specific shRNA (inducible B-myb (KD)) treated with 5-FU (0.38 mm, 30 h) were cultured in the presence or absence of 1 μg/ml of doxycycline (Dox) for 72 h to determine the effect of B-MYB on the expression of p53 target genes (right). Inducible expression of endogenous B-MYB by doxycycline was assessed by immunoblotting with an anti-B-MYB antibody. Equal amounts of protein in each lane were confirmed by immunoblotting with an anti-β-actin antibody. The relative level of the expression of p53 target genes was quantified by densitometric analysis. The fold-increase relative to untreated MCF7 or HEK293 cells was calculated.
To further investigate the stimulatory effect of B-MYB on p53 signaling, the effect of B-MYB-specific siRNA on p53-mediated gene responses was examined. Transfection of MCF7 cells with B-MYB-specific siRNA resulted in a remarkable decrease in the expression of p53 target genes p53, p21, MDM2, and BAX, which are normally up-regulated by p53 activators such as 5-FU (Fig. 8B, left). To physiologically confirm the role of B-MYB in p53 signaling, a stable system for the tetracycline-inducible expression of wild-type B-MYB (inducible B-myb(OE)) or B-MYB shRNA (inducible B-myb(KD)) in HEK293 cells was also generated. B-MYB-knockdown cells showed a lower level of p53 target gene expression in the presence of doxycycline, a tetracycline analog, compared with parental HEK293 cells (Fig. 8B, right). A similar trend was also observed in the analysis of the mRNAs of p53 targets (supplemental Fig. S1B). These results indicate that B-MYB contributes to the positive regulation of STRAP-mediated p53 signaling through direct interaction.
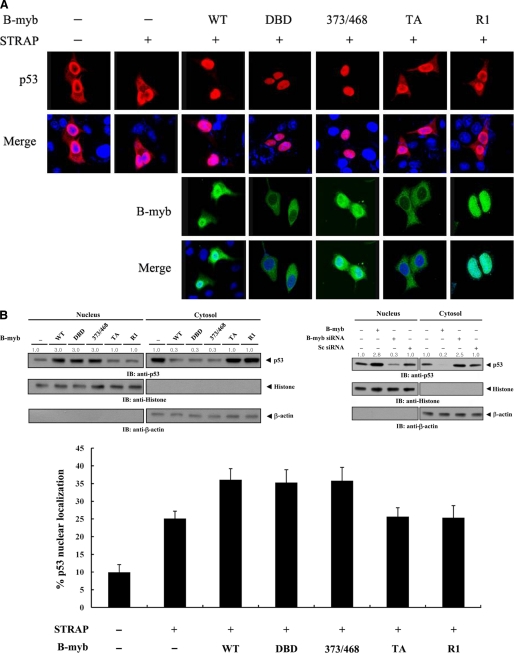
B-MYB Stimulates p53 Nuclear Translocation by Destabilizing the Association between p53 and MDM2
To investigate whether B-MYB stimulates p53-mediated transcription by affecting p53 nuclear translocation, MCF7 cells were transiently transfected with the indicated expression vectors encoding wild-type or deletion mutants of B-MYB, together with p53. Cells expressing p53 in the absence of B-MYB exhibited predominantly nuclear staining with some cytoplasmic staining, but the coexpression of wild-type B-MYB or its deletion mutants DBD and 373/468 markedly stimulated the nuclear translocation of p53 (Fig. 9A, top, second lane versus third to fifth lanes). The localization of p53 with coexpression of B-MYB deletion mutants TA and R1 was similar to that without B-MYB (Fig. 9A, top, second lane versus sixth and seventh lanes). Analysis of B-MYB subcellular localization indicates that the direct interaction between B-MYB and STRAP in the cytoplasm may contribute to the B-MYB-mediated stimulation of p53 nuclear translocation (Fig. 9A, third). To verify this, immunoblot analysis was performed using cytoplasmic and nuclear fractions of cell extracts transfected with expression vectors encoding wild-type and deletion forms of B-MYB. Higher levels of p53 were detected in the nucleus than in the cytoplasm upon expression of wild-type B-MYB or its deletion mutants DBD and 373/468 (Fig. 9B, upper left, Nucleus). In addition, the expected corresponding decrease in the accumulation of cytoplasmic p53 was observed (Fig. 9B, upper left, cytosol). Also as expected, the subcellular localization of p53 did not differ between control cells and cells expressing B-MYB deletion mutants TA or R1 (Fig. 9B, upper left, first lane versus fifth and sixth lanes). Furthermore, to determine whether the knockdown of endogenous B-MYB could modify the subcellular localization of p53, MCF7 cells transfected with a B-MYB-specific siRNA or a nonspecific scrambled siRNA were separated into cytoplasmic and nuclear fractions for immunoblot analysis. The accumulation of p53 in the nuclear fraction was noticeably decreased in the B-MYB-knockdown cells compared with control cells transfected with a nonspecific scrambled siRNA (Fig. 9B, upper right). A similar trend was also observed in the analysis of the percentage of p53 nuclear localization using a confocal microscopy (Fig. 9B, lower).
FIGURE 9.
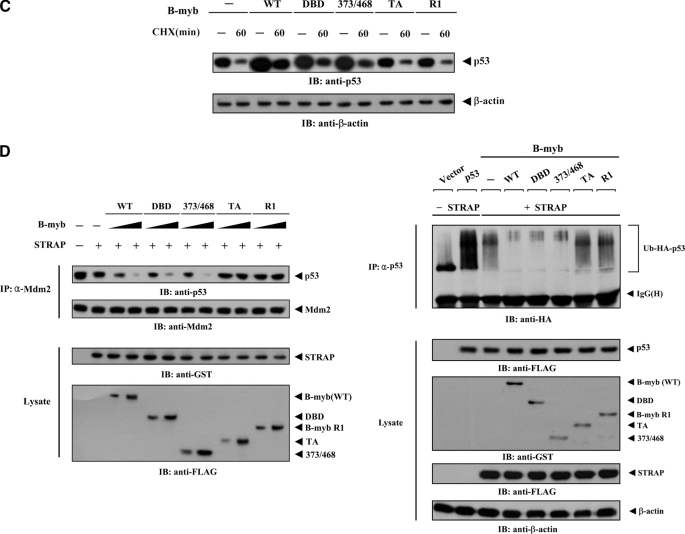
Modulation of p53 localization by B-MYB. A, MCF7 cells were transiently transfected with the indicated expression vectors for wild-type (WT) and deletion mutants (DBD, 373/468, TA, and R1) of B-MYB, together with or without STRAP, in the presence of p53. Cells were immunostained with anti-p53 antibody, followed by Alexa Fluor-594 anti-rabbit secondary antibody (red, for p53), and analyzed by confocal microscopy. B-MYB subcellular localization was determined by immunostaining with anti-FLAG and Alexa Fluor-488 anti-mouse secondary antibodies (green, for B-MYB). Nuclei (blue) were stained with DAPI. B, confirmation of endogenous p53 localization by immunoblot analysis. Cytoplasmic and nuclear fractions of MCF7 cells transfected with the indicated expression vectors or a B-MYB-specific siRNA #1 were analyzed by immunoblot using the indicated antibodies (upper panels). Nucleus and Cytosol indicate the nuclear and cytoplasmic fractions, respectively. The relative level of p53 expression was quantified by densitometric analysis. The fold-increase relative to untransfected samples in parental MCF7 cells was calculated. The percentage of p53 nuclear localization was calculated as the number of p53-positive cells with a nuclear localization divided by the total number of p53-positive cells (lower panel). About 300 cells were analyzed per sample using a confocal microscopy. C, measurement of p53 stability by immunoblot analysis. MCF7 cells were transiently transfected with the indicated expression vectors. Time intervals indicate the number of minutes after 20 μg/ml of cycloheximide (CHX) treatment. D, effect of B-MYB on p53·MDM2 complex formation and p53 ubiquitination. HCT116 cells were transfected with increasing amounts (0.8 and 1.6 μg) of wild-type or deletion mutants of B-MYB in the presence or absence of STRAP. The cells were lysed, immunoprecipitated with an anti-MDM2 antibody (IP: α-Mdm2), and immunoblotted (IB) with an anti-p53 antibody to determine the endogenous levels of p53·MDM2 complex formation (left, top panel). To measure B-MYB-mediated modulation of p53 ubiquitination, p53-null HCT116 cells were transfected with vectors expressing HA-tagged ubiquitin (Ub), p53, and wild-type or deletion mutants of B-MYB as indicated (2 μg each), in the presence or absence of STRAP. Cell lysates were immunoprecipitated (IP) with an anti-p53 antibody (IP: α-p53) and immunoblotted with an anti-HA antibody to determine the level of p53 ubiquitination (right, top panel).
To ensure the positive role of B-MYB in the regulation of p53 activity, the stability of p53 was also measured using immunoblot analysis in MCF7 cells. Expression of wild-type B-MYB and its deletion mutants DBD and 373/468 considerably increased the stability of p53 compared with the control in the absence of B-MYB, but this effect was not observed in the presence of the B-MYB deletion mutants TA and R1, which are unable to bind to STRAP (Fig. 9C).
To examine how B-MYB acts as a positive regulator of p53, the effect of B-MYB on endogenous p53·MDM2 interaction was also tested in the presence or absence of B-MYB using in vivo binding assays. Coexpression of wild-type B-MYB or its deletion mutants DBD and 373/468 markedly inhibited complex formation between endogenous p53 and MDM2 compared with untransfected control cells (Fig. 9D, left). However, coexpression of B-MYB deletion mutants TA and R1 had no effect on the formation of the endogenous p53·MDM2 complex. Because MDM2 is known to be an ubiquitinase (26), ubiquitination of p53 was then determined in the presence of wild-type B-MYB and deletion mutants. To this end, an in vivo ubiquitination assay was performed in which p53-null HCT116 cells were transfected with p53 and HA-tagged ubiquitin, together with the indicated plasmids. In agreement with the results obtained from the analysis of p53 stability (see Fig. 9C), coexpression of wild-type B-MYB and its deletion mutants DBD and 373/468 inhibited p53 ubiquitination compared with the control expressing STRAP alone, whereas B-MYB deletion mutants TA and R1 had no effect on p53 ubiquitination (Fig. 9D, right). These results suggest that B-MYB stimulates the nuclear accumulation of p53 by enhancing its stability.
B-MYB Potentiates p53-mediated Apoptosis and Cell Cycle Arrest Induced by STRAP
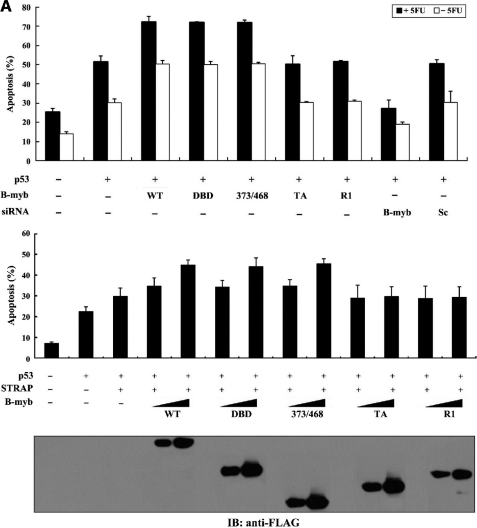
To determine whether binding of B-MYB to STRAP has a functional consequence for p53 activity, the effect of wild-type and deletion mutants of B-MYB on p53-mediated apoptosis was first examined using a GFP system in MCF7 cells. Approximately 51% of the MCF7 cells untransfected with B-MYB were apoptotic following 5-FU treatment. Transfection with wild-type B-MYB and its deletion mutants DBD and 373/468 increased apoptosis by about 41% relative to untransfected control MCF7 cells (Fig. 10A, upper, second lane versus third to fifth lanes) and this effect was dose-dependent (Fig. 10A, lower, third lane versus fourth to ninth lanes). However, the stimulatory effect of B-MYB on p53-mediated apoptosis was not detected in cells expressing B-MYB deletion mutants TA and R1. Furthermore, knockdown of endogenous B-MYB with B-MYB-specific siRNA resulted in a significant decrease in p53-mediated apoptosis (Fig. 10A, upper, second lane versus eighth lane).
FIGURE 10.
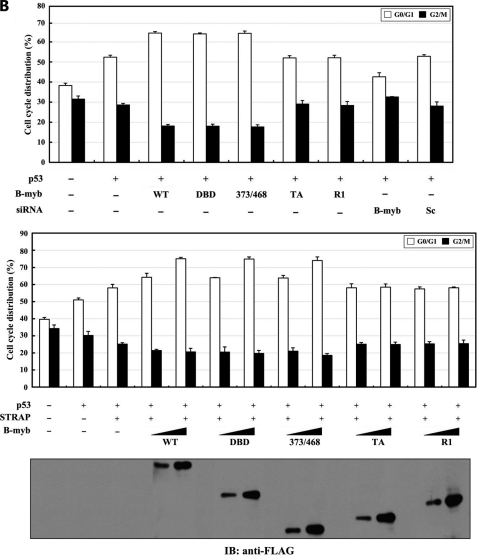
Effect of B-MYB on p53-induced apoptosis and cell cycle arrest. A, effect of B-MYB on p53-induced apoptosis. MCF7 cells were transiently transfected with 0.8 μg (upper) or increasing amounts (0.2 and 0.4 μg, lower) of the indicated expression vectors for wild-type (WT) and deletion mutants (DBD, 373/468, TA, and R1) of B-MYB and 200 nm B-MYB-specific (B-myb) or control scrambled (Sc) siRNAs, together with 0.5 μg of p53 and 1 μg of GFP, in the presence (black bars) or absence (white bars) of 0.38 mm 5-FU. Apoptotic cell death was determined using the GFP expression system. B, effect of B-MYB on p53-induced cell cycle arrest. Transfected MCF7 cells, as described above in A, were treated with doxorubicin (100 ng/ml) for 24 h. The indicated percentages represent the cells in G0/G1 (white bars) and G2/M (black bars) arrest. As a negative control, MCF7 cells expressing pFLAG-CMV-2 empty vector that were not transfected with p53 are indicated (first lanes). The expression level of FLAG-tagged wild-type B-MYB and its deletion mutants was determined by anti-FLAG antibody immunoblot.
Our analysis was extended to examine whether B-MYB affected p53-mediated cell cycle arrest. Under the same conditions, p53-mediated G0/G1 arrest was higher in cells expressing wild-type B-MYB or deletion mutants DBD and 373/468 than in control cells expressing p53 alone (Fig. 10B, upper, second lane versus third to fifth lanes), but expression of B-MYB deletion mutants TA and R1 had no effect. In addition, the opposite trend was observed in B-MYB-knockdown cells compared with control cells expressing p53 alone (Fig. 10B, upper, second lane versus eighth lane). Similarly, the stimulatory effect of B-MYB on p53-mediated cell cycle arrest was also dose-dependent (Fig. 10B, lower). Together, these results suggest that B-MYB has the ability to potentiate the STRAP-mediated stimulation of p53-induced apoptosis and cell cycle arrest, probably through physical interaction with STRAP.
DISCUSSION
In the present study, we investigated the regulatory mechanism of the activity of STRAP, which is involved in TGF-β and p53 signaling, and provide evidence that B-MYB, a transcription factor that belongs to the MYB gene family, physically interacts with STRAP and stimulates STRAP activity, suggesting that B-MYB is a positive regulator of STRAP.
TGF-β1 had been shown to induce rapid growth arrest and apoptosis in the murine myeloid leukemia cell line Ml (27, 28), and this process was accelerated by c-myb and c-myc oncogenes (27). In addition, B-myb contributed to accelerated programmed cell death in TGF-β1-treated Ml cells without affecting the regulation of c-myb and c-myc expression (17). Thus, it is possible that a functional link between B-MYB and TGF-β signaling pathways can occur in cells. Furthermore, cross-talk between TGF-β and p53 signaling has been reported previously (29). Based on these, to investigate the functional role of the B-MYB·STRAP complex in the cross-talk between TGF-β and p53 signaling, we compared the effect of B-MYB on TGF-β signaling in the presence of p53/Mdm2 double null mouse embryonic fibroblasts with that in intact mouse embryonic fibroblasts. The enhancing effect of B-MYB on STRAP-mediated inhibition of TGF-β signaling in the p53/Mdm2 double null mouse embryonic fibroblasts was much higher than in the intact mouse embryonic fibroblasts (supplemental Fig. S2A). In addition, SMAD7-knockdown cells strengthened the B-MYB effect on STRAP-mediated stimulation of p53 signaling compared with the control in the absence of doxycycline (supplemental Fig. S2B). These data suggest that the B-MYB·STRAP complex functions dependently on the TGF-β and p53 signaling pathways. We also employed an inducible STRAP shRNA system in HEK293 cells (5) to investigate the role of STRAP in the B-MYB-mediated regulation of TGF-β and p53 signaling. The knockdown of endogenous STRAP apparently attenuated the B-MYB-mediated down-regulation of TGF-β targets or up-regulation of p53 targets compared with either parental HEK293 cells or control STRAP(KD) cells untreated with doxycycline (supplemental Fig. S3), indicating the possible involvement of B-MYB·STRAP complex in the potential cross-talk between TGF-β and p53 signaling.
B-MYB has been shown to localize mainly to the nucleus for its transcriptional activity (30, 31). However, our present results indicate that B-MYB interacts with STRAP in the cytoplasm to regulate STRAP-mediated TGF-β and p53 signaling, suggesting that B-MYB may have a functional role in the cytoplasm separate from its transcriptional activity in the nucleus. Therefore, identification of B-MYB as an interacting partner of STRAP reveals a previously unrecognized function of B-MYB, independent of its transcriptional activity, which may contribute to its role in TGF-β and p53 signaling.
To gain insight into the mechanism(s) by which B-MYB positively regulates STRAP activity, reporter assays (Figs. 4 and 8), apoptosis assays (Figs. 7 and 10), FACS analyses (Figs. 7 and 10), and confocal microscopy (Figs. 5 and 9) were used to examine whether the direct interaction between B-MYB and STRAP is a critical determinant for the regulation of STRAP-mediated TGF-β and p53 signaling. Our results showed that B-MYB deletion mutants TA and R1, which are unable to bind to STRAP, have no effect on the STRAP-mediated inhibition of TGF-β signaling or stimulation of p53 signaling, whereas a marked increase in STRAP function was detected in the presence of wild-type B-MYB and its deletion mutants DBD and 373/468, which have the capacity to interact with STRAP (see Fig. 3B). These results strongly indicate that B-MYB stimulates STRAP function through direct interaction with STRAP. The functional role of B-MYB in the regulation of STRAP-mediated TGF-β and p53 signaling in part explains the similar phenotypes of knock-out mice for B-myb (32), Strap (33), Smads (12, 34–37), and p53 (14), which show early embryonic lethality or tumorigenicity due to impaired cellular proliferation or death.
In a previous report (4), we demonstrated that PDK1 enhances STRAP-mediated inhibition of TGF-β signaling by stabilizing the association between the TGF-β receptor and SMAD7. Thus, the mechanism by which B-MYB inhibits TGF-β signaling likely modulates complex formation between the TGF-β receptor and SMAD proteins, which are essential for TGF-β signaling. To test this hypothesis, we examined the effects of wild-type B-MYB and deletion mutants including DBD, 373/468, TA, and R1 on SMAD3 and SMAD7 binding to the TGF-β receptor (Fig. 6). Our results showed that the ability of B-MYB to inhibit TGF-β signaling correlates with B-MYB-mediated modulation of SMAD3 and SMAD7 binding to the TGF-β receptor. Wild-type B-MYB and its deletion mutants DBD and 373/468 remarkably decreased the association between the TGF-β receptor and SMAD3 and correspondingly increased the association between the TGF-β receptor and SMAD7, leading to the inhibition of TGF-β signaling. In addition, the B-MYB-mediated modulation of SMAD3 and SMAD7 binding to the TGF-β receptor was only detected in the cytoplasm. We were unable to detect any physical association of TGF-β receptor with SMAD3 or SMAD7 in the nucleus under the same conditions (supplemental Fig. S4 and data not shown). These data indicate that the direct interaction of B-MYB with STRAP in the cytoplasm is essential for regulating STRAP-mediated TGF-β signaling. Based on these findings, we hypothesize that B-MYB, like PDK1 (4), may enhance STRAP-mediated inhibition of TGF-β signaling by assisting STRAP in the recruitment of SMAD7 to the TGF-β receptor and stabilizing the association between the TGF-β receptor and SMAD7. However, the possibility that B-MYB directly modulates the association between the TGF-β receptor and SMAD proteins through physical interaction with SMAD proteins, instead of via STRAP binding, cannot be ruled out because B-MYB also interacted with SMAD proteins including SMAD3 and SMAD7 in cells (data not shown).
Several interacting partners of p53, including NM23-H1 (6), STRAP (6), and MIF (21), have been shown to modulate the interaction between p53 and MDM2, a known negative regulator of p53. Therefore, to investigate how B-MYB affects STRAP-mediated activation of p53 signaling pathways such as apoptosis and cell cycle arrest (6), the effect of B-MYB on the stability and ubiquitination of p53 was analyzed. An in vivo binding assay was performed in HCT116 cells transfected with wild-type B-MYB or its deletion mutants (Fig. 9D, left). When wild-type B-MYB or its deletion mutants DBD and 373/468 were expressed in these cells, they destabilized the association between p53 and MDM2 in a dose-dependent manner. However, expression of B-MYB deletion mutants TA and R1 did not alter the level of association. Correspondingly, B-MYB also interfered with p53 ubiquitination in an in vivo ubiquitination assay (Fig. 9D, right). These data suggest that B-MYB enhances p53 function by stabilizing the STRAP effect on the p53·MDM2 complex (6), probably through direct interaction with STRAP.
It is also conceivable that STRAP exerts its effect on cells through its phosphorylation, in addition to protein-protein interaction with intracellular molecules. Previous works showed that STRAP could be phosphorylated at its 57 C-terminal amino acids by the type 1 TGF-β receptor (1), a receptor-associated serine/threonine kinase, and at Thr-175 and Ser-179 in the middle region of STRAP by ASK1 (5). Furthermore, the T175A/S179A STRAP mutant that, unlike wild-type STRAP, is not phosphorylated by ASK1 had no effect on ASK1-mediated signaling, indicating that phosphorylation also plays a critical role in regulating STRAP-mediated biological functions.
In summary, our results suggest that B-MYB positively regulates STRAP-mediated TGF-β and p53 signaling through the direct binding of STRAP. The role of B-MYB as a novel stimulator of STRAP may provide a basis for clarifying the regulatory mechanism(s) of STRAP-mediated signaling in cells. Moreover, our identification and characterization of the B-MYB·STRAP complex will contribute to understanding the unrecognized role of B-MYB independent of its transcriptional activity.
Supplementary Material
Acknowledgment
We gratefully thank Dr. Haiyoung Jung for technical assistance.
This work was supported by National Research Foundation (NRF) of Korea Grant R0A-2007-000-20006-0.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- STRAP
- serine-threonine kinase receptor-associated protein
- PDK1
- 3-phosphoinositide-dependent protein kinase-1
- ASK1
- apoptosis signal-regulating kinase 1
- SMAD
- Sma and Mad-related protein
- TβR1(TD)
- activated type 1 transforming growth factor-β receptor
- 5-FU
- 5-fluorouracil.
REFERENCES
- 1. Datta P. K., Moses H. L. (2000) Mol. Cell. Biol. 20, 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Halder S. K., Anumanthan G., Maddula R., Mann J., Chytil A., Gonzalez A. L., Washington M. K., Moses H. L., Beauchamp R. D., Datta P. K. (2006) Cancer Res. 66, 6156–6166 [DOI] [PubMed] [Google Scholar]
- 3. Anumanthan G., Halder S. K., Friedman D. B., Datta P. K. (2006) Cancer Res. 66, 10824–10832 [DOI] [PubMed] [Google Scholar]
- 4. Seong H. A., Jung H., Choi H. S., Kim K. T., Ha H. (2005) J. Biol. Chem. 280, 42897–42908 [DOI] [PubMed] [Google Scholar]
- 5. Jung H., Seong H. A., Manoharan R., Ha H. (2010) J. Biol. Chem. 285, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jung H., Seong H. A., Ha H. (2007) J. Biol. Chem. 282, 35293–35307 [DOI] [PubMed] [Google Scholar]
- 7. Sala A., Calabretta B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10415–10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raschellà G., Negroni A., Sala A., Pucci S., Romeo A., Calabretta B. (1995) J. Biol. Chem. 270, 8540–8545 [DOI] [PubMed] [Google Scholar]
- 9. Sala A., Casella I., Bellon T., Calabretta B., Watson R. J., Peschle C. (1996) J. Biol. Chem. 271, 9363–9367 [DOI] [PubMed] [Google Scholar]
- 10. Lipsick J. S. (1996) Oncogene 13, 223–235 [PubMed] [Google Scholar]
- 11. Arsura M., Introna M., Passerini F., Mantovani A., Golay J. (1992) Blood 79, 2708–2716 [PubMed] [Google Scholar]
- 12. Chang H., Huylebroeck D., Verschueren K., Guo Q., Matzuk M. M., Zwijsen A. (1999) Development 126, 1631–1642 [DOI] [PubMed] [Google Scholar]
- 13. Lin D., Fiscella M., O'Connor P. M., Jackman J., Chen M., Luo L. L., Sala A., Travali S., Appella E., Mercer W. E. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10079–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr., Butel J. S., Bradley A. (1992) Nature 356, 215–221 [DOI] [PubMed] [Google Scholar]
- 15. Lang G., Gombert W. M., Gould H. J. (2005) Immunology 114, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santilli G., Schwab R., Watson R., Ebert C., Aronow B. J., Sala A. (2005) J. Biol. Chem. 280, 15628–15634 [DOI] [PubMed] [Google Scholar]
- 17. Bies J., Wolff L. (1995) Cancer Res. 55, 501–504 [PubMed] [Google Scholar]
- 18. Liu D. X., Biswas S. C., Greene L. A. (2004) J. Neurosci. 24, 8720–8725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jung H., Seong H. A., Ha H. (2008) J. Biol. Chem. 283, 34541–34553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seong H. A., Kim K. T., Ha H. (2003) J. Biol. Chem. 278, 9655–9662 [DOI] [PubMed] [Google Scholar]
- 21. Jung H., Seong H. A., Ha H. (2008) J. Biol. Chem. 283, 20383–20396 [DOI] [PubMed] [Google Scholar]
- 22. Seong H. A., Jung H., Kim K. T., Ha H. (2007) J. Biol. Chem. 282, 12272–12289 [DOI] [PubMed] [Google Scholar]
- 23. Yao K., Shida S., Selvakumaran M., Zimmerman R., Simon E., Schick J., Haas N. B., Balke M., Ross H., Johnson S. W., O'Dwyer P. J. (2005) Clin. Cancer Res. 11, 7264–7272 [DOI] [PubMed] [Google Scholar]
- 24. Kim T., Jung H., Min S., Kim K. T., Ha H. (1999) FEBS Lett. 460, 363–368 [DOI] [PubMed] [Google Scholar]
- 25. Wieser R., Wrana J. L., Massagué J. (1995) EMBO J. 14, 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ryan K. M., Phillips A. C., Vousden K. H. (2001) Curr. Opin. Cell Biol. 13, 332–337 [DOI] [PubMed] [Google Scholar]
- 27. Selvakumaran M., Lin H. K., Sjin R. T., Reed J. C., Liebermann D. A., Hoffman B. (1994) Mol. Cell. Biol. 14, 2352–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lotem J., Sachs L. (1990) Blood 76, 1315–1322 [PubMed] [Google Scholar]
- 29. Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C., Piccolo S. (2003) Cell 113, 301–314 [DOI] [PubMed] [Google Scholar]
- 30. Sala A. (2005) Eur. J. Cancer 41, 2479–2484 [DOI] [PubMed] [Google Scholar]
- 31. Joaquin M., Watson R. J. (2003) Cell. Mol. Life Sci. 60, 2389–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanaka Y., Patestos N. P., Maekawa T., Ishii S. (1999) J. Biol. Chem. 274, 28067–28070 [DOI] [PubMed] [Google Scholar]
- 33. Chen W. V., Delrow J., Corrin P. D., Frazier J. P., Soriano P. (2004) Nat. Genet. 36, 304–312 [DOI] [PubMed] [Google Scholar]
- 34. Lechleider R. J., Ryan J. L., Garrett L., Eng C., Deng C., Wynshaw-Boris A., Roberts A. B. (2001) Dev. Biol. 240, 157–167 [DOI] [PubMed] [Google Scholar]
- 35. Nomura M., Li E. (1998) Nature 393, 786–790 [DOI] [PubMed] [Google Scholar]
- 36. Zhu Y., Richardson J. A., Parada L. F., Graff J. M. (1998) Cell 94, 703–714 [DOI] [PubMed] [Google Scholar]
- 37. Sirard C., de la Pompa J. L., Elia A., Itie A., Mirtsos C., Cheung A., Hahn S., Wakeham A., Schwartz L., Kern S. E., Rossant J., Mak T. W. (1998) Genes Dev. 12, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.