Abstract
A high intrapulmonary protease burden is characteristic of cystic fibrosis (CF), and the resulting dysregulation of the protease/anti-protease balance has serious implications for inflammation in the CF lung. Because of this inflammation, micro-bleeds can occur releasing hemoglobin into the lung. The aim of this study was to investigate the effect of the protease-rich environment of the CF lung on human hemoglobin and to assess the proinflammatory effect of heme on CF bronchial epithelium. Here, we show that the Pseudomonas proteases (Pseudomonas elastase and alkaline protease) and the neutrophil proteases (neutrophil elastase (NE) and proteinase-3) are capable of almost complete degradation of hemoglobin in vitro but that NE is the predominant protease that cleaves hemoglobin in vivo in CF bronchoalveolar lavage fluid. One of the effects of this is the release of heme, and in this study we show that heme stimulates IL-8 and IL-10 protein production from ΔF508 CFBE41o− bronchial epithelial cells. In addition, heme-induced IL-8 expression utilizes a novel pathway involving meprin, EGF receptor, and MyD88. Meprin levels are elevated in CF cell lines and bronchial brushings, thus adding to the proinflammatory milieu. Interestingly, α1-antitrypsin, in addition to its ability to neutralize NE and protease-3, can also bind heme and neutralize heme-induced IL-8 from CFBE41o− cells. This study illustrates the proinflammatory effects of micro-bleeds in the CF lung, the process by which this occurs, and a potential therapeutic intervention.
Keywords: Cystic Fibrosis, Epithelial Cell, Heme, Protease, Toll-like Receptors (TLR), Epidermal Growth Factor Receptor, Meprin, alpha-1 Antitrypsin, Interleukin-8
Introduction
Cystic fibrosis (CF)2 is a chronic inflammatory disease, which is associated with high protease levels in the lung. The normal fine balance between the positive physiological function of the proteases and their deleterious effect is dysregulated in the CF lung. This results in unopposed protease activity that can cause lung damage.
In the CF lung, proteases are secreted by the host and also by bacteria that colonize the lung. The host secretes serine, metallo, cysteine, and aspartyl proteases that include elastases, matrix metalloproteases, cathepsins, prolyl endopeptidases, and napsins. However, it is the elastases that are the main contributor to the total protease activity found in the CF lung. Given the abundance of neutrophils in CF, the principal lung proteases are secreted by the neutrophil. These include neutrophil elastase (NE), protease-3 (PR-3), and cathepsin G. NE is thought to contribute 90% of the CF sputum elastinolytic activity (1) with PR-3 contributing 7% and the balance of activity derived from the remaining proteases. NE cleaves a broad spectrum of proteins, including immunoglobulins, plasma proteins, matrix proteins, cytokines, and protease inhibitors. In addition, NE can amplify inflammatory signaling by direct and transcriptional (2) regulation or activation of other proteases and induction of IL-8 from airway epithelium (3–5).
Although often disregarded, bacteria within the lung also contribute to the protease load in the CF lung. The range of bacteria that colonize the CF lung include Pseudomonas aeruginosa, Haemophilus influenzae, Burkholderia cepacia complex, and Prevotella, Veillonella, Rothia, Streptococcus, and Staphylococcus species. The most common bacteria to colonize the CF lung is P. aeruginosa, which is known to secrete the metalloproteases Pseudomonas elastase (PsE), alkaline protease (APR), and the less abundant staphylolysin. It is known that PsE and APR are capable of degrading a broad range of host proteins and of modifying the physiology of the CF lung (6–9).
Pseudomonas like many bacteria requires iron for growth, and degradation of host iron-containing proteins is an excellent source of such iron. Both the Pseudomonas proteases PsE and APR and the host protease NE are capable of cleaving transferrin and lactoferrin (7). In addition, a recent paper by Fischer et al. (10) showed that NE could cleave the iron-containing protein ferritin to produce an increase in lung iron levels. In addition to free iron, bacteria also utilize heme, which can be released from hemoglobin. This is of particular significance in cystic fibrosis due to the frequent occurrence of micro-bleeds, which leads to the presence of hemoglobin at the delicate CF lung epithelia in the presence of both host and bacterial proteases.
Hemoglobin is the main hemoprotein of the blood and accounts for 97% of the dry weight of the red blood cells. Hemoglobin is tetrameric, consisting of two α- and two β-globin chains (α2β2). In the center of each globin chain is a large central cavity where the heme group is bound by noncovalent forces (reviewed in Ref. 11). Hemoglobin when contained within the red blood cells is in the tetrameric form; however, on release it can dissociate into its dimeric form. Ferrous hemoglobin (Fe2+) may then be subject to autoxidation or by reaction with free radicals (12) converted to ferric (Fe3+) hemoglobin (methemoglobin). Formation of methemoglobin can lead to subsequent heme release (12). In addition to heme release by oxidation of oxyhemoglobin, heme has also been shown to be released by proteolytic activity of the protease interpain from the anaerobe Prevotella, which is associated with periodontal diseases (13). To date, there are no studies on the proteolytic effect of the principal CF lung proteases PsE, APR, NE, PR-3, and cathepsin G on the structure of hemoglobin. In this work, we study the ability of these proteases to degrade hemoglobin and in addition evaluate the effect of the liberated heme on CF bronchial epithelia.
Heme has been found to be an activator of G protein-coupled receptors where by it can activate neutrophil migration and reactive oxygen species production (14). In addition, in macrophages heme can induce the cytokine tumor necrosis factor α in an MyD88-, CD-14-, and TLR4-dependent manner (15). This is of particular interest as TLR and EGFR (3) pathways in CF bronchial epithelium are important mediators of inflammation. This study set out to examine the effect of the CF lung and lung proteases on hemoglobin. We further investigated the role of heme in the TLR/EGFR pathway and its effect on proinflammatory cytokine expression. Finally, we assessed the inhibitory capacity of AAT against the heme-induced effects.
EXPERIMENTAL PROCEDURES
Chemicals
Chemicals and reagents were purchased from Sigma unless indicated otherwise. Neutrophil elastase, PR-3, and Pseudomonas elastase were purchased from Elastin Products Co. (Owensville, MO).
Bronchoalveolar Lavage (BAL) Fluid Sample Collection
CF BAL was obtained from people with CF or non-CF controls undergoing bronchoscopy as part of routine care. During the procedure, 30 ml of sterile 0.9% NaCl was instilled into the right or left sub-segmental bronchi and then immediately collected and placed at 4 °C. The samples were centrifuged at 2000 rpm for 10 min at 4 °C. Supernatants were removed, aliquoted, and maintained at −80 °C until use in experiments. Prior to use in experiments, equal volumes from five separate CF BAL and non-CF samples were pooled together in equal volumes, and pooled CF BAL and non-CF BAL were used in experiments.
SDS-PAGE
Proteins were separated by electrophoresis in 12% acrylamide (30% w/v)/bisacrylamide (0.8% w/v) SDS gels. The separating and stacking gel buffers were pH 8.9 and 6.8, respectively. The gels were run in a Tris (25 mm), glycine (0.2 m) buffer, pH 8.9, with a continuous SDS concentration of 0.1% (w/v). In addition, gels were also run using the NuPAGE® Novex 12% BisTris gel system. The samples were treated with reducing sample buffer and boiled for 5 min prior to electrophoresis. The gels were stained with Coomassie Blue. For Western blot analysis, proteins were transferred to polyvinylidene difluoride membrane at 100 mA for 1 h using a semidry electrophoretic blotting system and blocked with 3% (w/v) marvel and 1% (w/v) BSA in PBST (phosphate-buffered saline/Tween). Specific rabbit polyclonal Abs were used to detect meprin (Dr. Erwin E. Sterchi), TACE (Santa Cruz Biotechnology, Heidelberg, Germany), and GAPDH (Santa Cruz Biotechnology) with a 1:1000, 1:250, and 1:1000 (respectively) dilution in the blocking solution. The nitrocellulose was washed three times for 10 min in PBS containing 0.01% (v/v) Tween 20 (PBST). The nitrocellulose was incubated with anti-rabbit IgG HRP-linked Ab (Santa Cruz Biotechnology), which was diluted 1:1000 in the blocking reagent. The nitrocellulose was washed again, and the immunoreactive protein bands were visualized employing Immobilon Western chemiluminescent substrate (Millipore). Immunoreactive bands were quantified by densitometry using the Syngene G:BOX Chemi XL gel documentation system (Syngene, Cambridge, UK). For 3,3′,5,5′-tetramethylbenzidine (TMB) gel analysis, the samples were treated with nonreducing sample buffer (0.25% bromphenol blue, 50 mm Tris-HCl, pH 7.5, 40% glycerol, and 1% SDS) prior to electrophoresis in 12% acrylamide (30% w/v)/bisacrylamide (0.8% w/v) SDS gel. The gel was then incubated with 10 ml of TMB ultrasensitive substrate (Millipore) at 37 °C in the dark for between 30 min and 1 h, and the image was captured using the Syngene G:BOX Chemi XL gel documentation system.
Cell Culture and Treatment
All cell lines were maintained in a 37 °C humidified CO2 incubator in minimal essential medium (MEM) supplemented with 10% fetal calf serum (FCS), 1% l-glutamine, 1% penicillin/streptomycin (Invitrogen). 16HBE14o− cells are an SV40-transformed human bronchial epithelial cell line, and CFBE41o− cells are human ΔF508 homozygote bronchial epithelial cells. The cell lines were obtained as a gift from D. Gruenert (California Pacific Medical Center Research Institute, San Francisco) (16). Prior to treatment, cells were placed in serum-free media overnight, and immediately prior to treatment with heme, cells were placed in fresh serum-free media. The exception to this is when treated with P. aeruginosa LPS the media contained 1% FCS. All cell culture experiments were carried out in triplicate. Heme treatment in all cases was for 24 h in serum-free media.
Heme Quantification
Hemin chloride was dissolved in 20 mm sodium hydroxide and centrifuged at 14,000 × g prior to quantification using the Nanodrop 8000 spectrophotometer (Thermo Scientific) using 385nm = 58,400 m−1 cm−1 (17). Prior to cell culture experiments, heme was diluted in serum-free media, and the pH was adjusted to pH 7.5, and the solution was filter-sterilized using a sterile Millex GP 0.22-μm syringe-driven filter unit (Millipore).
Protoporphyrin IX (PIX) Quantification
PIX was prepared in the same way as heme except using 408nm = 262,000 m−1 cm−1 (18).
Heme Spectrophotometric Assay
Heme was quantified by a modified version of that of Huy et al. (19). Briefly, 100 μl of sample or heme standard was added to 400 μl of TMB-ultrasensitive substrate (Millipore) for 30 min. 2 m sulfuric acid (500 μl) was added, and the absorbance was quantified at 450 nm using Multiskan Ex (Labsystems).
Hemoglobin-Agarose Experiment
To measure heme release from hemoglobin subunits, hemoglobin-agarose (6 mg/ml) was incubated with or without NE (1 μm) or with or without Pefabloc (2 mm, 1 h of NE pretreatment at room temperature) at 37 °C. Samples were removed at the specified times, and Pefabloc (2 mm) was added to samples not already containing Pefabloc, and the samples were stored at −20 °C. Heme release in the supernatants was measured by heme spectrophotometric assay (as described above).
IL-8 and IL-10 ELISA
IL-8 and IL-10 protein concentrations in the cell supernatants were determined by sandwich ELISA (R & D Systems).
Preparation of Polymyxin B-treated Heme
Heme (2.5 mm) and control were treated with polymyxin B sulfate (50 μg/ml) at room temperature for 30 min. The samples were then centrifuged at 14,000 × g for 15 min at 4 °C. The supernatant was then used to stimulate the CFBE41o− cells in triplicate overnight.
Transfection
Transfection of CFBE41o− cells with pCDNA3.1 and ΔMyD88 was carried out as described previously (3).
Heme and EGFR Inhibition
CFBE41o− cells (1 × 105) were treated in triplicate with media or DMSO, the EGFR inhibitor AG1478 (500 nm), or an anti-EGFR-neutralizing antibody (2 μg, Calbiochem) for 1 h prior to treatment with heme (5 μm, 24 h). IL-8 levels in supernatants were quantified by ELISA. CFBE41o− cells (1 × 105) were incubated in triplicate with a mixture of EGFR agonist-neutralizing antibodies. The neutralizing mixture of antibodies contained anti-TGFα (10 μg), anti-EGF (16 μg), anti-heparin-binding epidermal growth factor-like growth factor (5 μg), anti-amphiregulin (2 μg), anti-epiregulin (3 μg), and anti-betacellulin (0.16 μg). The CFBE41o− cells were pretreated for 1 h prior to stimulation with heme (8.75 μm, 24 h). IL-8 levels in supernatants were quantified by ELISA.
CFBE41o− Cells Treated with Metalloprotease Inhibitors
CFBE41o− cells (1 × 105) were incubated in triplicate with and without GM6001 (10 μm) or actinonin (100 μm) for 1 h prior to treatment with heme (8.75 μm, 24 h). IL-8 levels in supernatants were quantified by ELISA.
Meprin and TACE RT-PCR
Total RNA was extracted from 16HBE14o− cells and CFBE41o− cells using TRI Reagent. qRT-PCR was carried out using the LC480 and SYBR Green I Master with TACE (Roche Applied Science) (forward, 5′-GGTGGTGGATGGTAAAAACG-3′, and reverse, 5′-GCCCCATCTGTGTTGATTCT-3′), meprin α (forward, 5′-CATGAGAAGTATGACAACAGCCT-3′, and reverse, 5′-AGTCCTTCCACGATACCAAAGT-3′) or GAPDH primers (MWG Biotec). Relative expression was determined using the 2(2ΔΔCt) method. Meprin and TACE RT-PCR was also carried out in patient bronchial brush samples. Eight individuals were recruited into this study; four had CF (confirmed by sweat testing and/or genotyping) and four were non-CF controls. Bronchial brushings were collected as described previously (20, 21). Total RNA was isolated using TRI Reagent, and meprin RT-PCR was carried out as described above.
Meprin and TACE Knockdown Using siRNA
CFBE41o− cells (0.8 × 105) were transfected with 50 nm of an siRNA-targeting meprin α (siRNA ID 104082, Ambion), TACE (siRNA ID 104029, Ambion), or a scrambled control and GAPDH (silencer GAPDH siRNA control, Ambion) for 24 h. Cells were placed in serum-free medium overnight and in fresh serum-free medium prior to treatment with heme (7.5 μm for 24 h). Total RNA was extracted for qRT-PCR using TRI Reagent (Sigma) and meprin, TACE, and GAPDH; RT-PCR was carried out as described above to quantify knockdown. Cell supernatants were retained for IL-8 ELISA.
AAT De-salting and Quantification
AAT was dissolved in ultra pure water, desalted on an illustra NAP-5 column (GE Healthcare), and then quantified spectrophotometrically using 280 nm = 4.33 w/v cm−1 and using the nanodrop 8000 spectrophotometer (Thermo Scientific). AAT used in cell culture was diluted in SFM and filter-sterilized using a sterile Millex GP 0.22-μm syringe-driven filter unit (Millipore).
AAT ELISA
AAT (20 μg/ml) purified from human plasma (Athens Research and Technology, Athens, GA) was immobilized on a 96-well plate overnight at 4 °C before treatment with heme (50 μm, 4 h). AAT-bound heme was detected spectrophotometrically by direct heme reaction with TMB ultrasensitive substrate (Millipore) (100 μl/well) at 650 nm using Multiskan Ex (Labsystems). To investigate the specificity of the interaction of heme and AAT, heme binding to other proteins was examined. AAT (20 μg/ml), trypsin (20 μg/ml), porcine pancreatic elastase (PPE, 20 μg/ml), and elastin (20 μg/ml) were immobilized on a 96-well plate overnight before treatment with heme (50 μm). Protein-bound heme was detected spectrophotometrically by direct heme reaction with TMB-ultrasensitive substrate (Millipore) (100 μl/well) at 650 nm using Multiskan Ex (Labsystems).
AAT Neutralization of Heme-induced IL-8
CFBE41o− cells (1 × 105) were treated with heme (8.75 μm, 24 h) in the presence and absence of filter sterile AAT (20 μm). IL-8 levels in supernatants were quantified by ELISA.
EGFR ELISA
A soluble human EGFR recombinant fragment that contains the heme-binding motif (20 μg/ml) (Abcam, 330 Cambridge Science Park, Cambridge, CB40FL, UK) was immobilized on a 96-well plate overnight at 4 °C before treatment with heme (50 μm, 4 h). EGFR-bound heme was detected spectrophotometrically by direct heme reaction with TMB-ultrasensitive substrate (Millipore) (100 μl/well) at 650 nm using Multiskan Ex (Labsystems).
Statistical Analysis
All analyses were performed using GraphPad Prism 4.0 software package (San Diego). Results are expressed as the mean ± S.E. and were compared by Student's t test (nonparametric, one-tailed). Differences were considered significant at p ≤ 0.05.
RESULTS
Effect of Pseudomonas and Neutrophil Proteases on Hemoglobin
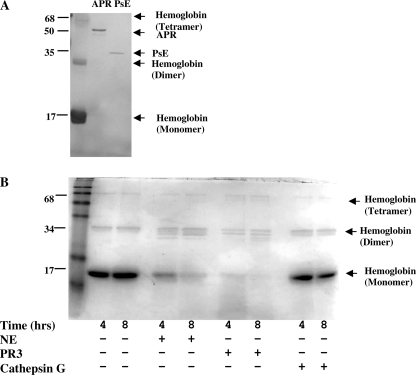
To examine if hemoglobin can be degraded by proteases found in the CF lung, hemoglobin was first incubated with the most abundant Pseudomonas proteases, PsE and APR. After 24 h of incubation at 37 °C, the hemoglobin is almost fully degraded (Fig. 1A). In addition to the bacterial proteases, a high concentration of the neutrophil proteases NE, PR-3, and cathepsin G are present in the CF lung. Incubation of hemoglobin at 37 °C for 4 and 8 h shows that the hemoglobin control does not degrade over the experiment time course (Fig. 1B). After 4 h, NE shows significant degradation of hemoglobin, and after 8 h the majority of the hemoglobin has been degraded. PR-3 is also capable of hemoglobin degradation and shows almost total degradation after 4 h without significant further degradation occurring between 4 and 8 h. In contrast, hemoglobin incubated with cathepsin G showed no detectable cleavage at either 4 or 8 h.
FIGURE 1.
Effect of Pseudomonas and neutrophil proteases on hemoglobin. Representative polyacrylamide gels were stained with Coomassie Brilliant Blue (R-250) of hemoglobin degradation by alkaline protease (1.6 μm) and Pseudomonas elastase (2.6 μm) after 24 h at 37 °C (n = 3) (A), and the effect of neutrophil proteases (1 μm) elastase (NE), protease-3, and cathepsin G on hemoglobin was at 37 °C (n = 2) (B).
Effect of CF BAL on Cleavage of Hemoglobin
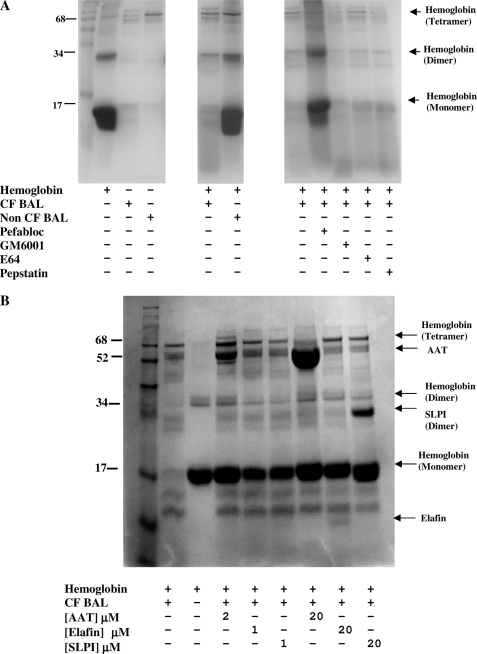
To investigate if hemoglobin is degraded in the CF and/or normal lung, hemoglobin was incubated at 37 °C for 48 h with CF and non-CF BAL (Fig. 2A). CF but not non-CF BAL degraded the hemoglobin. To determine the proteases that degrade hemoglobin in vivo in the CF lung, a range of protease inhibitors was included in the hemoglobin and CF BAL incubations. Degradation of hemoglobin by CF BAL was inhibited by the serine protease inhibitor Pefabloc but not by the metalloprotease inhibitor GM6001, the cysteine protease inhibitor E64, or the aspartyl protease inhibitor pepstatin A. The principal protease responsible for the cleavage of hemoglobin in CF BAL is a serine protease.
FIGURE 2.
Effect of CF BAL on cleavage of hemoglobin. A, hemoglobin (1.2 mg/ml) was incubated with CF and non-CF BAL (standardized by protein concentration) for 48 h at 37 °C in the presence and absence of protease inhibitors Pefabloc (10 mm), GM6001 (0.1 mm), E64 (2.8 mm), or pepstatin A (3 mm) (n = 2). B, hemoglobin (0.9 mg/ml) was incubated in the presence of CF BAL (1st lane), in the absence of CF BAL (2nd lane), in the presence of physiological concentrations of endogenous protease inhibitors AAT, elafin, or SLPI (2, 1, and 1 μm) (3rd to 5th lanes) and excess endogenous protease inhibitors AAT, elafin, or SLPI (20 μm) (6th to 8th lanes) for 24 h at 37 °C (n = 3).
Hemoglobin cleavage by CF BAL proteases was also inhibited by the endogenous inhibitors AAT, elafin, and SLPI (Fig. 2B) after 24 h. The bands of the endogenous inhibitors themselves can be seen at 52, 24, and 6 kDa due to AAT, SLPI dimers, and elafin, respectively. Equimolar concentrations of the endogenous inhibitors were used (at 20 μm); therefore, the inhibitors have different band intensities due to their different molecular weights. The hemoglobin and BAL were incubated with both physiological concentrations of the endogenous inhibitors and in excess because when there is bleeding in the CF lung the inhibitor levels will lie somewhere between the lung physiological levels and that of the blood. A large decrease in the intensity of the 17-kDa hemoglobin band is evident in the presence of CF BAL after 24 h. Pretreatment with AAT, elafin, or SLPI inhibits this degradation at both physiological lung concentrations and when the inhibitors are in excess. NE is strongly inhibited (1.8 × 10−11 m) by SLPI; however, PR3 is not significantly inhibited by SLPI (22). At both concentrations of SLPI (1 and 20 μm) after 24 h, the SLPI-inhibited BAL shows little cleavage indicating that NE and not PR3 is the predominant protease involved in in vivo hemoglobin degradation.
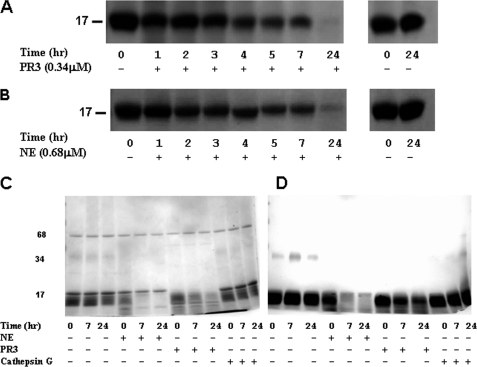
Differential Cleavage Patterns of Hemoglobin by the Neutrophil Proteases NE and PR-3
NE and PR-3 show different patterns of cleavage of hemoglobin. At equimolar concentrations, PR-3 degrades hemoglobin more rapidly than NE (Fig. 1B). However, NE cleavage of hemoglobin causes release of heme from the hemoglobin at an earlier stage of degradation than PR-3 cleavage. After NE cleavage of hemoglobin, a large amount of hemoglobin protein remains uncleaved as shown by the Coomassie gel (Fig. 3C). However, NE has released a proportionally large amount of heme as shown by the lighter bands on the TMB gel (to detect heme) (Fig. 3D). The NE-liberated heme is not bound to large fragments of hemoglobin and probably gets lost by diffusion. In contrast, in the Coomassie gel of PR-3 cleavage of hemoglobin, the majority of the hemoglobin monomers have been proteolyzed, but the TMB gel shows that PR-3-mediated hemoglobin monomer degradation releases larger degradation products that still retain the heme moiety.
FIGURE 3.
Differential cleavage patterns of hemoglobin by the neutrophil proteases NE and PR3. Representative polyacrylamide gels stained with Coomassie Brilliant Blue (R-250) of hemoglobin (1 mg/ml) incubated with PR3 (0.34 μm) (A) and NE (0.68 μm) (B) at 37 °C. At the specified times, samples were removed; Pefabloc (4 mm) was added and stored at −20 °C before electrophoresis. C, polyacrylamide gels stained with Coomassie Brilliant Blue (R-250); D, polyacrylamide gels stained with TMB of hemoglobin (3 mg/ml) incubated with NE (0.3 μm) or PR3 (0.03 μm) or cathepsin G (0.17 μm) for 0 (sample taken ∼2 min after protease addition), 7m and 24 h. Data shown are representative of two experiments.
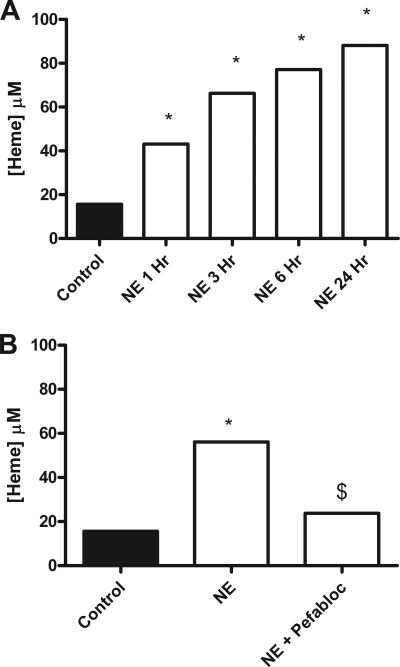
Effect of NE on Heme Release from Hemoglobin-Agarose in the Presence and Absence of Pefabloc
To investigate heme release from hemoglobin subunits, hemoglobin-agarose beads were incubated with NE in the presence and absence of Pefabloc. Incubation of NE with hemoglobin-agarose beads causes release of heme into the supernatants in a time-dependent manner (Fig. 4A). Heme release is due to the proteolytic activity of NE as incubation of NE with Pefabloc inhibits the heme release (Fig. 4B). TMB gel analysis of the 24-h sample is negative for heme, illustrating that the heme released from the beads by the NE is <4 kDa in size. Hemoglobin-agarose beads were incubated with NE and the control (Pefabloc-inhibited NE). Preceding the incubation, Pefabloc was added to the NE-hemoglobin-agarose supernatant to ensure all samples contained equal NE and Pefabloc concentrations prior to treatment of CFBE41o− cells. The cells were treated with supernatant that had been diluted in serum-free media and filter-sterilized. The supernatants from the incubation of Pefabloc-inhibited NE with hemoglobin-agarose beads neither caused significant heme release (Fig. 4B), as measured by the heme assay, nor IL-8 release from CFBE41o− cells. In contrast, the supernatants from active NE incubated with hemoglobin-agarose beads contained NE-liberated heme (as measured by the heme assay) (Fig. 4B) and caused significant IL-8 release as determined by IL-8 ELISA (data not shown).
FIGURE 4.
Effect of NE on heme release from hemoglobin-agarose in the presence and absence of Pefabloc. A, hemoglobin-agarose (6 mg/ml) was incubated with NE at 37 °C with samples removed at the specified times, and Pefabloc (2 mm) was added. Heme release (0–24 h) in the supernatants was measured by heme spectrophotometric assay. B, NE (1 μm) was incubated with and without Pefabloc (1 h, room temperature, 2 mm) and then hemoglobin-agarose (6 mg/ml) (37 °C, 24 h) was added. Heme release into the supernatants was measured by heme spectrophotometric assay. All heme assay experiments were performed in triplicate, and the data are represented as mean ± S.E. and are representative of two (A) and three (B) experiments. A, *, p ≤ 0.05, NE versus control; B, *, p ≤ 0.05, NE versus control; $, p ≤ 0.05, NE versus NE + Pefabloc.
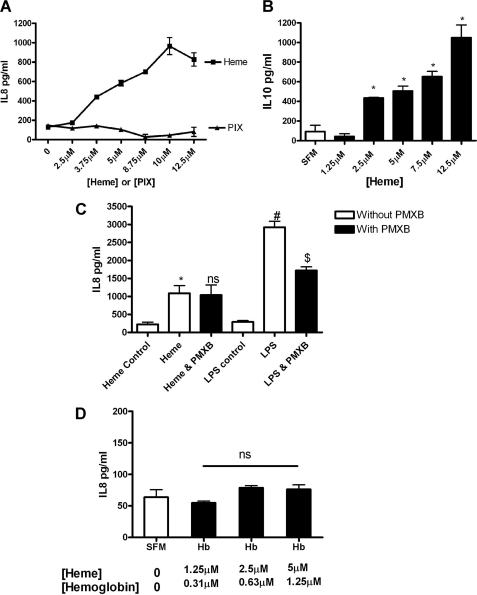
Effect of Heme on IL-8 and IL-10 Expression by CFBE41o− Cells
Heme induces dose-dependent IL-8 and IL-10 expression in CFBE41o− cells as shown by ELISA (Fig. 5, A and B). To rule out any endotoxin contamination, polymyxin B (PMXB), which binds endotoxin, was used to treat the heme. There is no significant difference in the IL-8 response between heme in the presence of PMXB versus in its absence, whereas an LPS-induced effect is significantly decreased by PMXB (Fig. 5C). To demonstrate that cleavage of hemoglobin to release heme is required for IL-8 release from CFBE41o− cells, the cells were treated with hemoglobin (Fig. 5D). Equal concentrations of heme in the form of hemoglobin do not cause IL-8 release. To characterize the molecular determinants of the heme-induced IL-8 secretion, CFBE41o− cells were treated with the heme precursor PIX (Fig. 5A). PIX differs from heme by the absence of the iron group. PIX was unable to induce IL-8 over a broad concentration range; therefore, heme-induced IL-8 was found to require the iron moiety of the heme compound.
FIGURE 5.
Effect of heme on IL-8 and IL-10 expression by CFBE41o− cells. CFBE41o− cells (1 × 105) were treated with heme (0–12.5 μm) or PIX (0–12.5 μm) for 24 h and IL-8 (A), and IL-10 secretion into supernatant was measured by ELISA (B). C–D, IL-8 levels were quantified by ELISA in response to heme or heme + PMXB (50 μg) or LPS (50 μg) or LPS + polymyxin B (C) or hemoglobin (0–5 μm, [heme]) (D). ELISAs were preformed in triplicate; the data are represented as mean ± S.E. and are representative of three (A), one (B), three (C), and three (D), experiments. *, p ≤ 0.05, heme versus SFM control; B, *, p ≤ 0.05, heme versus SFM control; C, *, p ≤ 0.05, heme versus heme control; #, p ≤ 0.05, LPS versus LPS control; $, p ≤ 0.05, LPS versus LPS and PMXB (D); p = not significant (ns), hemoglobin versus SFM control.
ΔMyD88 Inhibits Heme-induced IL-8 Expression
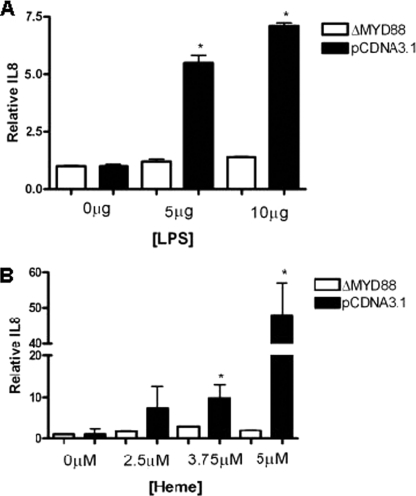
TLR4 has previously been reported to be activated by heme in the macrophage (15). To determine the role of TLRs in heme-induced IL-8 expression in CFBE41o− cells, we investigated the effect of a dominant negative MyD88 transgene (ΔMyD88). MyD88 is an adaptor protein for all of the TLR excluding TLR3. ΔMyD88 is a dominant negative form that contains only a functional TIR domain and lacks the death domain required for downstream signaling. Compared with empty vector-transfected cells, overexpression of ΔMyD88 inhibited LPS-induced (Fig. 6A) and heme-induced (Fig. 6B) IL-8 expression, implicating TLR signaling in these events.
FIGURE 6.
ΔMyD88 inhibits heme-induced IL-8 expression. CFBE41o− cells (1 × 105) were co-transfected for 24 h with pRLSV40 and an empty plasmid pCDNA3.1 or a vector expressing ΔMyD88 and then stimulated for 24 h with LPS (5–10 μg/ml) (A) or heme (2.5–5 μm) (B). Cells were lysed, and transfection efficiency was quantified by luminometry. IL-8 secretion in supernatants was measured by ELISA, and relative IL-8 levels in supernatants are shown. All ELISA experiments were performed in triplicate; the data are represented as mean ± S.E. Data shown are representative of three experiments. A, *, p ≤ 0.05, ΔMyd88 versus pCDNA3.1 with 5 μg or 10 μg of LPS; B, *, p ≤ 0.05, ΔMyD88 versus pCDNA3.1 with 3.75 or 5 μm heme.
Heme-induced IL-8 Occurs via Activation of EGFR by EGFR ligand in CFBE41o− Cells
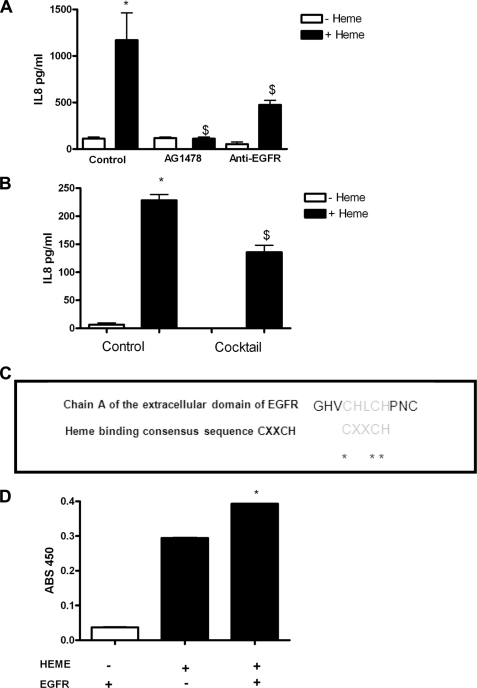
Heme-mediated IL-8 production in CFBE41o− cells in the presence of the EGFR inhibitor AG1478 or an EGFR-neutralizing antibody is reduced (Fig. 7A). This indicates that heme-mediated IL-8 production involves EGFR. Heme-mediated activation of EGFR is mediated by endogenous EGFR agonists that can be blocked by a mixture of EGFR agonist-neutralizing antibodies (anti-TGFα, anti-EGF, anti-heparin-binding epidermal growth factor-like growth factor, anti-amphiregulin, anti-epiregulin, and anti-betacellulin) (Fig. 7B). When individual EGFR agonists are neutralized, there is no significant decrease in IL-8 production (data not shown). Thus heme-mediated EGFR activation is dependent on more than one individual EGFR agonist, and therefore, when the agonists are neutralized individually, another compensates.
FIGURE 7.
Heme-induced IL-8 occurs via activation of EGFR by EGFR ligands in CFBE41o− cells. CFBE41o− cells (1 × 105) were treated with media or DMSO- (control), AG1478- (500 nm), or an EGFR-neutralizing antibody (2 μg,) for 1 h prior to treatment with heme (5 μm, 24 h) (A). IL-8 levels in supernatants were quantified by ELISA. CFBE41o− cells (1 × 105) were incubated with anti-TGFα- (10 μg), anti-EGF- (16 μg), anti-heparin-binding epidermal growth factor-like growth factor- (5 μg), anti-amphiregulin- (2 μg), anti-epiregulin- (3 μg), and anti-betacellulin (0.16 μg)-neutralizing antibodies (mixture, 1 h) and then stimulated with heme (8.75 μm, 24 h) (B). IL-8 levels in supernatants were quantified by ELISA. All IL-8 ELISA experiments were performed in triplicate; the data are represented as mean ± S.E. IL-8 ELISA data shown are representative of three experiments. C, alignment of chain A of an extracellular EGFR domain (UniProtKB/Swiss-Prot code P00533 (EGFR_HUMAN)) with the heme-binding motif. The asterisks indicate conserved residues within the motif. D, soluble human EGFR recombinant fragment containing the heme-binding motif binds heme. 20 μg/ml soluble human EGFR recombinant fragment was immobilized on a 96-well plate overnight before treatment with heme (50 μm). EGFR-bound heme was detected spectrophotometrically by direct heme reaction with TMB ultrasensitive substrate (100 μl/well). EGFR ELISA experiments were performed in duplicate; the data are represented as mean ± S.E. EGFR ELISA data shown are representative of two experiments. A, *, p ≤ 0.05, control versus heme; $, p ≤ 0.05, AG1478 + heme and anti-EGFR + heme versus control + heme; B, *, p ≤ 0.05, control versus heme; $, p ≤ 0.05, mixture + heme versus control + heme; D, *, p ≤ 0.05, heme or EGFR alone versus heme and EGFR.
Heme-induced IL-8 Is Involves a Metalloprotease
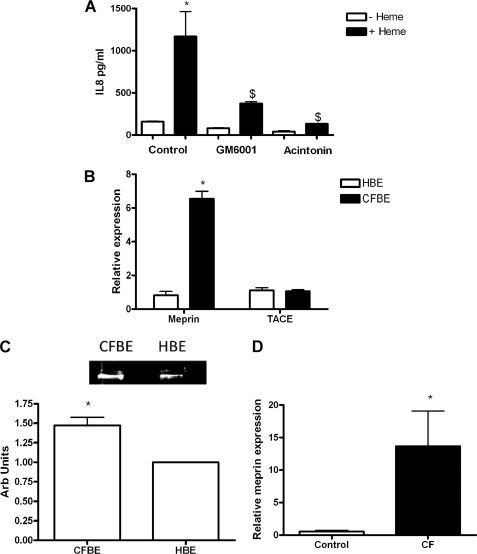
Pretreatment of CFBE41o− cells with either the generic metalloprotease inhibitor GM6001 or the meprin inhibitor actinonin decreases heme-mediated IL-8 production (Fig. 8A), indicating that a metalloprotease may be involved in the generation of the EGFR ligand. The metalloproteases meprin and TACE have both been implicated in the EGFR signaling pathway (3, 23). These were investigated as the metalloproteases involved in this pathway.
FIGURE 8.
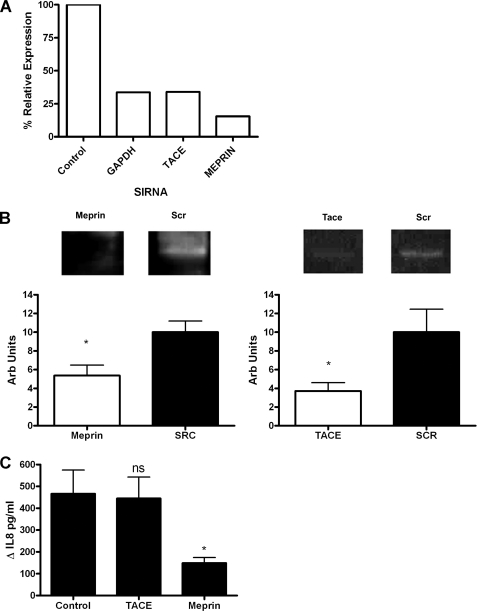
Meprin and TACE RNA expression in vitro and in vivo and in vitro meprin protein expression relative to GAPDH. A, CFBE41o− cells (1 × 105) were incubated with and without GM6001 (10 μm) or acintonin (100 μm) for 1 h prior to treatment with heme (8.75 μm, 24 h). IL-8 levels in supernatants were quantified by ELISA. B, relative meprin and TACE expression in CFBE41o− and 16HBE14o− cell lines (n = 3) was measured by qRT-PCR. C, meprin protein was analyzed by Western blot (n = 3) using anti-meprin and anti-GAPDH antibodies in CFBE41o− and 16HBE14o− cell lines. Representative densitometry of meprin relative to GAPDH. CFBE, cystic fibrosis bronchial epithelial cells; HBE, human bronchial epithelial cells. D, relative meprin expression in CF versus non-CF bronchial brushings (n = 4) was measured by qRT-PCR. All qRT-PCR experiments were performed in triplicate and included no-template controls; the data are represented as mean ± S.E. A, *, p ≤ 0.05, control versus heme; $, p ≤ 0.05, GM6001 + heme and acintonin + heme versus control + heme; B, *, p ≤ 0.05, meprin relative expression in CFBE41o− versus 16HBE14o− cell lines; D, *, p ≤ 0.05, meprin relative expression in CF versus non-CF bronchial brushings.
Meprin Mediates Heme-induced IL-8 Expression in CFBE41o−
Meprin exists in two forms as follows: membrane-bound β subunits and non-membrane-bound α subunits (24, 25). We have previously shown that meprin α is found in both 16HBE14o− human bronchial epithelial cells and in CF bronchial lavage fluid (3). Meprin and TACE expression were examined using RT-PCR in vitro in CFBE41o− and 16HBE14o− cell lines (Fig. 8B). In CFBE41o− cells, meprin was expressed at ∼6-fold higher levels than in 16HBE14o− cells. When meprin protein was examined by Western blot, CFBE41o− cells showed a 32 ± 7% greater amount of meprin protein (Fig. 8C). This is in agreement with the zymograph data of Bergin et al. (3). In vivo meprin expression was then examined by RT-PCR in bronchial brushings from people with CF and controls (Fig. 8D). Meprin expression in people with CF was significantly higher than in controls. This indicated that meprin most likely has a more central role in CF airway than in control airways. To investigate this in the context of heme-induced IL-8 expression, meprin and TACE were knocked down via siRNA in CFBE41o− cells, and these were then treated with heme (Fig. 9C). The knockdown of meprin and TACE was validated at both RT-PCR (Fig. 9A) and at protein level (Fig. 9B).
FIGURE 9.
Meprin mediates heme-induced IL-8 expression in CFBE41o−. A, CFBE41o− cells (0.8 × 105) were untransfected (control) or transfected with GAPDH, meprin α, or TACE (50 nm) siRNAs, and knockdown was quantified by qRT-PCR comparing siRNA target gene to β-actin expression, % relative expression is shown. B, meprin and TACE protein knockdown was analyzed by Western blot using anti-meprin, anti-TACE, and anti-GAPDH antibodies in CFBE41o− cell lines that had been transfected with meprin and TACE siRNA. Representative densitometry of meprin and TACE knockdown relative to GAPDH is shown (n = 5 and 6, respectively). The data are represented as mean ± S.E. *, p ≤ 0.05, meprin versus control and TACE versus control. These cells were stimulated with heme (7.5 μm, 24 h), and IL-8 levels in supernatants were quantified by ELISA (C). The graph shows ΔIL-8 (pg/ml) versus untreated cells. All ELISA and qRT-PCR experiments were preformed in triplicate; the data are represented as mean ± S.E. Data shown are representative of three experiments. *, p ≤ 0.05, meprin versus control.
When TACE was knocked down, there was no significant difference in the ΔIL-8 between control and heme-treated cells (Fig. 9C). In contrast, when meprin was knocked down there was a significant difference in the ΔIL-8 between control and heme-treated cells (Fig. 9C). These results indicate that meprin is involved in the heme-induced IL-8 pathway.
Heme-induced IL-8 May Involve Another Pathway in Addition to Meprin-activated EGFR
Although a significant decrease in IL-8 is seen when the mixture of EGFR ligand-neutralizing antibodies is used to inhibit heme-induced IL-8 release, there is some residual IL-8 secretion. We hypothesize that heme may also bind directly to EGFR to activate it. We have identified a heme-binding motif in chain A of an extracellular EGFR domain (UniProtKB/Swiss-Prot code P00533 (EGFR_HUMAN)) (Fig. 7C). The heme-binding motif identified “CXXCH” is a conserved domain that is also found in the heme-binding molecule cytochrome and in the recently identified heme-bound and functionally modified channel Slo1 (26). In support of this interaction when a soluble human EGFR recombinant fragment containing the heme-binding motif is coated on a 96-well plate and then treated with heme, there is significantly more heme bound by the EGFR-coated wells than uncoated wells (Fig. 7D). This shows that the EGFR chain and heme physically interact. We propose that in addition to the EGFR and meprin pathway that heme can also bind directly to EGFR and induce signaling.
AAT Binds Heme and Neutralizes Heme-induced IL-8 Secretion by CFBE41o−
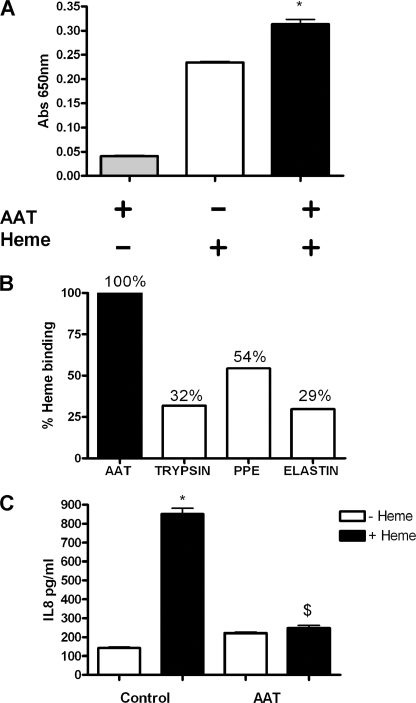
To examine whether AAT represents a possible therapeutic to treat heme-mediated inflammation in the lung, the ability of AAT to bind and neutralize heme was studied. AAT is an anti-protease found in the lung that has been found to have potent anti-inflammatory effects (27). AAT augmentation therapy is currently given to patients with AAT deficiency, a condition that is characterized by low AAT levels and higher than normal protease burden, predominantly NE. In this study, we examine the anti-inflammatory effect of AAT with respect to heme-mediated inflammation and assess AAT augmentation therapy in CF as a viable therapeutic for micro-bleed-induced inflammation. When AAT is coated on a 96-well plate and then treated with heme, there is significantly more heme bound by the AAT-coated wells than uncoated wells (Fig 10A). To investigate the selectivity of AAT binding to heme, wells were also coated with equal amounts of trypsin, PPE, or elastin. Heme binds to these other proteins to a much lesser degree than AAT showing that AAT selectively binds heme (Fig. 10B). At approximately physiological AAT concentrations (20 μm), AAT can neutralize heme-induced IL-8 production in CFBE41o− cells (Fig. 10C). CFBE41o− cells were simultaneously treated with heme and AAT, and the IL-8 values showed that AAT significantly decreased the heme-induced IL-8 expression.
FIGURE 10.
AAT binds heme and neutralizes heme-induced IL-8 secretion by CFBE41o−. A, 20 μg/ml of AAT was immobilized on a 96-well plate overnight before treatment with heme (50 μm). AAT-bound heme was detected spectrophotometrically by direct heme reaction with TMB-ultrasensitive substrate (100 μl/well). B, AAT (20 μg/ml), trypsin (20 μg/ml), PPE (20 μg/ml), and elastin (20 μg/ml) were immobilized on a 96-well plate overnight before treatment with heme (50 μm). Heme binding was quantified to show % binding of heme to AAT, trypsin, PPE, and elastin. C, CFBE41o− cells (1 × 105) were treated with heme (8.75 μm, 24 h) in the presence and absence of AAT (20 μm). IL-8 levels in supernatants were quantified by ELISA. All ELISA experiments were performed in triplicate; the data are represented as mean ± S.E. Data shown are representative of three experiments. A, *, p ≤ 0.05, AAT + heme versus AAT control and heme control; C, *, p ≤ 0.05, control versus heme; $, p ≤ 0.05, AAT + heme versus control + heme.
DISCUSSION
In this study we have demonstrated that hemoglobin is cleaved by the Pseudomonas proteases, PsE and APR, and the neutrophil proteases, NE and PR-3, but not by cathepsin G. We also demonstrate that NE is the principal protease responsible for hemoglobin degradation in vivo. Heme was also shown to stimulate the proinflammatory cytokine IL-8 utilizing a novel pathway involving MyD88, EGFR, and meprin. This study also highlights the importance of meprin in CF by showing both in vitro and in vivo that RNA and protein levels of meprin are elevated in CF. As a possible therapeutic, AAT was investigated as a binding partner and neutralizing agent of heme, and it was shown to be effective in doing so, suggesting that AAT not only has an ability to inhibit neutrophil proteases at the airway epithelial surface but also to dampen the damaging inflammatory effects of unopposed neutrophil proteases.
The role of lung proteases has long been considered to have significant effects on the CF lung (28, 29). Unopposed proteases can cause degradation of host proteins resulting in various degrees of loss of function (9, 30) in addition to release of degradation products that can affect the inflammatory status of the lung (31). We have shown for the first time that PsE and APR can degrade hemoglobin, thus extending the known mechanisms of iron/heme acquisition methods of the Pseudomonas spp. This has significance not only for CF but also in cases of Pseudomonas spp. infection associated with hemolysis.
For the first time, we have also shown the effect of the neutrophil proteases on the structural integrity of hemoglobin. NE and PR-3 degrade the hemoglobin units in all forms. This extends the previous data (32) that showed that NE and PR-3 released protein fragments from hemoglobin using the rudimentary hemoglobin assay of Anson (33). Of particular interest is the finding that the mechanism of NE cleavage of hemoglobin is more effective that PR-3 at liberating the heme, as demonstrated by the TMB gels. When this is coupled with the finding that in vivo, in CF BAL, NE is the predominant protease responsible for hemoglobin degradation, it extends the role of NE in the cleavage of iron and heme-containing proteins. In addition to NE-mediated heme release, the neutrophil produces reactive oxygen species that causes formation of methemoglobin that can lead to subsequent heme release (12). This illustrates how the neutrophil in vivo can use dual mechanisms to amplify heme-induced inflammation. The effect of the liberated heme in CF bronchial epithelia has been shown here to cause release of the proinflammatory cytokines IL-8 and IL-10, illustrating again the neutrophil as an amplifier of inflammation.
Prior to this work, the role of heme in the increased expression of proinflammatory cytokines in CF bronchial epithelial cells was unknown. The amphipathic nature of heme was once regarded as the method by which it exerted its biological effects, including its proinflammatory effects. In recent times our understanding of the heme mechanism of action has been extended significantly, and it is clear that heme may have multiple effects on CF lung epithelia. In addition to its role as a prosthetic group of many proteins and enzymes, heme is now known to act as a cellular messenger utilizing different methods of regulation. For example, heme is known to regulate its own synthesis and catabolism, and its other protean properties are reviewed by Furuyama et al. (34). Impressively, heme can target translation by heme-regulated inhibitor of translation that targets the eukaryotic initiation factor 2 (35). Heme can also directly target transcription by heme binding domains on the transcriptional repressor Bach1, inhibiting its function and resulting in transcription of its regulated genes, including heme oxygenase 1 (reviewed in Ref. 34). Recent research has illustrated that the ever increasing roles of heme also extend to regulation of miRNA (36), ion channels (37), and cell surface receptors, including G protein-coupled receptors (14) and TLR4 (15).
Given that heme-mediated IL-8 production in the macrophage has been reported to occur via TLR4, in this work we examined and found that heme-mediated IL-8 production was MyD88-dependent in CFBE41o− cells, and therefore we hypothesize that CFBE41o− cells also use TLR4. In support of this, we have previously shown that TLR4 and EGFR can co-localize on the surface of isogenic non-CF 16HBE14o− bronchial epithelial cells (3). In this study, in addition to MyD88, heme-induced IL-8 was found to require EGFR and an EGFR agonist. Therefore, the dual TLR4/EGFR requirement may be due to co-localization of the receptors upon activation, similar to what is observed when bronchial epithelial cells are stimulated with NE (3).
In addition to the meprin-activated EGFR, we have also identified another possible direct method of EGFR activation. We have identified that an extracellular domain of EGFR contains a heme-binding motif and that heme and EGFR can physically interact.
The importance of the role of meprin in CF is another pertinent finding of this work. Both the in vitro and in vivo data indicate that meprin is up-regulated at a gene and protein level in CF bronchial epithelium. This is of particular interest in this work as heme-mediated IL-8 was found to require meprin. Previously, NE-activated meprin was found to generate TGFα (3); however, here anti-TGFα-neutralizing antibodies alone did not significantly inhibit heme-induced IL-8, rather a pool of EGFR ligand antibodies was required. Thus heme-mediated meprin EGFR ligand generation shows a redundancy that was not seen in NE-mediated generation, showing that ligand generation is stimulus-specific.
In the CF lung where micro-bleeds commonly occur in a protease-rich environment, it is clear that endogenous anti-heme neutralization mechanisms can be overwhelmed. In such cases, the increased heme levels can provide an iron source possibly fueling microbial growth but also, as shown by this study, stimulating proinflammatory cytokines. This study suggests that in the CF lung heme levels may act as a new proinflammatory factor. With a view to identifying a potential therapeutic to treat bleeding into the CF lung, we investigated the ability of AAT to bind and neutralize heme. In CF, the natural protease anti-protease balance is over-ridden mainly due to the massive NE burden; unopposed NE action results in epithelial surface anti-protease degradation and inactivation. It has been suggested that AAT therapy may have therapeutic benefit to patients with lung disease characterized by an increased protease burden such as CF (38). The principle behind the therapeutic benefit of AAT is by negating the effect of NE. More recently, however, AAT has been shown to have other potentially beneficial properties. In this work, we found that AAT binds heme, and given the positive charge of heme and the negative charge of AAT, this interaction is likely to be relatively stable. This observation was shown to have functional relevance by the capability of AAT to neutralize heme-induced IL-8 in CFBE41o− cells. Endogenous AAT is lost in the protease-rich environment of the CF lung where AAT is cleaved, which further potentiates the effects of heme. Thus, neutralization of heme and its proinflammatory effects represent a new property of AAT, in addition to its known anti-protease, anti-apoptotic, and immuno-modulatory effects.
In this work, we focused on the effect of the liberated heme on the CF bronchial epithelial cell. To avoid the complication of the globin fragments, commercially available heme was used in the cell culture experiments. Hemoglobin is known to produce bioactive fragments preceding proteolysis reviewed by Ivanov et al. (39), which are not subject to rapid host neutralization. Another area of future work is the examination of the biological effects of these globin fragments. The protease-rich environment of the CF lung doubtlessly produces a unique and diverse tissue-specific pool of bioactive fragments. Previous studies have found the hemoglobin fragment neokyotorphin in the rat lung (40), which has been found to regulate Ca2+/K+ currents (39). Mapping the CF lung-specific pool of bioactive hemoglobin fragments presents a fascinating challenge that, in addition to the discovery of new bioactive molecules and their functions, may also provide a patient-specific biomarker pattern that could be used as a noninvasive clinical assessment technique.
Acknowledgments
Pseudomonas alkaline protease was gratefully received as a gift from Prof. Efrat Kessler (Goldschleger Eye Research, Institute Tel-Aviv, University Sackler Faculty of Medicine, Israel). Meprin primary antibody was obtained as a gift from Dr. Erwin E. Sterchi (Institute of Biochemistry and Molecular Medicine, University of Bern, Buehlstrasse 28, CH-3012 Bern, Switzerland).
This work was supported by Science Foundation Ireland, Health Research Board Grant SFI/08/US/B1676, Medical Charities Research Group, and United States Alpha One Foundation.
- CF
- cystic fibrosis
- PsE
- Pseudomonas elastase
- APR
- Pseudomonas alkaline protease
- NE
- neutrophil elastase
- BAL
- bronchoalveolar lavage fluid
- AAT
- α1-antitrypsin
- TMB
- 3,3′,5,5′-tetramethylbenzidine
- PIX
- protoporphyrin IX
- PMXB
- polymyxin B sulfate
- SLPI
- secretory leucoprotease inhibitor
- PPE
- porcine pancreatic elastase
- TLR
- Toll-like receptor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- qRT
- quantitative RT
- EGFR
- EGF receptor
- SFM
- serum-free media
- TACE
- tumor necrosis factor-α-converting enzyme.
REFERENCES
- 1. Rees D. D., Brain J. D., Wohl M. E., Humes J. L., Mumford R. A. (1997) J. Pharmacol. Exp. Ther. 283, 1201–1206 [PubMed] [Google Scholar]
- 2. Geraghty P., Rogan M. P., Greene C. M., Boxio R. M., Poiriert T., O'Mahony M., Belaaouaj A., O'Neill S. J., Taggart C. C., McElvaney N. G. (2007) J. Immunol. 178, 5871–5878 [DOI] [PubMed] [Google Scholar]
- 3. Bergin D. A., Greene C. M., Sterchi E. E., Kenna C., Geraghty P., Belaaouaj A., Belaaouaj A., Taggart C. C., O'Neill S. J., McElvaney N. G. (2008) J. Biol. Chem. 283, 31736–31744 [DOI] [PubMed] [Google Scholar]
- 4. Nakamura H., Yoshimura K., McElvaney N. G., Crystal R. G. (1992) J. Clin. Invest. 89, 1478–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walsh D. E., Greene C. M., Carroll T. P., Taggart C. C., Gallagher P. M., O'Neill S. J., McElvaney N. G. (2001) J. Biol. Chem. 276, 35494–35499 [DOI] [PubMed] [Google Scholar]
- 6. Alcorn J. F., Wright J. R. (2004) J. Biol. Chem. 279, 30871–30879 [DOI] [PubMed] [Google Scholar]
- 7. Britigan B. E., Hayek M. B., Doebbeling B. N., Fick R. B., Jr. (1993) Infect. Immun. 61, 5049–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dulon S., Leduc D., Cottrell G. S., D'Alayer J., Hansen K. K., Bunnett N. W., Hollenberg M. D., Pidard D., Chignard M. (2005) Am. J. Respir. Cell Mol. Biol. 32, 411–419 [DOI] [PubMed] [Google Scholar]
- 9. Guyot N., Bergsson G., Butler M. W., Greene C. M., Weldon S., Kessler E., Levine R. L., O'Neill S. J., Taggart C. C., McElvaney N. G. (2010) Biol. Chem. 391, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fischer B. M., Domowicz D. A., Zheng S., Carter J. L., McElvaney N. G., Taggart C., Lehmann J. R., Voynow J. A., Ghio A. J. (2009) Clin. Transl. Sci. 2, 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schechter A. N. (2008) Blood 112, 3927–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balla J., Jacob H. S., Balla G., Nath K., Eaton J. W., Vercellotti G. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9285–9289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Byrne D. P., Wawrzonek K., Jaworska A., Birss A. J., Potempa J., Smalley J. W. (2010) Biochem. J. 425, 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Porto B. N., Alves L. S., Fernández P. L., Dutra T. P., Figueiredo R. T., Graça-Souza A. V., Bozza M. T. (2007) J. Biol. Chem. 282, 24430–24436 [DOI] [PubMed] [Google Scholar]
- 15. Figueiredo R. T., Fernandez P. L., Mourao-Sa D. S., Porto B. N., Dutra F. F., Alves L. S., Oliveira M. F., Oliveira P. L., Graça-Souza A. V., Bozza M. T. (2007) J. Biol. Chem. 282, 20221–20229 [DOI] [PubMed] [Google Scholar]
- 16. Kunzelmann K., Schwiebert E. M., Zeitlin P. L., Kuo W. L., Stanton B. A., Gruenert D. C. (1993) Am. J. Respir. Cell Mol. Biol. 8, 522–529 [DOI] [PubMed] [Google Scholar]
- 17. Liebau E., De Maria F., Burmeister C., Perbandt M., Turella P., Antonini G., Federici G., Giansanti F., Stella L., Lo Bello M., Caccuri A. M., Ricci G. (2005) J. Biol. Chem. 280, 26121–26128 [DOI] [PubMed] [Google Scholar]
- 18. Sil S., Bose T., Roy D., Chakraborti A. S. (2004) J. Biosci. 29, 281–291 [DOI] [PubMed] [Google Scholar]
- 19. Huy N. T., Xuan Trang D. T., Uyen D. T., Sasai M., Harada S., Kamei K. (2005) Anal. Biochem. 344, 289–291 [DOI] [PubMed] [Google Scholar]
- 20. Chotirmall S. H., Greene C. M., Oglesby I. K., Thomas W., O'Neill S. J., Harvey B. J., McElvaney N. G. (2010) Am. J. Respir. Crit. Care Med. 182, 62–72 [DOI] [PubMed] [Google Scholar]
- 21. Oglesby I. K., Bray I. M., Chotirmall S. H., Stallings R. L., O'Neill S. J., McElvaney N. G., Greene C. M. (2010) J. Immunol. 184, 1702–1709 [DOI] [PubMed] [Google Scholar]
- 22. Zani M. L., Baranger K., Guyot N., Dallet-Choisy S., Moreau T. (2009) Protein Sci. 18, 579–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sunnarborg S. W., Hinkle C. L., Stevenson M., Russell W. E., Raska C. S., Peschon J. J., Castner B. J., Gerhart M. J., Paxton R. J., Black R. A., Lee D. C. (2002) J. Biol. Chem. 277, 12838–12845 [DOI] [PubMed] [Google Scholar]
- 24. Norman L. P., Matters G. L., Crisman J. M., Bond J. S. (2003) Curr. Top. Dev. Biol. 54, 145–166 [DOI] [PubMed] [Google Scholar]
- 25. Sterchi E. E., Stöcker W., Bond J. S. (2008) Mol. Aspects Med. 29, 309–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horrigan F. T., Heinemann S. H., Hoshi T. (2005) J. Gen. Physiol. 126, 7–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelly E., Greene C. M., Carroll T. P., McElvaney N. G., O'Neill S. J. (2010) Respir. Med. 104, 763–772 [DOI] [PubMed] [Google Scholar]
- 28. Greene C. M., McElvaney N. G. (2009) Br. J. Pharmacol. 158, 1048–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taggart C. C., Greene C. M., Carroll T. P., O'Neill S. J., McElvaney N. G. (2005) Am. J. Respir. Crit. Care Med. 171, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 30. Guyot N., Butler M. W., McNally P., Weldon S., Greene C. M., Levine R. L., O'Neill S. J., Taggart C. C., McElvaney N. G. (2008) J. Biol. Chem. 283, 32377–32385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gaggar A., Jackson P. L., Noerager B. D., O'Reilly P. J., McQuaid D. B., Rowe S. M., Clancy J. P., Blalock J. E. (2008) J. Immunol. 180, 5662–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kao R. C., Wehner N. G., Skubitz K. M., Gray B. H., Hoidal J. R. (1988) J. Clin. Invest. 82, 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anson M. L. (1938) J. Gen. Physiol. 22, 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Furuyama K., Kaneko K., Vargas P. D. (2007) Tohoku J. Exp. Med. 213, 1–16 [DOI] [PubMed] [Google Scholar]
- 35. Kramer G., Cimadevilla J. M., Hardesty B. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 3078–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Faller M., Matsunaga M., Yin S., Loo J. A., Guo F. (2007) Nat. Struct. Mol. Biol. 14, 23–29 [DOI] [PubMed] [Google Scholar]
- 37. Tang X. D., Xu R., Reynolds M. F., Garcia M. L., Heinemann S. H., Hoshi T. (2003) Nature 425, 531–535 [DOI] [PubMed] [Google Scholar]
- 38. McElvaney N. G., Hubbard R. C., Birrer P., Chernick M. S., Caplan D. B., Frank M. M., Crystal R. G. (1991) Lancet 337, 392–394 [DOI] [PubMed] [Google Scholar]
- 39. Ivanov V. T., Karelin A. A., Philippova M. M., Nazimov I. V., Pletnev V. Z. (1997) Biopolymers 43, 171–188 [DOI] [PubMed] [Google Scholar]
- 40. Blishchenko E. Y., Mernenko O. A., Yatskin O. N., Ziganshin R. H., Philippova M. M., Karelin A. A., Ivanov V. T. (1997) FEBS Lett. 414, 125–128 [DOI] [PubMed] [Google Scholar]