Abstract
Single-cell gene expression levels show substantial variations among cells in seemingly homogenous populations. Astrocytes perform many control and regulatory functions in the central nervous system. In contrast to neurons, we have limited knowledge about functional diversity of astrocytes and its molecular basis. To study astrocyte heterogeneity and stem/progenitor cell properties of astrocytes, we used single-cell gene expression profiling in primary mouse astrocytes and dissociated mouse neurosphere cells. The transcript number variability for astrocytes showed lognormal features and revealed that cells in primary cultures to a large extent co-express markers of astrocytes and neural stem/progenitor cells. We show how subpopulations of cells can be identified at single-cell level using unsupervised algorithms and that gene correlations can be used to identify differences in activity of important transcriptional pathways. We identified two subpopulations of astrocytes with distinct gene expression profiles. One had an expression profile very similar to that of neurosphere cells, whereas the other showed characteristics of activated astrocytes in vivo.
INTRODUCTION
Brain contains three neuroectoderm-derived cell types: astrocytes, neurons and oligodendrocytes. They all originate from the same multipotent neural stem cells. Traditionally, astrocytes were viewed as a homogeneous cell population that predominantly supports neuronal functions. Recent findings point to many additional functions of astrocytes in health and disease, including control of the number and the function of neuronal synapses (1).
Cell diversity is commonly studied with immunohistochemical analysis and gene expression profiling. Both methods have several limitations. Immunohistochemical and immunocytochemical analyses are restricted to few markers and cannot be used in a truly quantitative manner. Cell types are often defined by the presence or absence of specific markers. Such binary approach to define cell types or functional states is coarse and thus not suitable to detect subpopulations differing only in the degree of expression by individual genes. For example, the hallmark of activated astrocytes is the upregulation of the intermediate filament proteins glial fibrillary acidic protein (GFAP), vimentin (Vim) and nestin (Nes) (2). Gene expression profiling can in principle be applied on the whole transcriptome. Such measurements are in general limited to large cell populations and thus only reflect global transcript levels. Consequently, any important heterogeneity among the cells remains undetected.
With single-cell gene expression profiling we can study heterogeneity among and within cell types in a precise manner. The main obstacle to single-cell measurements has been the absence of sensitive and reproducible methods to measure small numbers of molecules. Single-cells can be collected by microaspiration, flow cytometry and laser capture microdissection (3–8). Transcript levels are then measured using microarrays or reverse transcription quantitative real-time PCR (RT-qPCR). Microarray measurements require a pre-amplification step (9,10), while RT-qPCR has the sensitivity to detect a single mRNA molecule. However, pre-amplification is also needed for RT-qPCR if many transcripts are to be quantified. To characterize well-defined cell types, cells can be enriched/selected for using specific antibodies. Antibody based enrichment is compatible with all cell collection methods, while morphology can only be used as a selection criterion when collecting cells with laser capture microdissection and microaspiration from tissues. Single-cell analysis is refining cell type characterization (11–13). Most single-cell studies so far have relied on preexisting knowledge about the analyzed cells. For instance, hematopoietic subpopulations can be isolated by flow cytometry using well-established surface markers (3,14). Specific types of neurons can be collected based on localization and/or immunohistochemistry using laser capture microdissection or microaspiration (4–7). Single-cell gene expression profiling can also be used to identify new subpopulations of cells from heterogeneous cell populations. This approach is still largely unexplored and tools for identification and classification of subpopulations are missing. Furthermore, transcription takes place in bursts in mammalian cells (15,16). Consequently, mRNA levels are highly variable even within a homogeneous cell population. Thus, gene expression levels between cells cannot be analyzed in the same way as in conventional cell population studies.
In this study, we have developed a strategy to identify and characterize subpopulations of cells. We show how subpopulations of primary astrocytes can be identified and defined by differences in correlated expression levels rather than by binary on/off responses from selected genes. Further, we show how transcriptional correlations can be used to reveal biologically important interactions between genes at a cellular level. Based on this platform, we identified two subpopulations of astrocytes, one with features commonly ascribed to activated astrocytes in vivo and one astrocyte subpopulation sharing characteristics with neurosphere cells.
MATERIALS AND METHODS
Animals and cell cultures
Primary astrocyte and neurosphere cultures were generated from mouse brains. The mice were housed in standard cages in a barrier animal facility with a 12-h light/dark cycle and feed ad libitum. All experiments were conducted according to protocols approved by the Ethics Committee of the University of Gothenburg.
Primary astrocytes were prepared from post-natal day (P) 1 mouse brains and cultured in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) containing 10% fetal calf serum (FCS), 2 mM L-glutamine, 100 U/ml of penicillin and 0.1 mg/ml streptomycin (all Invitrogen) as described (17). After 10–11 days in vitro, almost confluent astrocyte cultures were harvested for gene expression profiling.
Neurosphere cultures were generated from P4 brains with cerebellum removed. These were dissected in Leibovitz medium (Invitrogen) and digested enzymatically [0.1% trypsin, 0.5 mM EDTA in Hank’s balanced salt solution; (Sigma-Aldrich)] and mechanically dissociated into a single-cell suspension. Cells (∼105) were cultured in Neurobasal medium (Invitrogen) containing 2 mM L-glutamine, 100 U/ml of penicillin, 0.1 mg/ml streptomycin, 1X B27, 20 ng/ml basic FGF (all Invitrogen), 20 ng/ml EGF (Stemcell Technologies), 1 U/ml heparin (Sigma-Aldrich) and 0.25 µg/ml Fungizone (Bristol-Meyers Squibb). After 9 days in vitro the cells were used for gene expression profiling.
For cell population measurements, mice were killed at P1, P4 and P60. Whole brains were dissected (P4 with cerebellum removed) and stored at −80°C. Total RNA was extracted using RNeasy Lipid Tissue Mini Kit, including DNase treatment (Qiagen).
Single-cell isolation and cDNA synthesis
Astrocytes were washed twice in PBS and treated with 0.25% Trypsin/EDTA (Invitrogen) for 2 min to dissociate cells. Single-cells were kept in either PBS supplemented with 2.5% FCS or in astrocyte culture medium and kept on ice. The difference in cell medium had a negligible effect, so the astrocyte data were pooled for analysis. Neurospheres were enzymatically dissociated into single-cell suspensions with TrypLE (Invitrogen) and kept in neurosphere medium on ice until cell sorting. Cell aggregates were removed by filtering with 40 µm cell strainer (Becton Dickinson). Single cells were sorted with a BD FACSAria (Becton Dickinson) into 96-well plates (Sarstedt) containing 5 µl mQ water per well. Samples were frozen at −80°C until subsequent analysis. Single-cell sorting for gene expression profiling using flow cytometry has been described elsewhere (18).
SuperScript III RT (Invitrogen) was used for RT. Lysed single cells in 6.5 µl water containing 0.5 mM dNTP (Sigma-Aldrich), 5.0 µM oligo(dT15) (Invitrogen) and 5.0 µM random hexamers (Invitrogen) were incubated at 65°C for 5 min; 50 mM Tris–HCl, 75 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol, 20 U RNaseOut and 100 U SuperScript III (all Invitrogen; final concentrations) were added to a final volume of 10 µl. RT was performed at 25°C for 5 min, 50°C for 60 min, 55°C for 10 min and terminated by heating to 70°C for 15 min. All samples were diluted to 30 µl with water before qPCR.
qPCR
LightCycler 480 (Roche Diagnostics) was used for all qPCR measurements. To each reaction (10 µl) containing iQ SYBR Green Supermix (Bio-Rad) and 400 nM of each primer (Eurofins MWG Operon), we added 2–4 µl of diluted cDNA. Primer sequences used are listed in Supplementary Table S1. All primers were designed with Primer3 (http://frodo.wi.mit.edu/primer3/input.htm) and Netprimer (Premier Biosoft International). The temperature profile was 95°C for 3 min followed by 50 cycles of amplification (95°C for 20 s, 60°C for 20 s and 72°C for 20 s). The formation of expected PCR products was confirmed by agarose gel electrophoresis. All samples were analyzed by melting curve analysis. cDNA concentrations were determined by qPCR relative to standard curves based on purified PCR products (MinElute PCR Purification Kit, Qiagen). The concentration of purified PCR products was determined spectroscopically (NanoDrop ND-1000, Nanodrop Technologies). qPCR data were analyzed as described (19). Limit of detection was determined for all single-cell assays by serial dilution of known cDNA copy numbers. Six replicates were analyzed at each concentration and level of detection was determined by the lowest cDNA copy number where all six replicates were positive (Supplementary Table S1). All data points below the limit of detection were excluded from further analysis. Potential reference genes for cell population data were evaluated using NormFinder. Cell population data were normalized against the geometric mean expression of Gapdh and B2m using assay specific PCR efficiencies (20).
Single-cell analysis
The number of genes that can be analyzed in a single cell is limited by the number of transcripts of the studied genes. Theoretically, only one molecule is needed for detection, but ∼20 target molecules per PCR are needed for accurate quantification (21). This requirement was fulfilled for most of the cells and genes analyzed in this study. All single-cell assays were optimized to be specific enough not to produce primer-dimer signals within 45 cycles of amplification. Highest reproducibility is achieved by minimizing the dilution between RT and qPCR and avoiding the usage of replicates (21). Data are shown as the number of cDNA molecules per cell. The RT efficiency is gene dependent and generally <100% (22). Hence, the number of cDNA molecules is a lower-limit estimate of the number of mRNA molecules that were present in the cell. Since cDNA is single-stranded, we subtracted one cycle from the measured value when calibrating against standard curves based on double-stranded PCR products (23). Most assays were designed to span introns (Supplementary Table S1). All assays were checked by BLAST for pseudogenes. Only GS revealed two potential pseudogenes. All single-cell assays were tested for amplification of genomic DNA. Five individual cells per assay were tested and no genomic DNA amplification was observed.
Statistical analysis
Spearman correlation and partial correlation calculations were performed in SPSS (16.0 or later, SPSS Inc.) software. We calculated first-order partial correlations for all observed Spearman correlations. Heat maps, principal component analysis (PCA), Kohonen self-organizing maps (SOM) and potential curve analysis were performed in GenEx software (MultiD). Expression of each gene was mean-centered for the heat map analysis, which was calculated using Ward’s algorithm and Euclidean distance measure. For Kohonen SOMs and potential curve analysis autoscaled gene expression values were used to give all genes equal weight in the clustering algorithms. Parameters for the Kohonen SOMs were: 2–3 × 1 map, 0.10 learning rate, 2–3 neighbors and 10 000 iterations. The resulting clusters did not depend on parameter settings. The data were analyzed as described (24). Neurosphere cells were classified by potential curve analysis using the two subpopulations of astrocytes as training set (25). The t-tests and Fisher’s tests were performed with Bonferroni correction for multiple testing.
RESULTS
Primary astrocyte cultures prepared from P1 mouse brains and neurospheres from P4 brains are routinely used in many experimental paradigms. Single cells were collected by flow cytometry, lysed and analyzed by RT-qPCR. Expression of glutamine synthase (GS), glial fibrillary acidic protein α (GFAP), GFAPδ, nestin (Nes), vimentin (Vim), synemin (Syn), SRY-box containing gene 2 (Sox2), endothelin type B receptor (ETBR), wingless-related MMTV integration site 3 (Wnt3), leukemia inhibitory factor (Lif) and neuronal pentraxin 1 (Nptx1) was profiled in 164 astrocytes and 83 neurosphere cells (Figure 1). A second set of 164 astrocytes was measured and analyzed independently; the results were comparable to those in the first set (Supplementary Tables S2 and S3 and Supplementary Figures S2 and S4). To characterize individual cells we performed descriptive statistics, correlation studies and subpopulation screens using unsupervised learning algorithms.
Figure 1.
Cell heterogeneity among primary astrocytes and neurosphere cells. Heat maps for 164 primary astrocytes (A) and 83 primary neurosphere cells (B) were constructed using Ward’s algorithm and Euclidean distance measure for all cells. Expression levels of all genes were mean-centered.
To characterize the purity of the cultures of primary astrocytes and neurospheres, we compared the gene expression profiles with those of brain tissue from P1, P4 and P60 mice at cell population level (Supplementary Figure S1). In primary astrocyte enriched cultures, markers for microglia [allograft inflammatory factor 1 (Aif1/Iba1)] (26), oligodendrocytes [myelin basic protein (Mbp) (27,28) and 2′-3′-cyclic nucleotide 3′ phosphodiesterase (Cnp) (27,28)], neurons [microtubule-associated protein 2 (Mtap2) (29,30), neurofilament, light polypeptide (Nefl) (28,31) and Nptx1 (32)] and endothelial cells [platelet/endothelial cell adhesion molecule 1 (Pecam1) (28,33), von Willebrand factor homolog (Vwf) (28,33) and angiopoietin 2 (Angpt2) (33)] were downregulated, while markers for astrocytes [GFAP (2,28,34,35), aldehyde dehydrogenase 1 family, member L1 (Aldh1l1) (28) and GS (36,37)] were upregulated compared to mature P60 brains. These data confirm that our primary astrocytes were enriched for astrocytes and astrocyte progenitors. The expression profile of neurospheres was more complex with high expression of some but not all astrocyte (Aldh1l1), neuronal (Mtap2 and Nptx1) and endothelial cell (Angpt2) markers, indicating that neurospheres are a more heterogeneous cell population.
Most cells in primary astrocyte cultures and neurosphere cells co-express astrocyte and stem cell markers
Glutamine synthase converts the neurotransmitter glutamate into glutamine and is expressed by astrocytes in the brain (36,37). Of 164 cells from primary astrocyte cultures, 153 (93%) expressed GS (Table 1). Among GS-positive cells, 78% expressed Nes, 75% expressed Sox2 and 65% expressed Nes and Sox2, both markers of stem/progenitor cells (38) and found also in some astrocytes (39,40). The intermediate filament protein genes GFAP and Vim were expressed by 139 (85%) and 161 (98%) cells, respectively.
Table 1.
Statistical parameters describing gene expression in 164 primary astrocytes and 83 neurosphere cells
| Gene | Cell type | na | Arithmetic meanb | Geometric meanc | Log10 geometric mean (SD) | Maximum expressiond |
|---|---|---|---|---|---|---|
| GS | A | 153 | 520 | 320 | 2.5 (0.40) | 2700 |
| NS | 75 | 300 | 250 | 2.4 (0.26) | 1100 | |
| GFAP | A | 139 | 2900 | 1000 | 3.0 (0.56) | 45 000 |
| NS | 71 | 640 | 560 | 2.7 (0.25) | 1600 | |
| GFAPδ | A | 63 | 110 | 79 | 1.9 (0.38) | 720 |
| NS | 8 | 33 | 33 | 1.5 (0.07) | 40 | |
| Vim | A | 161 | 7500 | 5000 | 3.7 (0.38) | 48 000 |
| NS | 83 | 2600 | 2500 | 3.4 (0.15) | 5600 | |
| Nes | A | 124 | 460 | 320 | 2.5 (0.41) | 3700 |
| NS | 74 | 260 | 170 | 2.3 (0.42) | 1200 | |
| ETBR | A | 70 | 390 | 320 | 2.5 (0.35) | 1600 |
| NS | 44 | 240 | 200 | 2.3 (0.30) | 750 | |
| Sox2 | A | 122 | 160 | 130 | 2.1 (0.34) | 560 |
| NS | 52 | 160 | 130 | 2.1 (0.25) | 380 | |
| Nptx1 | A | 4 | 120 | 130 | 2.1 (0.10) | 140 |
| NS | 31 | 430 | 230 | 2.4 (0.40) | 2700 | |
| Wnt3 | A | 12 | 200 | 200 | 2.3 (0.13) | 340 |
| NS | 5 | 670 | 630 | 2.8 (0.17) | 1100 | |
| Syn | A | 22 | 130 | 100 | 2.0 (0.39) | 650 |
| NS | 18 | 130 | 110 | 2.0 (0.34) | 280 | |
| Lif | A | 21 | 110 | 100 | 2.0 (0.31) | 340 |
| NS | 11 | 97 | 87 | 1.9 (0.24) | 160 |
A, astrocytes; NS, neurosphere cells.
aNumber of cells expressing a given gene.
bThe arithmetic mean was calculated as: 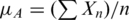 .
.
cThe geometric mean was calculated as: 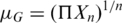 .
.
dHighest number of cDNA molecules of a gene in any cell.
Among neurosphere cells, 90% expressed GS; of these, 91% expressed Nes and 63% expressed Sox2. All cells that expressed GS and Sox2 also expressed Nes. GFAP, which is expressed in astrocytes and some progenitor cells (28,34,35), was expressed in 86% of the cells—the same proportion as in astrocyte cultures. All neurosphere cells expressed Vim.
GFAPδ, a splice form of GFAP found in neural progenitor cells (41), was expressed in 38% of primary astrocytes but in only 10% of neurosphere cells. Among GFAPδ-positive primary astrocytes, 92% also expressed GFAP. Syn, coding for the intermediate filament protein synemin, was expressed in 13% of astrocytes and 22% of neurosphere cells. Lif, a factor important for stem cell self-renewal and astrocyte differentiation (42), was expressed in 13% of primary astrocytes and 13% of neurosphere cells, and Wnt3, another regulator of self-renewal and neurogenesis (43), in 7 and 6%, respectively. The most prominent difference between the cultures was in the expression of Nptx1 that was found in 2.4% of astrocytes and 37% of neurosphere cells. We conclude that the majority of primary astrocytes and neurosphere cells co-express common astrocyte and stem/progenitor markers.
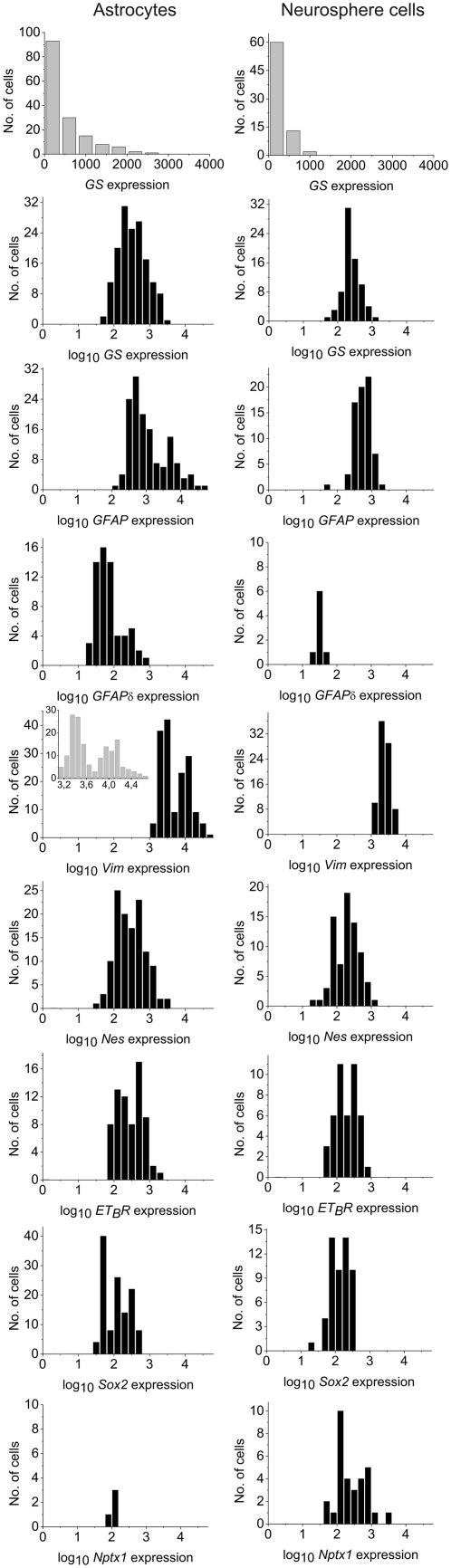
Transcript levels in primary astrocytes and neurosphere cells show lognormal features
The distributions of GS, GFAP, GFAPδ, Vim, Nes, ETBR, Sox2 and Nptx1 transcripts are shown in Figure 2 (see Supplementary Figure S2 for the second data set). Except for Vim, the distributions showed lognormal features, as described in other studies of mammalian cells (3,15,44). Lif, Syn and Wnt3 transcripts were detected in only few cells (Table 1 and Supplementary Table S2) and thus their distributions could not be reliably determined. The geometric and arithmetic means of the expression of individual genes are shown in Table 1. In a lognormal population, the geometric mean reflects the characteristic expression in a typical/median cell. The geometric mean is more conservative than the arithmetic mean. The latter overestimates the characteristic expression when expression levels are lognormally distributed.
Figure 2.
Gene expression levels in 164 primary astrocytes and 83 neurosphere cells. Gene expression is shown as the number of cDNAs per cell. GS expression is shown in both linear and log10 scales; other genes are shown in log10 scale. Inset shows a more detailed histogram of Vim expression in astrocytes.
The variations in gene expression levels between individual cells were substantial. Some astrocytes had ∼50 000 transcripts of Vim and GFAP per cell, while others had fewer than 100. The observed transcript variability is in agreement with transcriptional bursting (15,16). A consequence of the observed lognormallity is that a majority of the transcripts for a particular gene originate from a minority of the cells in a given population. For example, the 30 primary astrocytes with highest number of GFAP transcripts contributed to ∼75% of all transcripts for this gene.
Expression of genes upregulated in activated astrocytes correlates at the cellular level
Next, we looked for correlations between the mRNA levels of multiple genes in each cell. Table 2 shows Spearman correlation coefficients for all gene pairs. The correlation coefficient is a value between −1 and 1, where 1 reflects perfect positive correlation, −1 reflects negative correlation and 0 indicates no correlation. Interestingly, genes that are upregulated in activated astrocytes (GFAP, Vim, Nes, GS and ETBR) (45–49) showed positive correlations (P < 0.01) in individual cells collected from primary astrocyte cultures (Table 2). Also, expression of Sox2 and to some degree GFAPδ correlated with the expression of the intermediate filament genes GFAP, Vim and Nes, as well as with the expression of GS and ETBR (P < 0.01). The only negatively correlation observed was between Sox2 and Wnt3. In neurosphere cells, however, these genes showed little or no correlation, except for Vim, whose expression correlated with Nes and ETBR. In contrast, neurosphere cells expressed high levels of Nptx1, whose expression correlated positively with that of Sox2.
Table 2.
Spearman correlation coefficients for primary astrocytes and neurosphere cells
| Gene | Cell type | GS | GFAP | GFAPδ | Vim | Nes | ETBR | Sox2 | Nptx1 | Wnt3 | Syn | Lif |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GS | A | 1 | ||||||||||
| NS | ||||||||||||
| GFAP | A | 0.51 | 1 | |||||||||
| NS | 0.05 | |||||||||||
| GFAPδ | A | 0.35 | 0.57 | 1 | ||||||||
| NS | 0.09 | 0.26 | ||||||||||
| Vim | A | 0.59 | 0.56 | 0.23 | 1 | |||||||
| NS | 0.21 | 0.08 | 0.15 | |||||||||
| Nes | A | 0.50 | 0.39 | 0.06 | 0.68 | 1 | ||||||
| NS | 0.08 | 0.19 | 0.47 | 0.66 | ||||||||
| ETBR | A | 0.24 | 0.29 | 0.25 | 0.57 | 0.29 | 1 | |||||
| NS | 0.13 | 0.04 | 0.15 | 0.47 | 0.28 | |||||||
| Sox2 | A | 0.45 | 0.44 | 0.19 | 0.56 | 0.38 | 0.37 | 1 | ||||
| NS | −0.03 | 0.28 | 0.06 | 0.20 | 0.10 | −0.05 | ||||||
| Nptx1 | A | −0.33 | −0.43 | – | 0.43 | 0.60 | 0.310 | 0.60 | 1 | |||
| NS | 0.06 | −0.01 | – | 0.35 | 0.15 | 0.05 | 0.45 | |||||
| Wnt3 | A | −0.11 | −0.40 | 0.19 | −0.28 | −0.34 | 0.18 | −0.67 | – | 1 | ||
| NS | −0.50 | – | – | 0.05 | 0.30 | – | −0.20 | – | ||||
| Syn | A | 0.01 | 0.12 | −0.22 | −0.03 | 0.10 | 0.07 | 0.12 | – | – | 1 | |
| NS | −0.03 | 0.42 | – | −0.02 | 0.09 | −0.01 | −0.10 | −0.11 | – | |||
| Lif | A | 0.23 | −0.10 | −0.16 | 0.23 | 0.22 | 0.13 | 0.37 | – | – | – | 1 |
| NS | 0.03 | 0.38 | – | −0.39 | 0.17 | −0.71 | – | – | – | – |
All cells (328 single astrocytes and 83 dissociated neurosphere cells) were used for correlation calculations. Bold indicates ≥99% significance; underscore indicates ≥95% significance. Correlation coefficients were not calculated for gene pairs with fewer than five data points. A, astrocytes; NS, neurosphere cells.
To discriminate between direct and indirect interactions among the observed correlations, we calculated partial correlations between gene expression levels in individual cells. This was done by specifying a control gene that may interact with two other correlated genes and thus account for the observed correlation (50). The resulting partial correlation then becomes a unique correlation between the two initial genes that remains when the correlated variance explained by the control gene has been removed. Using partial correlations, we could determine if a measured correlation between two genes was unique or rather a consequence of the two genes both being dependent on a third gene (Figure 3A). Figure 3B shows the gene interaction map for Vim based on partial correlations. Vim interacts directly with ETBR and Nes. These interactions are independent of the other genes studied. The Vim interactions with GS, GFAP and Sox2 were partially direct, while its interaction with GFAPδ was indirect and could be explained as being a consequence of Vim’s interaction with either GFAP or ETBR. Interaction maps for the other genes are shown in Supplementary Figure S3. From the 20 statistically significant correlations in Table 2, nine interactions were direct while eleven could be fully explained by other genes using partial correlations. All direct correlations were dependent on Vim except for those that involved GFAPδ and the interaction between Sox2 and Wnt3. Figure 3C shows the complete interaction map based on the correlations in Table 2.
Figure 3.
Gene interactions. (A) Three different types of interaction between two genes can be identified using partial correlations. Case 1 shows a direct interaction between genes A and B. Case 2 represents a direct interaction that can be partly explained by a third gene, while case 3 represents an indirect interaction that can be fully explained by a third gene. We used a decrease of 0.15 in correlation as a cut off for partially explained interactions (Case 2) and a complete loss of significance for indirect correlation (Case 3). (B) A detailed interaction map for Vim. The interaction between Vim and Nes/ETBR is direct (Case 1), while the interactions with GFAP, GS and Sox2 can be partially explained by other genes (GS and Nes, Case 2). The interaction between Vim and GFAPδ was indirect and can be fully explained by interactions through GFAP or ETBR. See Supplementary Figure S3 for detailed interaction maps for other genes. (C) Nine of 20 observed correlations in Table 2 represented direct interactions that could not be explained by the other genes.
Primary astrocytes can be divided into two subpopulations
To identify possible subpopulations of cells based on their expression profile, we applied Kohonen SOMs (Figure 4A). SOM is an unsupervised learning algorithm that divides the cells into a given number of groups based on their characteristics. SOM uses random numbers to initiate and perform the classification. As a consequence reiterated SOM analysis may generate different classifications. If the same SOM is repeatedly produced, it evidences robust classification. The classification depends on gene expression levels. Highly expressed genes have greater influence than lowly expressed genes. This effect can be removed by subtracting the average of the expression level of each gene and dividing it by its standard deviation, i.e. performing autoscaling (24).
Figure 4.
Astrocyte subpopulations show distinct gene expression profiles. (A) Clustering of astrocyte subpopulations using Kohonen SOMs. Expression levels of all genes were autoscaled. Each dot represents one cell. (B) Principal component analysis confirmed the existence of two subpopulations with coloring according to the Kohonen SOMs classification. (C) Histograms of gene expression profiles (log10 scale) of the two astrocyte subpopulations. Descriptive statistics for the two astrocyte populations are shown in Table 3. PC, principal component.
The astrocytes were divided into two groups using a 2 × 1 SOM (Figure 4A). The SOM was based on the autoscaled expression levels of all eleven genes and was fully reproducible. The SOM classification was confirmed using principal component analysis (PCA), another unsupervised classification method based on different principles (Figure 4B) (24). To characterize the two subpopulations, we plotted the transcript distributions of the highly expressed genes: Vim, GS, GFAP, GFAPδ, Nes, Sox2 and ETBR (Figure 4C). This analysis revealed two, albeit overlapping, lognormal distributions. In the first subpopulation Vim, GS, GFAP, GFAPδ, Nes, Sox2 and ETBR were upregulated two- to five-fold (P < 0.01, Table 3) relative to the second subpopulation. In addition, more cells expressed GFAPδ, Syn and ETBR in the first subpopulation (P < 0.01). No significant differences for Lif, Nptx1 and Wnt3 were observed. The presence of two astrocyte subpopulations was confirmed in the independent data set (Supplementary Figure S4). The SOM analysis of the neurosphere cells revealed no distinguishable subpopulations (data not shown).
Table 3.
Statistical profile of subpopulations in primary astrocytes
| Gene | Statisticsa | Low expressing cellsb | High expressing cellsb | Ratioc |
|---|---|---|---|---|
| Vim | N | 89 | 72 | 4.3 |
| Geometric mean | 2600 | 11 000 | ||
| GS | N | 81 | 72 | 2.9 |
| Geometric mean | 210 | 600 | ||
| GFAP | N | 70 | 69 | 5.0 |
| Geometric mean | 500 | 2500 | ||
| GFAPδ | n | 13 | 50 | 2.4 |
| Geometric mean | 35 | 86 | ||
| Nes | n | 55 | 69 | 3.5 |
| Geometric mean | 140 | 510 | ||
| ETBR | n | 13 | 57 | 2.3 |
| Geometric mean | 150 | 330 | ||
| Sox2 | n | 53 | 69 | 2.4 |
| Geometric mean | 73 | 180 | ||
| Wnt3 | n | 4 | 8 | 0.81 |
| Geometric mean | 220 | 180 | ||
| Syn | n | 3 | 19 | 1.3 |
| Geometric mean | 74 | 96 | ||
| Lif | n | 12 | 9 | 1.1 |
| Geometric mean | 89 | 100 |
an, Number of cells expressing a given gene in the subpopulation defined by the two groups. Nptx1 was excluded because it was expressed by only four cells. Bold numbers indicate that the total number of cells among the cells with high expression of Vim, GFAP, GFAPδ, Nes, ETBR and Sox2 was increased compared to the cells with low expression. (P < 0.01, Fisher’s exact test with Bonferroni correction).
bSubpopulations defined by low/high expression of Vim, GFAP, GFAPδ, Nes, ETBR and Sox2.
cRatio of expression between cells with high and low expression of Vim, GFAP, GFAPδ, Nes, ETBR for a given gene in astrocytes. Bold numbers are statistically significant (P < 0.01, t-test with Bonferroni correction).
It was not possible to identify subpopulations of primary astrocytes based on the presence or absence of expression of any unique marker. All genes showed a unimodal distribution of transcript levels, except for Vim transcript levels, which had a bimodal distribution (Figure 2), implying two subpopulations of cells. Next, we tested if Vim can be used as a single classifier for the two subpopulations by indexing the cells using a threshold of 6300 transcripts, placed centrally between the two peaks in the distribution (Figure 2). Out of 164, 155 of astrocytes were accurately classified using Vim as a classifier (Supplementary Figure S5). Clearly, the Vim expression level is highly characteristic for these two astrocyte subpopulations. To test if Vim is the sole determinant we excluded it from the analysis. The resulting SOM classification correctly classified 159 of 164 astrocytes (Supplementary Figure S6). Furthermore, we also tested if the expression data support the presence of three astrocyte subpopulations using a 3 × 1 SOM (Supplementary Figure S7). Again, two of the groups were characterized by an overall high and low expression, respectively of Vim, GS, GFAP, GFAPδ, Nes, Sox2 and ETBR. The new third group had an intermediate expression pattern relative the other two, but most similar to the low expressing group. The intermediate group showed no unique features, suggesting the existence of only two distinct subpopulations of astrocytes. This conclusion was supported by PCA (Supplementary Figure S7B).
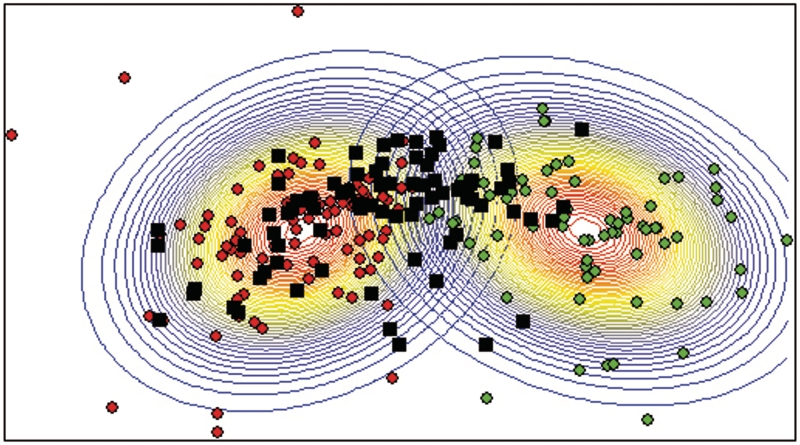
Finally, we determined if the neurosphere cells had gene expression profile similar to any of the astrocyte subpopulations using PCA and potential curve analysis (Figure 5). The principal component space was calculated using the expression profiles of the astrocytes only. When the neurosphere cells were positioned in this space based on their expression profiles, the classification revealed a high degree of similarity between gene expression profile of neurosphere cells and the astrocyte subpopulation characterized by low expression of Vim, GS, GFAP, GFAPδ, Nes, Sox2 and ETBR.
Figure 5.
Classification of neurosphere cells. Principal component and potential curve analysis were used to classify neurosphere cells (black) based on the expression profiles of the two subpopulations of astrocytes. Each dot represents one cell: red dots are cells with low expression of Vim, GFAP, GFAPδ, Nes, ETBR and Sox2; green dots are cells with high expression of Vim, GFAP, GFAPδ, Nes, ETBR and Sox2.
DISCUSSION
Most single-cell gene expression studies in the brain have focused on neurons and neuronal progenitors (4–7,11, 51,52). Here, we characterized primary astrocytes and neurosphere cells. Primary astrocyte cultures prepared from neonatal rodent brains have long served as an experimental system to study the properties of astrocytes (53). These cultures are derived from unidentified populations of proliferating precursor cells. Comparative microarray experiments have shown that primary astrocytes are similar, but not identical, to in vivo astrocytes (28). Here, we studied eleven different markers selected to reflect astrocyte properties, including astrocyte activation and stem/progenitor cell properties.
Our analysis of primary astrocytes showed prominent expression of established markers for astrocytes, while the expression of other cellular markers was low (Supplementary Figure S1). At a single-cell level, 94% of astrocytes expressed GS (Table 1), a marker of immature and mature astrocytes (36). Low expression of GS has also been reported in oligodendrocytes (37). GFAP has lower cell type specificity than GS but is not expressed in mature oligodendrocytes (28,34). Seventy nine percent of cells in astrocyte cultures co-expressed GFAP and GS. Furthermore, 69 of 70 ETBR-positive cells in astrocyte cultures co-expressed GS. Thus, since endothelial cells are ETBR-positive and GS-negative (54), our primary astrocyte cultures were not contaminated with endothelial cells. Only four cells in astrocyte cultures expressed Nptx1, a marker for neuron and neural progenitor cells (32). Our study shows that most cells in primary astrocyte cultures co-express the astrocyte marker GS and two genes expressed by neural progenitor/stem cells, namely Nes and Sox2. In summary, the primary astrocytes have expression pattern similar to that of stem/progenitor cells and astrocytes in vivo. However, we cannot exclude the possibility that other cell types exists in our cultures, but they should be present only in small fractions.
Cell population data for the neurospheres showed that these cells have less distinct expression pattern than the primary astrocytes (Supplementary Figure S1). Markers for endothelial cells (Angpt2) and neurons (Nptx1 and Mtap2) were highly expressed in neurospheres, while markers for astrocytes (Aldh1l1, GFAP and GS) were expressed to a lesser degree compared to primary astrocytes. Interestingly, 78% of the neurosphere cells co-expressed GFAP and GS at a single-cell level. Thirty seven percent of the neurosphere cells expressed Nptx1. Similar to the primary astrocytes, the majority of the neurosphere cells co-expressed GS, Sox2 and Nes. In summary, the neurosphere cells have some properties similar to primary astrocytes. They also show characteristics of several cell types, a finding consistent with the fact that they are a heterogeneous cell population.
High correlation between the numbers of transcripts per cell of GFAP, Vim, Nes, GS and ETBR suggests that these genes have common regulatory elements and might be transcribed in synchronized bursts (55). Indeed, they are all known to be upregulated in activated astrocytes (45–49). Surprisingly, the expression of a stem cell marker, Sox2 (38), correlated positively with the expression of intermediate filament proteins GFAP, Vim and Nes, as well as with GS and ETBR in the primary astrocytes. Thus, the activation of astrocytes may be linked to a transition into a more immature or stem cell-like state, as suggested by studies reporting that at least some astrocytes acquire stem cell properties after brain injury (56). Interestingly, all observed correlations were directly dependent on either Vim or GFAPδ, except the negative correlation between Sox2 and Wnt3. These data suggest that Vim and GFAPδ may have important role in cell fate determination in primary astrocytes (Figure 3C).
The bimodal distribution of Vim expression was apparent already from inspection of raw data, but this was not the case for the other genes (Figure 2). Only after cells were classified into two subpopulations did bimodal expression profiles become evident for ETBR, GS, GFAP, GFAPδ, Nes and Sox2 (Figure 4C). The distribution of gene expression levels among individual cells in the respective subpopulations was almost perfectly lognormal. Conventional cell type characterizations are generally based on presence or absence of well-established markers. This was not possible here, since no such marker is known. Instead, we applied multivariate methods to divide samples into groups. Using SOM and PCA analyses we were able to show that the primary astrocyte cultures are a mixture of two defined subpopulations with unique expression profiles. The most distinct single classifier, Vim, is expressed in both subpopulations, although to partially different levels. Based on Vim transcript levels alone, 95% of the primary astrocytes could be correctly classified. A drawback of using Vim alone as a classifier is that the threshold value is variable between different cell populations: 6300 and 2500 Vim transcripts, respectively were used in the two independent data sets. We do not know the underlying reason for the different thresholds. We conclude that for subpopulation classification, the use of multivariate SOM analysis is more accurate and robust than the use of Vim expression alone.
The two subpopulations of cells in primary astrocyte cultures may represent different cell states or different cell types, which may be reversible. From our data, we cannot discriminate between these alternatives. However, classification of astrocytes into three groups showed cells with an intermediate expression profile. It is conceivable that these astrocytes are in a transition state between the original two subpopulations. This would support the hypothesis of reversible cell states. Such subpopulations may also exist in the brain or be the result of in vitro culture conditions. In vivo studies have revealed different subpopulations of astrocytes (57,58)—none similar to those described here—classified by the expression or lack of expression of specific markers identified by immunostaining or in some cases by electrophysiological properties (58,59). We identified subpopulations of primary astrocytes not by the presence of specific markers but rather by expression levels of shared markers. Conceivably, the subpopulation of astrocytes with high transcript levels correspond to activated astrocytes in vivo, which are characterized by upregulation of GFAP, Nes, ETBR and Syn (48,60). The gene expression profile of astrocytes changes during brain development (Supplementary Figure S1) (28,61). We found that ETBR, GS, GFAP, GFAPδ, Nes, Sox2, Vim were co-regulated at single-cell level in vitro, whereas GS, GFAP and GFAPδ were upregulated and ETBR, Nes, Sox2 and Vim were downregulated in developing brains (Supplementary Figure S1) (61). Thus, the two subpopulations of cultured astrocytes are unlikely to reflect different stages of maturation.
In summary, we introduce how single-cell gene expression profiling can be applied as a novel research tool to identify and characterize distinct subpopulations of cells and how gene correlations can be applied to determine detailed gene interaction networks using this tool. We found that the majority of cells in primary astrocyte cultures and cells from dissociated neurospheres express mRNAs encoding markers characteristic of astrocytes as well as markers characteristic of neural stem/progenitor cells. The transcription of genes encoding proteins associated with astrocyte activation seems to be regulated by a common mechanism where Vim and GFAPδ have key functions. The population with high expression of Vim, GFAP, GFAPδ, Nes, ETBR and Sox2, has the gene expression profile of activated astrocytes, while the population with low expression has a profile similar to neurosphere cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Amlöv’s Foundation; Foundation Edit Jacobson’s Donation Fund; Frimurare Foundation; Grant Agency of the Academy of Science (IAA50520809 and IAA500970904); Grant Agency of the Czech Republic (P303/10/1338); Hjärnfonden; LUA/ALF Göteborg; Region of Västra Götaland (RUN); Sigurd and Elsa Goljes Memory Foundation; Socialstyrelsens Foundation; Swedish Stroke Foundation; Swedish Society for Medicine; Swedish Society for Medical Research (A.S.); Swedish Medical Research Council (project 11548 and 2005-5174 to A.S.); Trygg-Hansa; Torsten and Ragnar Söderberg Foundations; Wilhelm and Martina Lundgren’s Research Foundation. Funding for open access charge: LUA/ALF Göteborg.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Science. 2001;291:5657–5661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- 2.Pekny M, Nilsson M. Astrocyte activation and reactive gliosis. Glia. 2005;50:428–434. doi: 10.1002/glia.20207. [DOI] [PubMed] [Google Scholar]
- 3.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc. Natl Acad. Sci. USA. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamme F, Salunga R, Yu J, Tran DT, Zhu J, Luo L, Bittner A, Guo HQ, Miller N, Wan J, et al. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J. Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gründemann J, Schlaudraff F, Haeckel O, Liss B. Elevated α-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson’s disease. Nucleic Acids Res. 2008;36:e36. doi: 10.1093/nar/gkn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginsberg SD, Che S, Counts SE, Mufson EJ. Single cell gene expression profiling in Alzheimer’s disease. NeuroRx. 2006;3:302–318. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginsberg SD, Elarova I, Ruben M, Tan F, Counts SE, Eberwine JH, Trojanowski JQ, Hemby SE, Mufson EJ, Che S. Single-cell gene expression analysis: implications for neurodegenerative and neuropsychiatric disorders. Neurochem. Res. 2004;29:1053–1064. doi: 10.1023/b:nere.0000023593.77052.f7. [DOI] [PubMed] [Google Scholar]
- 8.Ståhlberg A, Bengtsson M. Single-cell gene expression profiling using reverse transcription quantitative real-time PCR. Methods. 2010;50:282–288. doi: 10.1016/j.ymeth.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Alldred MJ, Che S, Ginsberg SD. Terminal Continuation (TC) RNA amplification enables expression profiling using minute RNA input obtained from mouse brain. Int. J. Mol. Sci. 2008;9:2091–2104. doi: 10.3390/ijms9112091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat. Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- 11.Sul JY, Wu CW, Zeng F, Jochems J, Lee MT, Kim TK, Peritz T, Buckley P, Cappelleri DJ, Maronski M, et al. Transcriptome transfer produces a predictable cellular phenotype. Proc. Natl Acad. Sci. USA. 2009;106:7624–7629. doi: 10.1073/pnas.0902161106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altar CA, Vawter MP, Ginsberg SD. Target identification for CNS diseases by transcriptional profiling. Neuropsychopharmacology. 2009;34:18–54. doi: 10.1038/npp.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Edvardsson L, Olofsson T. Real-time PCR analysis for blood cell lineage specific markers. Methods Mol. Biol. 2009;496:313–322. doi: 10.1007/978-1-59745-553-4_21. [DOI] [PubMed] [Google Scholar]
- 15.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequnences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pekny M, Eliasson C, Chien CL, Kindblom LG, Liem R, Hamberger A, Betsholtz C. GFAP-deficient astrocytes are caple of stellation in vitro when cocultured with neurons and exhibit a reduced amount of intermediate filaments and an increased cell saturation density. Exp. Cell Res. 1998;239:332–343. doi: 10.1006/excr.1997.3922. [DOI] [PubMed] [Google Scholar]
- 18.Ståhlberg A, Bengtsson M, Hemberg M, Semb H. Quantitative transcription factor analysis of undifferentiated single human embryonic stem cells. Clin. Chem. 2009;55:2162–2170. doi: 10.1373/clinchem.2009.131433. [DOI] [PubMed] [Google Scholar]
- 19.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 20.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengtsson M, Hemberg M, Rorsman P, Ståhlberg A. Quantification of mRNA in single cells and modeling of RT-qPCR induced noise. BMC Mol. Biol. 2008;9:63. doi: 10.1186/1471-2199-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ståhlberg A, Kubista M, Pfaffl M. Comparison of reverse transcriptases in gene expression analysis. Clin. Chem. 2004;50:1678–1680. doi: 10.1373/clinchem.2004.035469. [DOI] [PubMed] [Google Scholar]
- 23.Ståhlberg A, Håkansson J, Xian X, Semb H, Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 2004;50:509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 24.Ståhlberg A, Elbing K, Andrade-Garda JM, Sjögreen B, Forootan A, Kubista M. Multiway real-time PCR gene expression profiling in yeast Saccharomyces cerevisiae reveals altered transcriptional response of ADH-genes to glucose stimuli. BMC Genomics. 2008;9:170. doi: 10.1186/1471-2164-9-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomás X, Andrade JM. Application of simplified potential curves to classification problems. Química Analítica. 1999;18:225–231. [Google Scholar]
- 26.Ahmed Z, Shaw G, Sharma VP, Yang C, McGowan E, Dickson DW. Actin-binding proteins coronin-1a and IBA-1 are effective microglial markers for immunohistochemistry. J. Histochem. Cytochem. 2007;55:687–700. doi: 10.1369/jhc.6A7156.2007. [DOI] [PubMed] [Google Scholar]
- 27.Brunner C, Lassmann H, Waehneldt TV, Matthieu JM, Linington C. Differential ultrastructural localization of myelin basic protein, myelin/oligodendroglial glycoprotein, and 2′,3′-cyclic nucleotide 3′-phosphodiesterase in the CNS of adult rats. J. Neurochem. 1989;52:296–304. doi: 10.1111/j.1471-4159.1989.tb10930.x. [DOI] [PubMed] [Google Scholar]
- 28.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christpherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417:39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- 30.Couillard-Despres S, Quehl E, Altendorfer K, Karl C, Ploetz S, Bogdahn U, Winkler J, Aigner L. Human in vitro reporter model of neuronal development and early differentiation processes. BMC Neurosci. 2008;9:31. doi: 10.1186/1471-2202-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trojanowski JQ, Walkenstein N, Lee VM. Expression of neurofilament subunits in neurons of the central and peripheral nervous system: an immunohistochemical study with monoclonal antibodies. J. Neurosci. 1986;6:650–660. doi: 10.1523/JNEUROSCI.06-03-00650.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlimgen AK, Helms JA, Vogel H, Perin MS. Neuronal pentraxin, a secreted protein with homology to acute phase proteins of the immune system. Neuron. 1995;14:519–526. doi: 10.1016/0896-6273(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 33.Bhasin M, Yuan L, Keskin DB, Out HH, Libermann TA, Oettgen P. Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genomics. 2010;11:342. doi: 10.1186/1471-2164-11-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casper KB, McCarthy KD. GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol. Cell Neurosci. 2006;31:676–684. doi: 10.1016/j.mcn.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969-2000) Neurochem. Res. 2000;25:439–451. doi: 10.1023/a:1007677003387. [DOI] [PubMed] [Google Scholar]
- 36.He Y, Hakvoort TB, Vermeulen JL, Labruyère WT, De Waart DR, Van Der Hel WS, Ruijter JM, Uylings HB, Lamers WH. Glutamine synthetase deficiency in murine astrocytes results in neonatal death. Glia. 2010;58:741–754. doi: 10.1002/glia.20960. [DOI] [PubMed] [Google Scholar]
- 37.Cammer W. Glutamine synthetase in the central nervous system is not confined to astrocytes. J. Neuroimmunol. 1990;26:173–178. doi: 10.1016/0165-5728(90)90088-5. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz PH, Nethercott H, Kirov II, Ziaeian B, Young MJ, Klassen H. Expression of neurodevelopmental markers by cultured porcine neural precursor cells. Stem Cells. 2005;23:1286–1294. doi: 10.1634/stemcells.2004-0306. [DOI] [PubMed] [Google Scholar]
- 39.Komitova M, Eriksson PS. Sox-2 is expressed by neural progenitors and astroglia in the adult rat brain. Neurosci. Lett. 2004;369:24–27. doi: 10.1016/j.neulet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 40.Baer K, Eriksson PS, Faull RL, Rees MI, Curtis MA. Sox-2 is expressed by glial and progenitor cells and Pax-6 is expressed by neuroblasts in the human subventricular zone. Exp. Neurol. 2007;204:828–831. doi: 10.1016/j.expneurol.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Roelofs RF, Fischer DF, Houtman SH, Sluijs JA, Van Haren W, Van Leeuwen FW, Hol EM. Adult human subventricular, subgranular, and subpial zones contain astrocytes with a specialized intermediate filament cytoskeleton. Glia. 2005;52:289–300. doi: 10.1002/glia.20243. [DOI] [PubMed] [Google Scholar]
- 42.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1998;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 43.Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 44.Bengtsson M, Ståhlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilhelmsson U, Li L, Pekna M, Berthold CH, Blom S, Eliasson C, Renner O, Bushong E, Ellisman M, Morgan TE, et al. Absence of GFAP and vimentin prevents hypertrophy of astrocytic processes and improves post-traumatic regeneration. J. Neurosci. 2004;24:5016–5021. doi: 10.1523/JNEUROSCI.0820-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishikawa N, Takemura M, Koyama Y, Shigenaga Y, Okada T, Baba A. Endothelins promote the activation of astrocytes in rat neostriatum through ET(B) receptors. Eur. J. Neurosci. 1997;9:895–901. doi: 10.1111/j.1460-9568.1997.tb01440.x. [DOI] [PubMed] [Google Scholar]
- 47.Koyama Y, Takemura M, Fujiki K, Ishikawa N, Shigenaga Y, Baba A. BQ788, an endothelin ET(B) receptor antagonist, attenuates stab wound injury-induced reactive astrocytes in rat brain. Glia. 26:269–271. doi: 10.1002/(sici)1098-1136(199905)26:3<268::aid-glia8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 48.Pekny M, Pekna M. Astrocyte intermediate filaments in CNS pathologies and regeneration. J. Pathol. 2004;204:428–437. doi: 10.1002/path.1645. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Lundkvist A, Andersson D, Wilhelmsson U, Nagai N, Pardo AC, Nodin C, Ståhlberg A, Aprico K, Larsson K, et al. Protective role of reactive astrocytes in brain ischemia. J. Cereb. Blood Flow Metab. 2008;28:468–481. doi: 10.1038/sj.jcbfm.9600546. [DOI] [PubMed] [Google Scholar]
- 50.de la Fuente A, Bing N, Hoeschele I, Mendes P. Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics. 2004;20:3565–3574. doi: 10.1093/bioinformatics/bth445. [DOI] [PubMed] [Google Scholar]
- 51.Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 52.Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 53.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lüscher TF. Endothelin, endothelin receptors, and endothelian antagonists. Curr. Opin. Nephrol. Hypertens. 1994;3:92–98. [PubMed] [Google Scholar]
- 55.Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439:861–864. doi: 10.1038/nature04281. [DOI] [PubMed] [Google Scholar]
- 56.Buffo A, Rite I, Tripathi P, Lepier A, Colak D, Horn AP, Mori T, Götz M. Origin and progeny of reactive gliosis: a source of multipotent cells in the injured brain. Proc. Natl Acad. Sci. USA. 2008;105:3581–3586. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K, Walz W. Unusual topographical pattern of proximal astrogliosis around a cortical devascularizing lesion. J. Neurosci. Res. 2003;73:497–506. doi: 10.1002/jnr.10683. [DOI] [PubMed] [Google Scholar]
- 58.Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–522. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Ambrosio R, Wenzel J, Schwartzkroin PA, McKhann GM, 2nd, Janigro D. Functional specialization and topographic segregation of hippocampal astrocytes. J. Neurosci. 1998;18:4425–4438. doi: 10.1523/JNEUROSCI.18-12-04425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jing R, Wilhelmsson U, Goodwill W, Li L, Pan Y, Pekny M, Skalli O. Synemin is expressed in reactive astrocytes in neurotrauma and interacts differentially with vimentin and GFAP intermediate filament networks. J. Cell Sci. 2007;120:1267–1277. doi: 10.1242/jcs.03423. [DOI] [PubMed] [Google Scholar]
- 61.Bramanti V, Tomassoni D, Avitabile M, Amenta F, Avola R. Biomarkers of glial cell proliferation and differentiation in culture. Front. Biosci. 2010;2:558–570. doi: 10.2741/s85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.