Abstract
Chromatin immunoprecipitation (ChIP) followed by high throughput sequencing (ChIP-seq) is rapidly becoming the method of choice for discovering cell-specific transcription factor binding locations genome wide. By aligning sequenced tags to the genome, binding locations appear as peaks in the tag profile. Several programs have been designed to identify such peaks, but program evaluation has been difficult due to the lack of benchmark data sets. We have created benchmark data sets for three transcription factors by manually evaluating a selection of potential binding regions that cover typical variation in peak size and appearance. Performance of five programs on this benchmark showed, first, that external control or background data was essential to limit the number of false positive peaks from the programs. However, >80% of these peaks could be manually filtered out by visual inspection alone, without using additional background data, showing that peak shape information is not fully exploited in the evaluated programs. Second, none of the programs returned peak-regions that corresponded to the actual resolution in ChIP-seq data. Our results showed that ChIP-seq peaks should be narrowed down to 100–400 bp, which is sufficient to identify unique peaks and binding sites. Based on these results, we propose a meta-approach that gives improved peak definitions.
INTRODUCTION
Chromatin immunoprecipitation (ChIP) followed by high throughput sequencing (ChIP-seq) is becoming the preferred method for genome wide mapping of interactions between DNA and proteins (1–3). Such genome-wide maps are essential tools for understanding gene regulation in multi-cellular organisms. The output of a ChIP-seq experiment is a library of short (25–35 bp) sequence tags mapped to the genome of interest. Protein-specific antibodies are used to pull down DNA fragments bound by the relevant protein, and the tag library is therefore enriched with sequences from interaction sites for this protein. This means that a considerable number of sequence tags will map to genome regions bound by the protein, leading to enriched regions or peaks in the tag profile along the genome. As tag profiles also contain spurious peaks, identifying true interaction sites within a tag profile is the main challenge when analysing ChIP-seq data.
Currently, two main research areas generate most ChIP-seq data; mapping of epigenetic information such as histone modifications (4–6) and mapping of transcription factor binding sites (TFBS) (7,8). Whereas histone modifications may span regions of several hundred kilobases (kb) (9), transcription factors bind short regions of DNA (typically 5–25 bp). Consequently, the ChIP-seq profiles of histone modifications and transcription factors usually are very different. Here, we will focus on transcription factors and discuss the main issues when identifying true TFBS in ChIP-seq data.
Although transcription factors bind short DNA sequences, the immunoprecipitated DNA fragments are fairly large and typically cover a region of 150–600 bp around the binding site (10). As the double-stranded fragments are sequenced from either 5′-end at random, binding sites will typically appear as shifted peaks in the tag profiles on the positive and negative DNA strands (Figure 1A). Despite that such shifted peaks are characteristic of true binding sites, finding the true peaks in the tag profiles is not trivial and at least three issues must be considered when planning ChIP-seq experiments and evaluating potential binding locations.
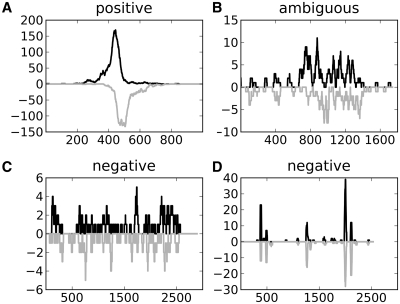
Figure 1.
Peak regions representing (A) a positive peak, (B) an ambiguous peak, (C) a negative region showing evenly distributed tags without a peak-profile and (D) a negative region with peaks lacking the characteristic shift-property on opposite strands.
First, all ChIP-seq data include a certain level of background tags. This background level is not constant, but has substantial local biases and also correlates with the true signal (11). Global and local background models can be estimated from sample data. However, it is more common to make independent samples of background data; for example, by sequencing fragmented DNA before immunoprecipitation (10). Such background data can reveal local or sequencing biases, and can be used to filter out false peaks.
Second, sequencing depth—the number of DNA fragments sequenced—will in general influence ChIP-seq sensitivity and specificity. Increasing the sequencing depth can give more tag profile peaks. However, it is often difficult to decide whether these new peaks are true binding sites or artefacts created by randomly aggregating tags (10,12). Using both background samples and data from replicate experiments can help separate true from false peaks. Given limited sequencing resources, however, it is unclear whether increased sequencing depth or replicated experiments will give the best results.
Third, ChIP-seq technology offers the possibility to discover binding sites with increased resolution compared to alternative technologies like ChIP-chip. It is thus important that the final results after data analysis produce peak regions that reflect this advantage in resolution, as narrow regions are often necessary to identify binding sites unambiguously. To illustrate, a significant binding site motif within a peak region can confirm true peaks, but a major problem with motif analyses is the high number of false positive discoveries. For longer regions, the increased probability of including false motif occurrences by chance generally reduces the value of motif analyses. Another challenge with the motif approach is that not all observed peaks should be expected to contain an instance of the motif; for example, because of indirect binding (13).
Currently, there are several available programs for identifying peaks in tag profiles and these programs use different strategies for peak prediction in relation to background, replicates and sequencing depth. Because of this, programs will have different performance characteristics and strengths. Existing studies have evaluated peak-finder programs and algorithms by comparing peak prediction reproducibility (14), or using false discovery rates (FDR) (11), frequencies of motif occurrences close to identified peaks (11,12,14–17), or experimentally confirmed qPCR sites (14,16). Counting the number of peaks scoring above a certain threshold in real versus background data will give a reasonable estimate of FDR, but will also focus on clear and strong peaks. As for motif-based benchmarks, these have the weaknesses of motif analyses mentioned earlier: high false positive and false negative rates. Binding sites confirmed by qPCR are good measures for true binding affinities, but such confirmed sites are few and may be biased towards specific types of sites. Consequently, such benchmarks likely fail to uncover strengths and weaknesses at analysing the broad spectrum of strong and weak peaks typically found in tag profiles. Another problem with previous program comparisons is the limited ChIP-seq and test data that were available. The consequence of this is that the same data-sources were used by several publications, and that these sources sometimes are identical to those that were used to optimize the programs in the first place, making an unbiased comparison difficult.
Because of the weaknesses in FDR, qPCR and motif-based benchmarks, performance of ChIP-seq peak-finder programs are best evaluated on data sets with known true and false peaks. However, for ChIP-seq peaks, no such data set currently exists. To address this deficiency, we have created a benchmark set of positive and negative peak regions for three transcription factors, neural restrictive silencer factor (NRSF also known as REST), serum response factor (SRF) and Max. The data sets consist of a selection of 400–500 regions representing the different types of peak patterns found in the complete data set from each factor. By visually inspecting the peak profiles and potential binding sites in these regions, and considering different combinations of background data, sequencing depth and replicates, we manually classified the data sets into positive peaks and negative peak-like regions, with associated positive and negative binding sites. We then used this benchmark set to evaluate and compare five peak-finder programs: MACS (11), SISSRs (13), FindPeaks (18), PeakSeq (19) and QuEST (20). MACS, SISSRs, PeakSeq and QuEST were chosen because they all performed reasonably well in recent program evaluations (14–16). MACS employs a sophisticated background estimation model, whereas SISSRs and QuEST take advantage of the peak-shift property to identify binding locations. PeakSeq is the method referenced for creating the tracks that can be viewed in the UCSC Genome Browser. FindPeaks and QuEST both offer options for sub-peak identification within the identified regions, but FindPeaks is the only one with an option for further peak-trimming to improve peak-resolution. Unfortunately FindPeaks is also the only program which currently offers no options for including background data. QuEST only returns single coordinate peak-summits within its subpeaks-regions, which makes it less relevant for peak resolution studies. The diversity of features incorporated into these programs makes all of them interesting for comparisons.
Comparisons of program performances and the manual evaluation highlighted areas of improvement for all issues introduced above. Based on these findings we created a simple meta-approach that used features and outputs from four of the programs. The meta-approach gave improved results when tested with the manually curated benchmark data sets, underlining the importance of including several features when analysing ChIP-seq data.
MATERIALS AND METHODS
The following is an overview of the central aspects of ‘Materials and Methods’ section. More details are given in the Supplementary Data.
Data sets
The three ChIP-seq data sets used in this study were downloaded from the UCSC collection of ChIP-seq data. Aligned tags for NRSF and Max were downloaded (NRSF: Encode project, Myers Lab at the HudsonAlpha Institute for Biotechnology, Max: Encode project, Michael Snyder’s Lab at Yale University) for the cell-line k562, while aligned tags for SRF were downloaded (Encode project, Myers Lab at the HudsonAlpha Institute for Biotechnology) for the cell-line Gm12878. A summary of the data sets are given in Supplementary Table S1. Position Weight Matrices (PWMs) for each factor were downloaded from the TRANSFAC public database (21) with accessions M00256, M00152 and M00118 for NRSF, SRF and Max, respectively.
Peak-finder programs
All programs were run with default or recommended parameters for transcription factor peak-finding. Exceptions were for MACS, where the mfold parameter was changed from default 32 to 12 for SRF and 15 for Max. This was necessary to create the initial peak-shift model, but does not influence the subsequent peak-identification. FindPeaks was run with recommended parameters for sub-peak identification and peak-trimming for evaluation purposes, but only sub-peaks were used to create regions for manual selection. In SISSRs the u option was used to include peaks with tags on only one strand during region selection, but the option was not used during program evaluation. The following program versions were used: Macs 1.3.7.1, SISSRs v1.4, FindPeaks 3.1.9.2, PeakSeq v1.01 and QuEST v2.4. A selection of parameter changes was also created to investigate the effect of alternative parameter settings on program performances. The parameter changes used for these evaluations are given in Supplementary Table S2.
Selection of benchmark regions
To ensure that the benchmark contained an unbiased selection of regions with respect to programs and peak features, we performed the following procedure for region selection.
MACS, SISSRs, FindPeaks and PeakSeq were run with default parameters, or recommended parameter settings for transcription factor binding. This produced lists of peak-regions for each transcription factor and each program sorted by genomic coordinates, and with a score associated with each region. QuEST returns peak positions rather than regions, making it less suitable for unbiased definition of benchmark regions. To reduce the possibility of false negatives, we used lists produced without using additional background data or replicates. These lists were then combined to a single list of potential enrichment regions by merging overlapping regions from all programs. Without background data, the number of tags mapped to each region (tag-count) is the most intuitive measure for a binding event. We therefore wanted our benchmark to include regions somewhat evenly distributed throughout the range of tag-counts. However, inspection of the distribution of tag-counts from all combined regions revealed a large bias towards regions containing less than 50 tags. Because of this, we initially split the combined regions for each transcription factor into three subsets containing regions with more than 200 tags, between 50 and 200 tags and less than 50 tags, respectively. Within each subset, we then wanted to sample peaks representing different characteristics. We therefore calculated parameters for each region, and used Principal Component Analysis (PCA) (Supplementary Data S3) to sample peaks displaying various combinations of these parameters. Only a few simple characteristics were used, to avoid biasing the selection towards any specific mathematical model. These parameters were region length, total number of tags in region, total number of unique tags in region (i.e. tags with different start positions), maximum tag-intensity, the ratio of unique tags relative to total tags and the ratio of tag-maximum to total tags. The two ratios were included to account for variations in peak-width. The PCA sampling procedure was used to produce somewhat equal number of peaks with diverse features from each tag-count category (Supplementary Figure S4). To avoid biasing of the benchmark towards regions produced by a specific program, we used a balanced number of regions from each program. Since FindPeaks and PeakSeq produced a considerably higher number of peaks than MACS and SISSRs, we used only the top 30 000 and 20 000 scoring peaks from these programs for the factors NRSF and Max respectively. To further avoid program-based biases we also included indicators for overlap between each combined region and the regions from each program as a parameter in the PCA model. To avoid selecting regions which certainly were noise, we excluded regions with length <25 bp (the length of a mapped tag), and regions containing less than four tags. We also excluded regions longer than 3000 bp, because these would be difficult to classify manually. At most 1% of the regions from each transcription factor were excluded using the last three criteria. A total of 1347 regions were selected for manual evaluation by this procedure, 480 for NRSF, 452 for SRF and 415 for Max.
Manual classification of peaks
As intended, the automatically generated set of 1347 regions contained many different tag profiles that ranged from regions with obvious peaks to regions with no apparent peaks. We classified the peaks with visual characteristics that obviously corresponded to transcription factor binding regions as positive regions and the regions that did not contain any such peaks as negative regions. Moreover, to reflect that some regions contained some, but not all characteristics of actual binding regions, we classified such regions as ambiguous regions (Figure 1 and Supplementary Data S3). For each positive and ambiguous region, we then manually identified the sub-region or regions representing visible peaks corresponding to true and possible transcription factor binding events. After this initial classification, we reclassified each region three times by including background data, by including replicate data and by considering a random subset of the reads in the relevant regions. The random subset emulated a less deeply sequenced data set. Finally, we did an overall classification by considering all the available information.
Manual classification of binding sites
An enrichment threshold for potential binding sites for each transcription factor was defined by selecting the top 1000 peak-regions with the highest tag-counts among the combined regions. All sub-sequences in these regions were then scanned by the PWM for this factor, as downloaded from TRANSFAC, giving a PWM-score for each sub-sequence. At the same time the number of tags associated with each sub-sequence was also counted. Sub-sequences were then grouped into bins according to their PWM-score, and the average tag-count over all sequences in each bin was calculated. A plot showing increasing PWM-score versus average tag-count in each bin was generated, and the enrichment threshold was decided as the PWM-score for the bin where an enrichment of tags is first observed compared to the average tag-count for all sequences (Supplementary Figure S4). This was done for all three transcription factors, resulting in an enrichment threshold of 0 for NRSF and Max and −2 for SRF. The intention of this procedure was to generate a low threshold, avoiding the exclusion of true sites at the cost of including a large amount of false positives. All potential binding sites in the manually selected regions where then classified as representing a binding event (positive site), not representing a binding event (negative site) or possibly representing a binding event (ambiguous site), according to their association with the previously classified peaks. When a region did not include a potential site, the highest scoring site in the region was identified and evaluated. A total of 3071 binding sites were classified, 775 for from NRSF, 927 from SRF and 1369 from Max. Because of the low threshold, most of the binding sites were classified as negative. Several positive sites with scores close to the threshold were also frequently observed, however (Supplementary Figure S5).
Downloadable files
Manually classified peaks and sites, all regions used for the evaluations, and tracks ready for upload to the UCSC Genome Browser can be downloaded from http://tare.medisin.ntnu.no/chipseqbenchmark/. A script implementing the meta-approach can also be downloaded from this site.
Evaluations of program performance
The evaluation curves for each program in Figure 2 were created by identifying overlaps between the program-defined regions and the manually evaluated regions. The program-defined regions were first sorted in descending order according to their score, meaning that the most confident regions appeared at the beginning of the list. The list was then traversed, and the level of false positives (program regions overlapping with evaluation regions not classified as true or ambiguous peaks) was calculated each time a new true positive (program region overlapping with a true peak) was encountered. The number of false positives thus accumulates as more true positives are found.
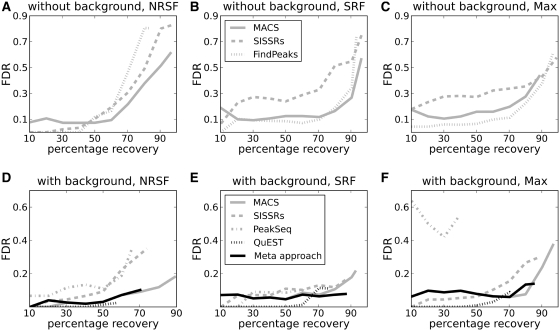
Figure 2.
Performance of the different programs in the manually evaluated regions for the three transcription factors NRSF (A and D), SRF (B and E) and Max (C and F). The plots show how the FDR increases with percentage recovery of true peaks. Note the difference in scale on the FDR-axes for plots (A–C) compared to (D–F). Plots (A–C) show results when no background data is used in the analysis, whereas D–F show results when additional background data is included. The latter plots also show results from the meta-approach described in this study.
Performance evaluations at the nucleotide level were calculated for peaks as follows: Nucleotides in regions where the program defined peaks showed overlap with the manually identified peaks were true positives, nucleotides in regions where the program-defined peaks had no overlap with the manually identified peaks were false positives and nucleotides in regions where the manually identified peaks did not overlap with the program-defined peaks were false negatives (Supplementary Figure S6). Site evaluations were defined in a similar way, but now only the nucleotides in the potential binding sequences were considered. Peak-regions overlapping with nucleotides from positive sites were true positives, regions overlapping with nucleotides in negative sites were false positives and nucleotides in positive sites not overlapping with the peak-region were false negatives (Supplementary Figure S7a).
Validation by motif discovery in MEME
The quality of the manually identified regions was validated by submitting the regions to the motif discovery program MEME (22). The performance of MEME was compared between the manually and program-defined peaks in the selected regions. For the program-defined peaks both the full regions and the region-maximum ±125 bp were used as input. The following parameters were specified as input to MEME: dna (sequences use DNA alphabet), mod = zoops (one or zero motif occurrences per region), w (motif width, 21 for NRSF, 18 for SRF and 14 for Max), nmotifs = 5 (number of different motifs to find) and revcomp (allow sites on both strands). All other parameters were set to default values. Visual inspection of the motif logo from MEME turned out to be sufficient to decide whether the motif resembled the TRANSFAC motif or not.
RESULTS
A manually curated data set for benchmarking ChIP-seq peak-finder programs
We had three major goals when creating a benchmark for evaluating peak-finder programs. First, the benchmark should assess the importance of background data, replicate data and sequencing depth for program performance. Second, in addition to assessing the programs’ performance at separating true from false peak regions, the benchmark should assess how well the peak predictions correspond with actual visible peaks and with likely TFBS. Third, the benchmark should cover the range of peak types and regions found in tag profiles. To create the benchmark we manually classified potential peaks and binding sites in selected regions as positive, ambiguous and negative as described in ‘Materials and Methods’ section. Table 1 lists the number of peaks in each category for each reclassification of the benchmark. Unless stated otherwise, evaluations are based on the overall classification.
Table 1.
Additional data resolve ambiguous regions
| Initial | Background | Replicate | Random | Overall | Sites | |
|---|---|---|---|---|---|---|
| NRSF | ||||||
| pos | 126 | 140 | 134 | 107 | 138 | 117 |
| amb | 77 | 31 | 54 | 29 | 31 | 43 |
| neg | 293 | 320 | 307 | 356 | 323 | 615 |
| sum | 496 | 491 | 495 | 492 | 492 | 775 |
| SRF | ||||||
| pos | 124 | 152 | 135 | 111 | 136 | 92 |
| amb | 67 | 27 | 27 | 41 | 16 | 24 |
| neg | 272 | 285 | 303 | 310 | 312 | 811 |
| sum | 463 | 464 | 465 | 462 | 464 | 927 |
| Max | ||||||
| pos | 184 | 265 | 209 | – | 226 | 335 |
| amb | 142 | 46 | 69 | – | 56 | 177 |
| neg | 184 | 186 | 208 | – | 201 | 857 |
| sum | 510 | 497 | 486 | – | 483 | 1369 |
The table shows the number of positive, ambiguous and negative peaks that were manually classified. All peaks were classified without using any additional information (initial), using background data (background), replicates (replicate), randomly samples subset (random, not for Max) and an overall evaluation (overall). The number of binding sites (sites) evaluated as positive, ambiguous and negative are also included. The difference in column sums is caused by multiple peaks in the same region. If ambiguous or positive, they are classified as individual peaks. Otherwise the whole region is classified as negative.
Most peaks can be classified by visual inspection without using additional background and replicate samples
Other studies have indicated that background and replicate data are important for correctly identifying false peaks (12,15). This was also true in our data set, as using tag counts adjusted to background was a better predictor of true peaks than using tag counts in sample data alone (Supplementary Figure S4). However, visual inspection of the selected regions revealed that most false peaks could be identified without additional information from background data or replicates. Over 80% of the false peaks lacked the expected visual appearance of a typical ChIP-seq peak, which made it possible to immediately classify them as false peaks. This included peaks with no shift property, peaks with high intensity spikes covering short 25–50 bp regions and longer regions that may be somewhat enriched but where no peak-shape was visible (Figure 1C and D). The latter regions are relevant when studying histone-modifications, but do not seem to be characteristic for transcription factor binding (9,10). Just an additional 5–15% of the false peaks in the manual evaluation could be correctly classified only by using background data and replicates. Consequently, information in the peak profile itself can be used to separate true peaks from most false peaks. As can be seen from Table 1, the most important contribution from background data and replicates was in separating initial ambiguous regions into negative or positive peaks.
Use of background data substantially improves peak-finder performance
Inclusion of background data was essential for the programs to separate most of the true peaks from artefacts and random noise (Figure 2 and Supplementary Figure S9). Generally, ChIP-seq peaks from all programs include a considerable amount of false positives, both when analysed with and without external background data. At the standard FDR-level of 0.05, no >60% of the true peaks can be recovered by any program, and an increase in recovery results in a dramatic increase in the number of false positives. The highest recovery (>90%) is achieved for data analysed without background, but the number of false positives at these recovery rates are intolerably high, with more than three out of five peaks being false positives (FDR > 0.6). The number of false positives is significantly reduced when external background data is used, with FDR dropping to 0.3 (on average) for the last identified peaks. A side effect of including background is that true positive peaks are also removed along with false peaks. However, the overall positive effect on the FDR leads to the conclusion that external background data is essential for programs to identify true peaks in ChIP-seq data. Note that though the FDR improves when data is analysed with background, it is still above the 0.05 FDR standard-level, and a cut-off on the peak-score is generally recommended to keep the number of false positives at an acceptable level. Considering that most false peaks could be identified without background data in the manual classification, these results also show that peak-appearance features are currently under-utilized by existing software.
Of the programs tested, MACS had the best performance with ∼80% of the true peaks identified at FDR ∼0.1. Without background data, the percentage of true peaks found at FDR ∼ 0.1 dropped to 60. The same trend was observed for the other programs. The stable performance by MACS can probably be attributed to its advanced statistical background model, which estimates different local backgrounds from tags 1000, 5000 and 10 000 bp from each peak-centre. QuEST showed a low FDR for NRSF (Figure 2D), but this came at the cost of reduced sensitivity; only 60% of the true positives were identified in the final list. For the other two factors, identifying additional true positives for QuEST also lead to increasing FDRs (Figures 2E and F). Generally, including background data functions as a filter on the results generated when data is analysed without background. The total number of identified peaks is considerably reduced and most of the removed peaks are false positives, which leads to an improved FDR. However, for the transcription factor Max analysed with MACS, the total number of peaks was not reduced when background data was included. Instead the program identified an alternative set of peaks with a reduced FDR compared to the peaks identified without background. This indicates that more advanced background models, such as the one used by MACS, can give improved results not only by removing peaks, but also by discovering new true peaks which remained hidden during the analysis without independent background.
SISSRs generally showed a higher level of false positives than MACS, FindPeaks and QuEST did. The reason for this is that the approach for defining peaks is different in SISSRs. Whereas MACS, FindPeaks and PeakSeq use a sliding window approach resulting in longer regions of a few hundred to over thousand basepairs in length, SISSRs localizes the precise shift point between peaks on the positive and negative strand, returning regions of >100 bp. Thus, several short SISSRs regions are often located within the longer regions returned by the other programs, potentially leading to a higher number of false positive regions. The advantage with SISSRs’ approach is of course that it can identify multiple true peaks within a longer region.
Using more sequence data improves identification of weak binding sites only when analysed with external background data
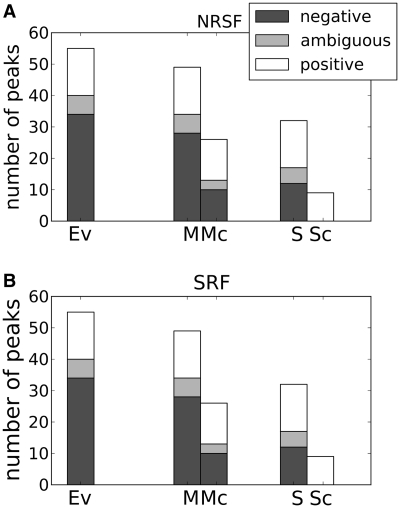
As high-throughput sequencing becomes more common, the number of deeply sequenced data sets together with an extensive use of replicates is expected to increase. However, how this may influence the identification of peaks and the performance of peak-finder programs is not clear (12,19). The idea behind using more sequence data is to identify more weak binding sites while making stronger sites appear more pronounced. Running the programs on the more deeply sequenced data sets did indeed produce more peaks compared to the smaller randomly sampled subsets, which is in accordance with previous studies (12,19). The question is, however, whether these additional peaks represent additional binding sites. Comparing the evaluation curves for the deeply sequenced sets to the randomly sampled subsets showed no obvious improvement in performance (with the exception of QuEST, which performed poorly on the randomly sampled subsets) (Supplementary Figure S10). To investigate this paradox further, we closely inspected a subset of the manually evaluated peaks. This subset included peaks with clearly improved visibility in the complete sets compared to the randomly sampled subsets. A total of 136 peaks satisfied this criterion from the NRSF and SRF data sets, where 33 of these were classified as true peaks, 29 as ambiguous peaks and 74 as noise features. When examining the program outputs on the selected regions, improved performance is observed in the more deeply sequenced sets only when an external background model is included (Figure 3). Without an external background model, the programs cannot identify more true peaks without including an intolerable number of false positives, and one is often better off using the random subset.
Figure 3.
New peaks identified for (A) NRSF and (B) SRF in deeply sequenced sets, compared to random subsets. The bars show the number of positive, ambiguous and negative peaks found by MACS (M), MACS with background (Mc), SISSRs (S) and SISSRs with background (Sc), together with the manual evaluation reference (Ev).
Improved resolution in ChIP-seq is not reflected by returned peak-regions, which makes it difficult to pinpoint the true binding sites
One potential advantage with ChIP-seq compared to alternative methods like ChIP-chip is the increased resolution. In our manually curated data set, most peaks could visually be narrowed down to 100–400 bp, which is a considerable improvement compared to most ChIP-chip experiments (peaks typically above 1 kb). However, this improvement in resolution was not reflected in the regions returned by most of the programs (Figure 4).
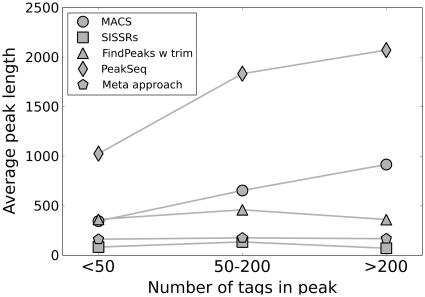
Figure 4.
Average lengths for program-defined peaks with increasing tag counts. For MACS and PeakSeq the average length increases with the number of tags in the peak.
To investigate how the programs performed with respect to peak-resolution, we selected a subset of the manually evaluated regions such that each region in the subset had been identified by all programs and contained only one true peak, with at least one binding site motif occurrence. A total of 125 peaks satisfied these criteria. We then asked to what extent the annotated peaks corresponded with the programs’ peak definitions at the nucleotide level Supplementary Figure S7).
As Figure 5A shows, the longer regions defined by MACS, FindPeaks and PeakSeq had a complete overlap with nearly every manually classified region, resulting in sensitivities close to 1. However, most of these regions also included several hundred base pair extensions, resulting in many false positive nucleotide predictions. To correct for this, FindPeaks offers a peak-trimming option to narrow down regions, which somewhat improved the results. As can be seen from Figure 4, the longest regions are often also the ones with the highest number of tags. In contrast to the other programs, SISSRs uses the peak-shift property to identify and define peaks. SISSRs peak-regions were therefore short—sometimes no more than 40 bp—but SISSRs also often predicted multiple peaks within the regions. Consequently, SISSRs had lower FDR, but also lower sensitivity, because the peaks were usually too short to represent the full peak-profile. In summary, none of the programs produced peak-regions that corresponded to the visual peak-profile.
Figure 5.
Program performance on (A) peak-region definitions and (B) binding-site identification. The plots show sensitivity versus FDR for factors NRSF (dark grey), SRF (light grey) and Max (white) and programs MACS (circle), SISSRs (square), FindPeaks (triangle up), FindPeaks with trim (triangle down), PeakSeq (diamond) and our meta-approach (pentagon).
A related but different question is whether the programs’ peak definitions encompassed likely TFBS (Supplementary Figure S8a). Again MACS, FindPeaks and PeakSeq identified most of the manually identified true binding sites within evaluated regions, but the programs also showed high FDR (Figure 5B), which again is an effect of the longer peak regions produced by these programs. Consequently, these regions alone can not distinguish true from false binding sites unless one considers local variations within the regions. Regions beyond a few hundred basepairs will generally contain too many false positive hits in addition to the true site when using scanning procedures with position weight matrices to predict binding sites. Narrowing down the regions is therefore essential to identify the exact binding site. By manually defining peak-boundaries we could usually define the binding regions in the range from 100 to 400 bp, which seems to be sufficient for finding the true binding sites, without including too many false positives. The exception is where several potential binding sites are clustered close together (Supplementary Figure S8b). Compared to the other programs, SISSRs produced shorter peak-regions with resolution of 40–150 bp that in most cases overlapped with the true binding site (Figure 5B). However, these regions did not always contain the site completely. To illustrate, SISSRs-defined peaks overlapped 85% of the NRSF sites, but only 47% of the sites were completely contained within the same peaks. Having the complete motif included in the region is important for example in motif discovery by programs like MEME.
Combining output from different programs gives improved peak definitions
The benchmarks of the different peak-finder programs showed that there is still considerable potential for improvement in both discovering and defining the correct peak-regions in ChIP-seq data. As the programs have different strengths, we hypothesized that combining the most promising features of each program could improve the overall result. To test this hypothesis, we developed a meta-analysis tool that consisted of four simple steps. First, we used the set of peaks from MACS as starting point, as MACS generally was the best performing program. Second, we used SISSRs to filter the MACS-regions because SISSRs takes advantage of the shift property not employed by MACS or other window-based approaches. For SISSRs peaks we included peaks with tags on only one strand, since these may be potential true positives. Third, if replicate data were available, we required that the same MACS regions were present in all the replicates. Fourth, as we started with regions defined by MACS, we performed peak-trimming similar to FindPeaks to reduce the region length to 100–400 bp. Compared with the existing programs, this meta-approach gave reduced false positives and improved peak-regions that better corresponded to binding site motif occurrences (Figures 2 and 5). The results were consistent for all three transcription factors. The main disadvantage was that some true positive peaks were removed because of the strict filtering criteria.
Results from motif discovery validates manual evaluations
A disadvantage of using manually classified data for benchmarking is the potential subjective bias introduced during evaluation. To validate the quality of the data sets, we used manually defined peak-regions as input to the motif discovery program MEME (22). We reasoned that in a high-quality data set, de novo motif discovery methods should easily find motifs similar to the transcription factor’s known binding site motif. Indeed, MEME unambiguously identified the correct binding site motif for all factors in the manually classified regions and the regions defined by our improved meta-approach (Table 2). We therefore concluded that these regions were good representations of true peaks and binding sites. In contrast, MEME could not recover all the correct motifs from the program-defined regions. Whereas MEME rarely found the correct motif in the long regions returned by MACS, FindPeaks and PeakSeq, MEME returned the correct motif for two of the three transcription factors in the shorter SISSRs regions. The failure to identify the last motif could be because the SISSRs regions do not cover all binding-sites completely (Figure 5B). We also tested an alternative approach for defining regions for motif discovery, where we used a fixed-sized region flanking the position in the peak-region with the highest number of tags. With this approach we could also test the peak-summits of sub-peaks identified by QuEST. Using a fixed extension of ±125 bp improved the results for most programs, and with data from QuEST, MEME could identify the correct motif for all three transcription factors.
Table 2.
Results from motif discovery by MEME
| Full length regions |
Maximum ± 125 bp |
|||
|---|---|---|---|---|
| Best score | Top 5 | Best score | Top 5 | |
| Macs | 0 | 1 | 1 | 1 |
| Macs w background | 0 | 1 | 2 | 1 |
| SISSRs | 0 | 2 | 0 | 0 |
| SISSRs w background | 2 | 0 | 2 | 0 |
| FindPeaks w trim | 0 | 0 | 0 | 1 |
| PeakSeq | 1 | 1 | 1 | 1 |
| QuEST | – | – | 3 | 0 |
| Meta-approach | 3 | 0 | – | – |
| Manually evaluated | 3 | 0 | – | – |
Both full length regions and the regions created by adding/subtracting 125 bp around the maximum tag-intensity in each region were tested. The table shows the number of times the correct motif was identified as the best scoring one or was among the top five scoring motifs returned by MEME. Both in the meta-approach and the manually classified regions the correct motif was returned as the top scoring one for all three transcription factors.
Changing program parameters gives improved performance for some of the peak-finder programs
We investigated whether changing the parameter setting from their default values gave improved peak-finder performance when used on the manually curated data sets. We did not perform a comprehensive parameter optimization, as this would increase the risk of overfitting, and it is also less relevant from a practical viewpoint. Rather we opted for adjusting a selection of parameters that a user would be likely to consider in an actual analysis, given the a priori knowledge of the data set. An overview of all parameters investigated, and their overall effect on the results are given in Supplementary Table S2. For MACS and QuEST default or recommended parameters gave the best performance. The results from MACS could only be marginally improved for NRSF and SRF by including the –futurefdr option. The importance of the background model for MACS was further underlined, as leaving it out using the –nolambda option gave consistently poorer performance. SISSRs and PeakSeq were both sensitive to parameter changes, but which parameters that gave improved results changed depending on the specific transcription factor. To illustrate, decreasing the max_threshold in PeakSeq from 100 to 50 gave considerably improved performance for NRSF and Max, whereas the effect was opposite for SRF. Increasing the window size from 1 million (default) to 2 million gave improved performance for all factors. SISSRs showed improved results when the scanning window size, –w, was increased from 20 (default) to 50, but the effect was much more pronounced in NRSF than in SRF and Max. FindPeaks showed a consistent decrease in false positive predictions when the minimum parameter was raised from 8 (recommended) to 12, but this improvement did not compensate for FindPeaks inability to include background data. An overview of the best performing parameters for each program on each transcription factor is shown in Table 3. Plots of true versus false positives for the best performing parameter setting for each program are shown in Supplementary Figure S11.
Table 3.
Parameter change that gave the best performance on each transcription factor for the different programs
| Program | NRSF | SRF | Max |
|---|---|---|---|
| MACS | Include futurefdr | Include futurefdr | default |
| SISSRs | Increase scan window size from 20 to 50 | Increase scan window size from 20 to 50 | Increase scan window size from 20 to 50 |
| FindPeaks | Increase Minimum from 8 to 12 | Increase Minimum from 8 to 12 | Increase Minimum from 8 to 12 |
| PeakSeq | Reduce Max Threshold from 100 to 50 | Increase Window Size from 1 to 2 mill. | Reduce Max Threshold from 100 to 50 |
| QuEST | default | default | default |
DISCUSSION
Do current peak finder programs have room for improvement?
One of our main observations is that most false peaks could be identified manually without using additional background and replicate data. Peak features visible in the sample data alone were sufficient to distinguish them from the true peaks. An important reason why this information is not used by current programs may be that these features are challenging to model. Nevertheless, our results suggest that to get further improvements, peak-finder software should focus on developing methods that use these features.
Many of the current methods use background data in the analysis to effectively identify and remove false positive peaks. MACS, using the most advanced background model, generally performed better than the other programs. Thus improved statistical background modelling may also further improve peak identification. This may especially be true for the more deeply sequenced data. Deeper sequencing has the potential to discover more weak binding locations, but our results show that background models are essential to avoid too many false peaks. Though the binding affinities of weak sites are small, they can still be biologically important (23,24), so identifying such sites is relevant.
None of the programs tested can include replicate data in their analyses. Instead, replicate data are typically analysed separately to create a second set of output peaks and only peaks that overlap in both sets are kept in the final list. Results from different replicates were generally consistent for all programs (Supplementary Figure S12), indicating that ChIP-seq data are reproducible. However, weak signals that may fall below the detection threshold within an individual replicate but that are consistent between replicates will be missed with this current approach. To identify such signals, future peak-finder software should likely analyse all replicates within a common statistical model.
Do region definitions matter?
Another important area of improvement is in the region-definitions provided by the programs, which are in general too long to identify binding-sites unambiguously. The reason for the increased length is that MACS, FindPeaks and PeakSeq use a sliding window approach to define the peak regions. Consecutive windows of a certain base-pair width are evaluated along the genome, according to the tag count. If the window has a significant sample tag signal compared to the background tags or background model, this window is marked as an enriched region. The windows themselves may be of high resolution. However, if consecutive regions are significant, they are merged into one single region. It is not uncommon to observe a general enrichment of tags, especially around larger peaks (Supplementary Figure S5a), meaning that these merged regions may extend beyond several thousand basepairs. The result is a loss of resolution, especially for the larger peaks. This is unfortunate, since the larger peaks are in most cases also the most certain binding locations. FindPeaks is the only program which compensates for this by offering a sub-peaks option, which splits long regions into smaller ones to capture overlapping peaks, and a trim-peak option which aims to shorten down the regions by identifying the peak-feature more precisely. Both these options gave improved results in the regions from FindPeaks. SISSRs does not use the sliding window approach, but rather takes advantage of the shift-property characteristic for true peak signals. In this way SISSRs manages to identify binding locations with considerable improved resolution compared to both the manual evaluation and the other programs. However, as can be seen from the MEME results, defining short regions has disadvantages in motif discovery when the regions are too short to cover the motifs completely.
Is the benchmark set representative of transcription factors in general?
When studying ChIP-seq data, some variation in peak appearance is expected depending on the transcription factor studied. Though the most prominent peak properties such as the shift between positive and negative strand peaks are characteristic for all factors, some smaller variations are also observed between the three factors selected for this study. Data for NRSF are characterized by peaks with a high average tag-count compared to the background, and usually only one peak per region. The latter is also true for SRF, but with the average peak being less pronounced compared to the background. Data for Max are more challenging, with a considerably higher proportion of less pronounced and overlapping peaks. Consequently, studies on a single factor are not always representative for transcription factors in general. Here this has been compensated for by including three factors displaying somewhat varying peak-properties. However, different peak characteristics may be expected for other transcription factors.
Are the manually defined regions representative of true binding sites?
The results of motif-discovery by MEME showed that MEME more often found the expected motifs within the manually defined regions than within the regions defined by the existing peak-finder programs. This result indicates that the manually defined regions give a good representation of actual binding sites and include few false positive peaks. Nevertheless, on some occasions the peak finders report more regions to contain sites with high PWM scores than those that were identified in the manual classification. Two different interpretations can explain this observation. First, the programs can detect tag-densities not visible by manual inspection, in which case there are more true peaks in the evaluated regions than what we found by visual classification. Second, the additional sites are from false positive peaks which by chance also included a false positive site. Since the number of false positive peaks and PWM sites scoring above our enrichment threshold are both considerable, chance overlaps will occur quite often. Consequently, only additional experiments can determine whether these cases represent true binding sites.
Are the performance estimates representative of current peak finders?
Finally it should be noted that the performance estimates displayed in Figure 1 are only valid for the manually selected regions and not for ChIP-seq data in general. The reason for this is that the region-samples were not drawn at random, but were sampled to span the variation in size and peak-features for each data set. Sampling at random would have produced a large number of small peaks more representative for the region tag-count distribution. However, we wanted to focus on the variation in peak-appearance rather than evaluating a large number of very similar peaks (Supplementary Figure S7), which is why we used a multivariate approach for peak-region selection. Moreover, we did not consider true peaks not discovered by any of the programs, but as we selected peaks based on four different programs, we expect that there were few such peaks. For the evaluations on region-definitions, we chose to use only regions identified by all programs in order to compare programs on an equal basis. Consequently, mostly high-quality single-peak regions were used in this comparison. More challenging regions, such as less pronounced or multiple peaks, were not evaluated in this part of the study. However, this bias towards easy regions only underscores the general conclusion that there is considerable potential for improvement in peak-region and binding site definitions.
Can the visual classification be used on other types of ChIP-seq data?
As mentioned in the introduction, there are mainly two types of data which are currently mapped by ChIP-seq: binding sites for transcription factors, and genome wide maps of histone modifications. Peak-profiles for histone modifications are more diffuse and often span wide regions of the genome, which make them more difficult to distinguish visually. In comparison, ChIP-seq peaks surrounding TFBS span a short region of the genome, and have characteristic features which make it simpler to distinguish true or false peaks. Alternative methods may therefore be necessary to create proper benchmark-sets for histone modification data.
CONCLUSIONS
ChIP-seq data, when used together with external background or control samples, have sufficient resolution and quality to discover unambiguous genome-wide transcription factor binding locations. However, focus must now be put on the subsequent data-analysis to fulfil this potential. Based on our findings, further improvements in peak-finder programs for ChIP-seq data should concentrate on the following four tasks: (i) separate true peaks from noise and artefacts by using both characteristic peak features and statistical distributions; (ii) include external background data, sequencing depth and replicates within a common model framework to guide this separation; (iii) narrow down identified peaks to 100–400 bp; and (iv) include options to identify multiple and overlapping peaks within the same region. Improved results for the introduced meta-approach are an indication that the potential for improvement in ChIP-seq data analysis is considerable.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Functional Genomics Program (FUGE) of the Norwegian Research Council. Funding for open access charge: NTNU.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 2.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 3.Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J. Cell. Biochem. 2009;107:11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Wallerman O, Motallebipour M, Enroth S, Patra K, Bysani MS, Komorowski J, Wadelius C. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic Acids Res. 2009;37:7498–7508. doi: 10.1093/nar/gkp823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laajala TD, Raghav S, Tuomela S, Lahesmaa R, Aittokallio T, Elo LL. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics. 2009;10:618. doi: 10.1186/1471-2164-10-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS ONE. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeva V, Surdez D, Guillon N, Tirode F, Fejes AP, Delattre O, Barillot E. De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis. Nucleic Acids Res. 2010;38:e126. doi: 10.1093/nar/gkq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 23.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 24.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.