Abstract
Ribosomal RNA, transcribed by RNA polymerase (Pol) I, accounts for most cellular RNA. Since Pol I transcribes rDNA repeats with high processivity and polymerase density, transcription termination is a critical process. Early in vitro studies proposed polymerase pausing by Reb1 and transcript release at the T-rich element T1 determined transcription termination. However recent in vivo studies revealed a ‘torpedo’ mechanism for Pol I termination: co-transcriptional RNA cleavage by Rnt1 provides an entry site for the 5′–3′ exonuclease Rat1 that degrades Pol I-associated transcripts destabilizing the transcription complex. Significantly Rnt1 inactivation in vivo reveals a second co-transcriptional RNA cleavage event at T1 which provides Pol I with an alternative termination pathway. An intact Reb1-binding site is also required for Rnt1-independent termination. Consequently our results reconcile the original Reb1-mediated termination pathway as part of a failsafe mechanism for this essential transcription process.
INTRODUCTION
RNA polymerase (Pol) I is responsible for ribosomal RNA transcription and synthesizes the great majority of RNA in every living cell. Efficient termination of transcription is crucial to coordinate Pol I transcription and allow polymerase recycling. Ribosomal DNA is organized in a tandem array of units comprising the pre-rRNA encoding DNA plus upstream and downstream regulatory elements; a schematic of a yeast rDNA repeat is represented in Figure 1A. The 35S pre-rRNA is transcribed and subsequently processed to produce mature 18S, 5.8S and 25S rRNAs. Each rDNA unit also contains the Pol III-transcribed 5S rRNA gene in the opposite orientation.
Figure 1.
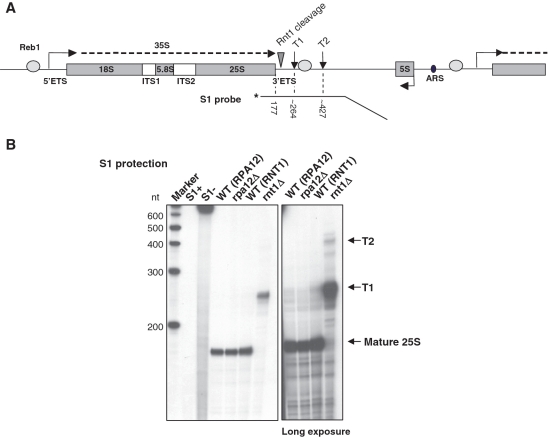
3′-extended 25S rRNA is produced in rnt1Δ cells. (A) Schematic of a S. cerevisiae rDNA repeat. In addition to the sequence encoding 18S, 5.8S and 25S rRNA (gray rectangles), the Pol I transcription unit includes External and Internal Transcribed Sequences (ETS and ITS); the 35S primary transcript is shown as a dashed line. Gray ovals represent binding sites for Reb1, triangle Rnt1 cleavage site and vertical arrows denote the T-rich elements of the terminator. 5S rDNA, transcribed by Pol III in opposite orientation, and Autonomously Replicating Sequence (ARS) are shown. The 3′-labeled probe used in S1 protection and size of the expected bands are indicated below. (B) S1 protection on total RNA from rpa12Δ, rnt1Δ and isogenic WT. S1+ and S1− controls show the probe alone after incubation with or without S1 nuclease. Arrows on the right indicate the position of mature 25S rRNA and transcripts extending to T1 or T2 terminator elements. Longer exposure is shown in the right hand panel.
Early experiments on the mechanism of transcription termination by Pol I relied on in vitro approaches, often using purified components. This led to the definition of the main elements of the rDNA terminator in the region immediately 3′ to the 35S pre-rRNA sequence (1). Both in yeast and mammals a DNA-binding protein (Reb1 in yeast, TTF1 in mammals) interacts in a sequence-specific manner with the rDNA terminator (2,3). Furthermore, binding of this protein was shown to cause polymerase pausing and to promote transcription termination at an upstream T-rich ‘release’ sequence (1). In Saccharomyces cerevisiae this T-rich element was identified as the main terminator (or T1) as opposed to a second T-rich tract located ∼157 bp downstream, named the failsafe (or T2) terminator (4). In mammals an additional Pol I-interacting factor, PTRF, was implicated as a release activity (5).
More recent in vivo studies have progressively added complexity to the process of Pol I termination and revealed close parallels between the mechanisms of transcription termination by Pol I and Pol II (6–8). Pre-RNA processing is coupled to transcription elongation, with the first event being cleavage of the transcript in the 3′ External Transcribed Sequence (ETS) by Rnt1 to produce the 35S pre-rRNA (9). Rnt1 is a RNase III-like endonuclease that recognizes and cleaves across a stem–loop structure in the pre-rRNA. Several lines of evidence show that RNA cleavage by Rnt1 is a co-transcriptional event (10,11). In particular co-transcriptional RNA cleavage by Rnt1 is required for Pol I to terminate through a ‘torpedo’ mechanism (7,8). The 5′–3′ exonuclease Rat1 is recruited to the 5′-end of the downstream Rnt1 cleavage product and cooperates with the helicase Sen1 to progressively degrade Pol I-associated transcripts. When Rat1 reaches the still transcribing polymerase, transcription complex destabilization promotes polymerase release from the template DNA.
The torpedo mechanism does not negate the previous model for termination as both mechanisms may coexist. However depletion of Reb1 did not reveal any significant loss of termination by Pol I in vivo (8). On the other hand the torpedo model postulates RNA cleavage by Rnt1 has a central role in transcription termination. Cells lacking Rnt1 do show a termination defect as assessed by transcriptional run-on (TRO) analysis (6,8), however this defect appears to be quite minor when compared with the profile obtained in cells lacking the small Pol I subunit Rpa12, which is known to be critical for Pol I termination (6). Overall the above account suggests that other activities may be involved in Pol I termination.
Here we investigate in vivo the consequence of loss of Rnt1 activity on Pol I transcription termination. We detect a second co-transcriptional RNA cleavage event that maps to the T1 terminator element. T1 cleavage provides Pol I with an alternative pathway to efficiently terminate transcription. Furthermore we dissect the relative contribution of the previously implicated Pol I terminator elements and observe that an intact Reb1-binding site is required when the Rnt1-dependent pathway is impaired. These observations reconcile previous data and reveal a failsafe mechanism for Pol I termination.
MATERIALS AND METHODS
Yeast strains and plasmids construction
Strains used in this study are shown in Supplementary Table S1. Standard media and growth conditions were employed. rat1-1 sen1-1 and isogenic WT were grown at 25°C then shifted at 37°C for 3 h prior to RNA extraction. pGAL-REB1 cells were grown in 1% Glu + 1% Gal or switched to 2% Glu for approximately five generations prior to RNA extraction.
Pol I minigene construction procedure is reported in Supplementary Data.
S1 protection
An amount of 10 µg of total RNA were analyzed by S1 protection analysis (12). The probe was prepared by digestion from the construct pGEM-3′rDNA (containing the 3′-region of rDNA) with NheI (inside 25S) and NaeI (on pGEM) followed by 3′-end labeling with Klenow in presence of [α-32P]dCTP. The annealing sequence protects the region −177 to +276 relative to the end of 25S rDNA.
Biotin labeling of RNA selection probes
Biotinylated RNA probes were prepared by in vitro transcription (13). For hybrid selection TRO, the probe was complementary to the BsrGI-EcoRI fragment in the 3′-region of rDNA (position −80 to +102 relative to the end of 25S rDNA). For hybrid selection cRACE, the probe was complementary to the NheI-BsrGI fragment (position −177 to −80 relative to the end of 25S rDNA).
Hybrid selection transcription run-on
hsTRO analysis (13) and M13 probes (8) as previously described. Signal intensity was measured with a phosphorimager and plotted relative to the signal obtained with probe 2 = 100%. The proportion of selected and non-selected signal for each probe is indicated in the same graphs.
Hybrid selection circular RACE
hscRACE was performed as in (14). After selection with the biotinylated probe, directed RNase H treatment with the oligo ‘RNaseH’ was used to release the selected transcripts from the magnetic beads. After circularization of the released RNA with T4 RNA ligase, the RT reaction was primed with the oligo ‘R-rev’ and PCR with the oligos ‘R-fw’ and ‘R-rev’. The PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced.
Primer extension
Primer extension was performed in standard conditions from 5 μg of RNA from cells transformed with the indicated constructs, priming the reaction with the 32P labeled oligo ‘Ext-rev’. The products were separated on 5% polyacrylamide/urea gel.
RT–PCR
All RT reactions were performed in standard conditions with Superscript III Reverse Transcriptase (Invitrogen) on 1.5 μg of RNA extracted from cells transformed with the indicated constructs and primed with the oligos A-rev or B-rev. Real-time PCR was performed with a Corbett Rotorgene system using Quantace SensiMix kit. Primer sequences for PCR A and B (Figures 3D and 4B) are indicated in Supplementary Table S2.
Figure 3.
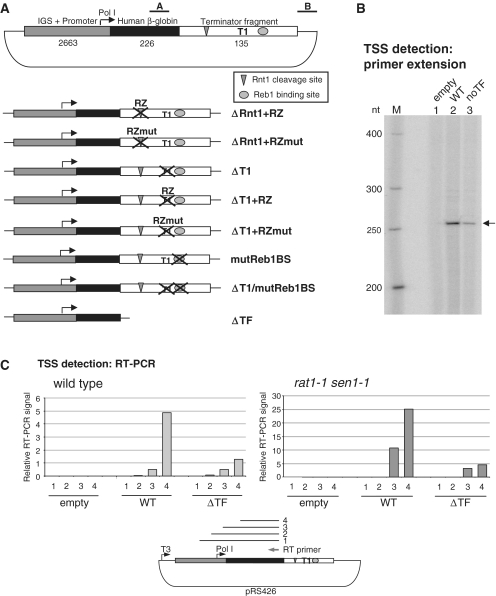
A Pol I minigene to study transcription termination. (A) Top: Scheme of the ribosomal minigene, including Pol I promoter plus upstream intergenic sequence (gray), selection fragment derived from the human β-globin gene (black) and Pol I terminator fragment (white) including Rnt1 cleavage site (triangle), T1 and Reb1-binding site (oval). Sizes in base pairs are indicated below. A and B show position of the PCR products for the RT–qPCR analyses in Figure 4B–D. Bottom: schematic of the terminator mutants incorporated into the minigene. (B) Primer extension showing authentic Pol I 5′-ends are produced from the minigene. Arrow indicates a single primer extension product corresponding to correctly initiated Pol I transcripts. (C) TSS detection on the Pol I minigene by RT–qPCR. The analysis was conducted in WT (left) or rat1-1 sen1-1 (right) cells, transformed with the indicated constructs. Reverse transcription was primed with an oligo selective for the plasmid-encoded transcripts, PCR with a communal reverse primer and different forward primers to generate products 1, 2, 3 and 4, shown in the scheme below. PCR efficiency was normalized to T3 transcript produced in vitro (T3 promoter is indicated).
Figure 4.
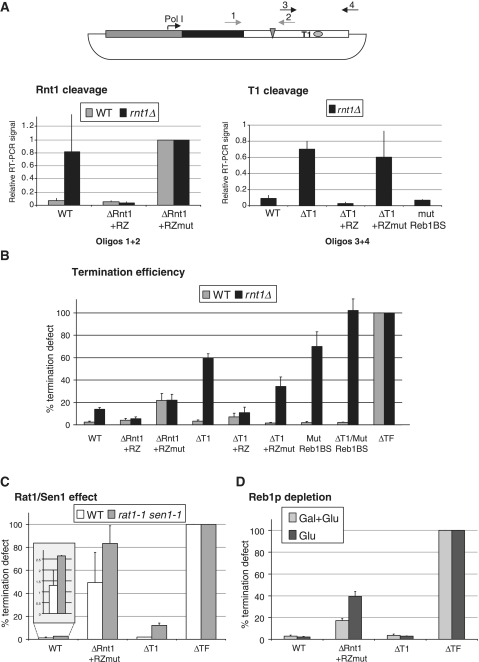
Co-transcriptional RNA cleavage and failsafe termination pathway in rnt1Δ cells. (A) Transcript cleavage analysis at the Rnt1 and T1 sites. cDNA was produced with a specific primer (B-rev, see Supplementary Table S2) downstream to the terminator fragment and subjected to PCR with oligos across Rnt1 cleavage site (1 + 2) or T1 element (3 + 4), as indicated. Left: RNA is cleaved by Rnt1 in WT but not rnt1Δ. Data are normalized to ΔRnt1 + RZmut (no cleavage). Right: signal across T1 is detectable in rnt1Δ. Cleavage is dependent on the presence of the T-rich element. The data are normalized towards the total amount of cDNA, detected downstream of the cleavage site. WT and mutant ribozyme provide positive and negative controls. An average of three independent experiments is shown, where error bars indicate SD. (B) Termination efficiency in different minigene mutants (outlined in Figure 3A) in WT and rnt1Δ cells. Termination was measured by RT–qPCR, and plotted relative to the construct lacking the whole terminator (100% termination defect). An average of three independent experiments is shown and error bars indicate SD. Raw data, not normalized toward ΔTF, are provided in Supplementary Figure S1. In WT (grey), termination is efficient so long as cleavage at the Rnt1 site is provided. In rnt1Δ (black), alternative cleavage at T1 and presence of an intact Reb1-binding site are crucial to terminate transcription efficiently. (C) Rat1 and Sen1 effect on termination in different minigene mutants. Termination defect plotted as in Figure 4B. rat1-1 sen1-1 shows higher than WT transcriptional read-through also in ΔRnt1 + RZmut where only T1 cleavage takes place. Data for the WT construct are magnified in the square above for better visualization. (D) Reb1 depletion effect on termination. pGAL-REB1 cells were grown in galactose + glucose or switched to glucose for approximately five generations to repress REB1 transcription. Termination was assessed as described for Figure 4B. Reb1 depletion causes a partial defect in ΔRnt1 + RZmut, where no Rnt1 cleavage takes place.
In Figure 3C an in vitro transcript was produced with T3 polymerase (Promega) and reverse transcribed with the same primer as the minigene-derived RNA. PCR was performed with the common reverse primer ‘A-rev’ and the forward primers ‘3C-1’, ‘3C-2’, ‘3C-3’ or ‘3C-4’. Values were corrected for PCR efficiency normalizing towards the results obtained with the T3 transcript.
PCR in Figure 4A was performed with the oligos ‘4A-1’, ‘4A-2’, ‘4A-3’ and ‘4A-4’.
Primer sequences are indicated in Supplementary Table S2.
RESULTS
3′-extended 25S rRNA is produced in rnt1Δ cells
We investigated the effect of Rnt1 depletion on Pol I termination and initially observed that rnt1Δ displays a slow growth phenotype but is still viable. We next mapped the 3′-ends of rRNA generated in the rnt1Δ strain versus isogenic wild-type (WT) using S1 protection analysis with a probe covering the 3′-end of 25S rRNA and the rDNA terminator region, including the previously described elements T1 and T2 (4) (Figure 1). In parallel we tested a strain lacking Rpa12, the small non-essential subunit of Pol I, previously shown to produce a significant defect in termination (6). As expected, S1 protection on total RNA from WT cells produced a strong band corresponding to the mature 25S rRNA; the same band is also visible in rpa12Δ. Unexpectedly, in rnt1Δ a higher band was visible, corresponding in size to transcripts ending at T1. We predict that this stable RNA is a 3′-extended form of 25S rRNA that must still be assembled into a functional though presumably suboptimal ribosome. In agreement with this observation, it has been shown that correct formation of 25S rRNA 3′-end is impaired in rnt1 mutant cells (9,15). With a longer exposure we also detected in rnt1Δ a minor fraction of transcripts ending at T2.
In a WT situation the 3′-end of 25S rRNA is generated by the exosome 3′–5′ exonuclease activity following initial cleavage by Rnt1 (9,16); it is apparent that the exosome does not detectably degrade the longer T1 transcript observed in rnt1Δ cells. We therefore sought to determine where exactly this new 3′-end maps and whether it is generated by RNA cleavage or termination.
Co-transcriptional RNA cleavage maps at the T-rich element of the Pol I terminator
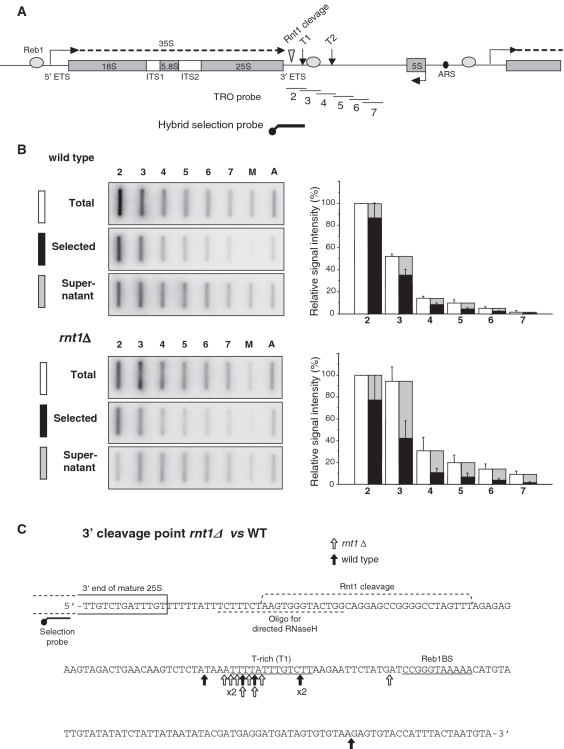
To test if the transcript RNA is cleaved at the 3′-ETS in rnt1Δ cells, we used hybrid selection transcription run-on (hsTRO) analysis (17). hsTRO monitors nascent transcripts and distinguishes whether they are continuous or have been cleaved downstream to the selection probe. We employed a probe spanning the Rnt1 cleavage site to monitor transcripts prior to Rnt1 cleavage (Figure 2A). In WT we obtained the expected TRO profile showing transcription termination beyond the Rnt1 cleavage site, with a strong signal over probe 2, followed by a significant drop over probes 3 and 4 and further reduction progressively downstream (Figure 2B). As previously described (6), the non-selected (Total) TRO profile for rnt1Δ is slightly altered with increased signal over probes 3–7, indicative of impaired termination. hsTRO analysis gave similar results in WT and rnt1Δ (Figure 2B). Thus full selection of the TRO signal was observed for probe 2 showing that the hybrid probe efficiently pulled down TRO labeled RNA. However for both strains a substantial fraction of the TRO signal was not selectable with probes 3–7, especially in rnt1Δ. This result indicates that nascent transcripts are still cleaved 3′ to the Rnt1 site, suggesting a secondary RNA cleavage process. Indeed in rnt1Δ, where no cleavage can occur at the Rnt1 hairpin sequence, all of the signal in the supernatant fraction results from this secondary cleavage process. Since TRO analysis detects nascent transcription this secondary cleavage must occur co-transcriptionally and is therefore likely to be related to transcription termination.
Figure 2.
Co-transcriptional RNA cleavage maps at the T1 element of the terminator. (A) Schematic of a rDNA repeat, as in Figure 1A, showing the position of the single stranded M13 DNA probes (2–7) used for the TRO assay (thin lines) and of the biotinylated probe used for transcripts hybrid selection (thick line). (B) Left: Hybrid Selection-TRO assay performed in WT and rnt1Δ. Transcription was performed in permeabilized cells in presence of α-32P-UTP; the extracted RNA was aliquoted in two parts and hybridized to the filter directly (Total) or after fractionation with streptavidin-coated beads and the biotinylated probe shown in A (Selected and Supernatant). M = M13 (negative control), A = Actin (positive control). Right: quantitation of the experiment shown on the left. Data normalized to probe 2 = 100%. Average of three independent experiments is shown, error bars indicate SD. (C) Hybrid Selection Circular-RACE detecting transcripts 3′-ends in WT and rnt1Δ. Hybrid selection followed by RNase H treatment selects the transcripts that extend beyond the Rnt1 cleavage site. The sequence of the rDNA terminator is shown. This includes the 3′-end of 25S rRNA sequence, the Rnt1 cleavage site region, the position of the probe used for hybrid selection and the oligo used for transcript release by RNase H treatment. Underlined are the T-rich element T1 and Reb1-binding site. Vertical arrows indicate the detected transcript 3′-ends in WT (black) and rnt1Δ (white) cells.
We next mapped the 3′-ends of selected transcripts by hybrid selection circular RACE (hscRACE) (14) (Figure 2C). Transcripts were selected with a biotinylated probe over the 25S rRNA then cleaved with RNase H using an oligo across the Rnt1 cleavage site to release 3′-transcripts. Both in WT and in rnt1Δ, hscRACE detected clusters of 3′-ends in the T1 region, just upstream to the Reb1-binding site (4). These products are generated by cleavage and not just by termination as a significant proportion of polymerases are still transcribing the downstream template (probes 4–7 in Figure 2B).
Overall we conclude that a RNA cleavage activity separate to Rnt1 recognizes and cleaves the nascent RNA specifically at T1 of the Pol I terminator, generating the 3′-end of the extended 25S rRNA observed in rnt1Δ cells (Figure 1).
Mutagenesis of the rDNA terminator in a Pol I minigene
We next investigated which Pol I terminator elements are required for this T1-associated cleavage to occur and aimed to establish their relative contribution to the termination process.
The study of rRNA transcription is complicated by the repetitive nature of the ribosomal DNA. In S. cerevisiae there are about 150 tandem rDNA repeats, ∼50% of which are silenced (18). We therefore designed a Pol I minigene (Figure 3A) which comprises the rDNA promoter plus upstream sequence [including the whole intergenic space (IGS) between two rDNA repeat] followed by a selection fragment (human β-globin sequence). This was inserted into a multicopy plasmid together with the rDNA terminator region containing the end of 25S rRNA and all the key downstream terminator elements (Rnt1 cleavage site, T1 and Reb1-binding site). The use of an exogenous sequence (human β-globin) allowed the plasmid-encoded transcripts to be distinguished from endogenous RNA. Mutagenesis of the terminator region allowed us to analyze the transcriptional effect of each terminator element (Figure 3A). As well as performing cis-mutagenesis on this Pol I minigene, we also transformed it into different genetic backgrounds to test the effect of different trans-acting factors.
We first established that the Pol I minigene is actually transcribed by Pol I. Total RNA from cells transformed with the Pol I minigene (plus or minus terminator) was subjected to primer extension using an antisense oligo annealing to the β-globin selection fragment (Figure 3B). We obtained a clear band corresponding to transcripts initiated at position +1 of the Pol I promoter and also observed reduced signal intensity with the ΔTF construct. We conclude that transcription initiated at the authentic Pol I transcription start site (TSS) and that impaired termination (see also Figure 4B) resulted in reduced signal likely caused by transcription interference. To exclude the presence of longer non-specific transcripts, possibly undetectable by primer extension, we confirmed the authentic TSS by RT–qPCR (Figure 3C). RNA from cells transformed with different constructs was reverse transcribed with an oligo inside the β-globin selection fragment and then PCR amplified using a common reverse primer and different forward primers producing progressively longer PCR products. Primers 1 and 2 anneal upstream and 3 and 4 downstream to the TSS. The PCR efficiency was normalized using a T3 transcript produced in vitro from the same template. A signal dependent on the presence of the Pol I minigene and initiating at the expected TSS was observed. Since oligo 3 anneals very close to the TSS, this may explain the lower RT–PCR signal detected here compared to using oligo 4. We also performed this analysis in a rat1-1 sen1-1 mutant strain and obtained a profile similar to the WT but quantitatively higher, due to transcript stabilization. Overall these data confirm the specificity and validity of the Pol I minigene system. However there are limits to its use due to low expression caused by competition for the Pol I machinery with the chromosomal rDNA repeats (19) and the instability of the transcripts. Even so RT–qPCR gave reproducible data.
Co-transcriptional RNA cleavage is dependent on the presence of the T-rich element
In Figure 2, using rnt1Δ cells, we have identified a second RNA cleavage site mapping at the terminator T-rich element T1. We therefore employed our Pol I minigene system to define the sequence requirements for T1 cleavage. We prepared constructs in which the Rnt1 cleavage site is substituted with a hammerhead ribozyme (20) (ΔRnt1 + RZ) or its mutant inactive form (ΔRnt1 + RZmut). Similarly, to study the co-transcriptional cleavage at T1, we either deleted the T-rich tract (ΔT1) or substituted it with the WT or mutant ribozyme (ΔT1 + RZ, ΔT1 + RZmut). Finally, we addressed the role of Reb1 by introducing two point mutations in its binding site known to abolish Reb1 binding (3) (Figure 3A).
RNA isolated from these minigene-transformed strains was assessed for RNA cleavage by RT–qPCR with oligo pairs across the Rnt1 and T-rich cleavage sites (see scheme in Figure 4A) in both WT and rnt1Δ cells. As expected cleavage by Rnt1 occurred in WT but not rnt1Δ (Figure 4A, left panel). Also the constructs with ribozyme or mutant ribozyme replacing Rnt1 cleavage gave expected results. We next investigated T1 co-transcriptional cleavage in rnt1Δ cells using the same RT–qPCR assay. As RT was primed with oligo 4, annealing downstream to the cleavage site, we excluded from the analysis any potential transcript terminated at T1. As shown (Figure 4A, right panel), RNA was cleaved at T1 when employing a WT template. However RNA cleavage was lost when T1 was deleted but not affected by mutation in the Reb1-binding site (compare WT, ΔT1 and mutReb1BS). Again WT and mutant ribozyme were used as controls. In a WT strain we failed to obtain significant PCR signal (data not shown), probably because when Rnt1 cleavage occurs the downstream transcripts are rapidly degraded by Rat1. It should be noted the distance between the two cleavage sites is very short, ∼35 nt.
Overall we conclude that T1 is the cis-acting element dictating co-transcriptional cleavage at the Pol I terminator as defined by our hsTRO analysis (Figure 2). We propose that T1 generates an alternative entry site for Rat1 so that when Rnt1 is missing it provides a failsafe pathway for Pol I transcription termination.
Defining the relative contribution of Pol I terminator sequences
To finally establish the role of RNA cleavage and of Reb1 binding on Pol I termination, we measured transcription termination efficiency in the different terminator mutants, outlined in Figure 3A, transformed into WT or rnt1Δ cells. To do so we employed RT–qPCR, using RT oligos that specifically select for plasmid-encoded transcripts, and measured the amount of transcript upstream or downstream to the termination fragment (over region A and B, see scheme in Figure 3A). We then determined termination efficiency as a ratio B/A.
In the WT strain (Figure 4B, gray bars) we observe that deletion of individual cis-acting elements does not significantly impair termination: neither cleavage at T1 nor mutagenesis of Reb1-binding site produced a significant termination defect (cfr. ΔT1, ΔT1 + RZ, ΔT1 + RZmut, mutReb1BS). Also the double mutation ΔT1/mutReb1BS did not affect termination in presence of Rnt1. It should be noted that while Reb1 binding to the terminator and its role in polymerase pausing have been extensively studied (1,3,4), ChIP analysis did not detect binding of Reb1 at the Pol I terminator in vivo (8). The only situation where termination was significantly affected in WT cells occurred for the ΔRnt1 + RZmut construct, where ∼20% of the polymerases produced read-through transcripts. This implies transcript cleavage by Rnt1 or by another activity like the ribozyme at the same location is critical to achieve efficient termination, providing an entry site for Rat1. Note that ribozyme cleavage was nearly as effective as cleavage by Rnt1 in promoting termination (compare WT with ΔRnt1 + RZ), despite the fact that different 5′-ends are produced in the downstream cleavage product. While cleavage by Rnt1 produces a 5′-phosphate end, that is a good substrate for the exonuclease Rat1 (21), the ribozyme produces a 5′-OH. We speculate that in this context a RNA kinase activity may be involved. To exclude an effect on our assay of transcript stability following RNA cleavage, we employed a strain lacking Rrp6, a nuclear exosome component. This resulted in a general stabilization of the transcripts but did not significantly affect termination (data not shown).
We obtained a different profile in rnt1Δ cells (black bars). While a small termination defect, consistent with the run-on data in Figure 2B, was detectable with a WT terminator, when the T-rich tract was deleted (ΔT1) a strong termination defect was observed, with ∼60% transcription read-through. This result points towards a key role in Pol I termination for T1 cleavage. Similarly, as observed for the Rnt1 cleavage site, replacement of T1 with the ribozyme restored efficient termination, while lack of cleavage as in ΔT1 + RZmut impaired termination. Significantly, an intact Reb1-binding site also proved crucial for efficient termination in rnt1Δ cells, since the Reb1 mutant construct strongly affected Pol I termination (mutReb1BS). This result is in agreement with the earlier model for Pol I termination involving polymerase pausing by Reb1 binding to the DNA template (1). Finally, the double mutant ΔT1/mutReb1BS completely failed to terminate, as expected since no cleavage or pausing elements remain.
These data point towards the presence of two distinct pathways for Pol I termination: a main one, dependent on RNA cleavage by Rnt1, and a ‘failsafe’ one, observed in the absence of Rnt1, dependent on both cleavage at T1 and presence of an intact Reb1-binding site at the terminator.
Rat1/Sen1 and Reb1 activities are involved in the Rnt1-independent pathway
As RNA cleavage at T1 occurs co-transcriptionally, we propose that this cleavage step provides an alternative entry site for Rat1 in the absence of Rnt1. To verify this hypothesis we tested our minigene constructs in a rat1-1 sen1-1 strain, as both the exonuclease Rat1 and the helicase Sen1 are implicated in Pol I termination via a torpedo mechanism (8). As shown in Figure 4C, in rat1-1 sen1-1 higher transcriptional read-through was detectable with a WT terminator, albeit termination remains efficient, confirming previous observations (8). The other mutants analyzed similarly gave higher read-through levels (increased termination defect) in the rat1-1 sen1-1 strain. Significantly, rat1-1 sen1-1 produced a transcriptional defect with ΔRnt1 + RZmut, where no cleavage takes place at the Rnt1 position and the only RNA cleavage site is T1. We conclude Rat1/Sen1 are involved in the Rnt1-independent termination pathway, most probably as T1 provides them with an alternative entry site.
We next considered the second factor potentially affecting this termination pathway, as predicted by mutation of the Reb1-binding site. In order to verify whether Reb1, and not just its binding site, influences termination efficiency, we employed a strain where REB1 is under the control of a GAL promoter (22). After glucose shift Reb1 mRNA levels dropped to ∼30% (data not shown). Even though deletion of RNT1 in this strain was lethal, we could anyway mimic Rnt1 absence on our minigene by analysis of the ΔRnt1 + RZmut construct and so measure termination efficiency following Reb1 depletion (Figure 4D). Confirming our previous observations, Reb1 did not significantly affect termination when cleavage by Rnt1 takes place (WT, ΔT1), while it did affect ΔRnt1 + RZmut, with a ∼2-fold increase in read-through transcripts. The smaller effect of Reb1 depletion with this construct as compared to mutation of the Reb1-binding site (cfr. Figure 4B, mutReb1BS), is possibly due to incomplete depletion of Reb1. Alternatively, these data could suggest some redundancy of Reb1 with other activities with similar DNA-binding specificity.
Overall our results provide a more complete picture of the Pol I termination mechanism and may reconcile previous in vitro results with recent in vivo studies. In a normal situation, transcription termination is coupled with rRNA processing: the primary transcript is cleaved by Rnt1 in the 3′-ETS and polymerase displacement from the template is mediated by the action of Rat1/Sen1; none of the other terminator elements (T1 or Reb1-binding site) significantly influences this process. However, when Rnt1 activity is missing, a failsafe termination pathway acts to promote Pol I termination. This involves co-transcriptional RNA cleavage at T1 and the presence of an intact Reb1-binding site. Rat1 and Sen1 involvement in this Rnt1-independent pathway suggests that T1 cleavage provides an alternative entry site for these activities to ‘torpedo’ Pol I; Reb1 is likely to be involved in this process even though its depletion causes only a partial termination defect.
DISCUSSION
We provide a unifying model for termination of transcription by Pol I and identify a new co-transcriptional cleavage activity associated with the rDNA terminator transcript (Figure 5). We have mapped this cleavage sequence to T1 in the 3′-ETS and demonstrate that it provides an alternative pathway for termination in the absence of Rnt1. Rat1 and Sen1 activities are involved in this pathway, suggesting that this secondary cleavage is an alternative or ‘failsafe’ entry site for the torpedo Rat1, to promote Pol I termination. We cannot determine whether this cleavage activity works together with or only independently of Rnt1. In WT cells it is not detectable since Rnt1 cleavage provides an entry site for rapid degradation of the downstream transcript by the exonuclease Rat1. In any case T1 co-transcriptional cleavage appears to be dispensable for termination in a WT situation where the transcript is cut at the upstream position. We have also shown that RNA cleavage at T1 generates an upstream 3′-end that is not a substrate for normal exosome processing, resulting in an extended 25S rRNA (Figure 1). The transcript ends detectable at T1 could arise from either RNA cleavage or from Pol I termination. However the hsTRO and Pol I minigene cleavage assays imply a cleavage process as they both detect read-through transcripts downstream of T1, arguing that Pol I is still engaged with the template. On the other hand the presence of an intact Reb1-binding site is required for efficient termination but does not seem to influence RNA cleavage in the rnt1Δ strain. This supports a combined termination mechanism, involving ‘torpedo’ and ‘pausing/release’. Early studies that suggested the pausing/release mechanism for Pol I termination (1) were conducted in vitro and therefore missed the connection between transcription and rRNA processing, in particular RNA cleavage by Rnt1. Here we show a clear defect in termination when we mutate the Reb1-binding site in rnt1Δ and partially reproduce the defect by depletion of Reb1. It is therefore puzzling that ChIP analysis fails to detect the presence of Reb1 over the Pol I terminator (8). We also performed Reb1 ChIP in rnt1Δ cells and obtained the same negative results (data not shown). A possible explanation for these results is the involvement of an additional DNA-binding protein with sequence specificity similar to Reb1. We propose that polymerase pausing by Reb1 (or an alternative DNA-binding protein) works in concert with co-transcriptional cleavage at T1, thereby enhancing an otherwise inefficient or kinetically slow event.
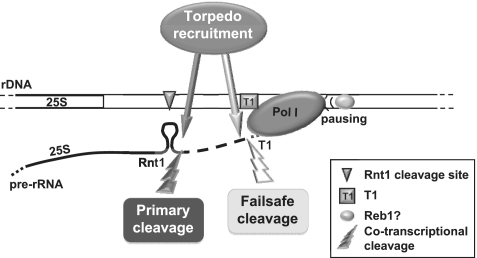
Figure 5.
Model. Two co-transcriptional cleavage events take place at the Pol I terminator. Cleavage of nascent RNA by Rnt1 provides the initial entry site for the ‘torpedo’ Rat1 to promote Pol I transcription termination. In the absence of Rnt1, the downstream transcript is stabilized and an additional cleavage event occurs at T1, providing an alternative entry site for Rat1. In this situation the specific interaction of Reb1 (or alternative DNA-binding protein) with the Reb1-binding site is required to pause the polymerase and promote transcription termination.
The activity responsible for co-transcriptional RNA cleavage at T1 is still unknown. However it should be noted that in the early in vitro termination studies (1,23) pure Pol I and Reb1 were often employed, indicating that T1 co-transcriptional cleavage may be an intrinsic Pol I activity. A possible candidate is Rpa12, the small subunit of Pol I shown to possess RNA cleavage activity in a backtracked elongation complex (24) and to cause a termination defect (6). However, deletion of RPA12 had no detectable effect on T1 cleavage (data not shown). Also Dis3, the core exosome subunit shown to possess both endo- and exo-nuclease activity (25), is a possible candidate. Finally the NRD complex, involved in sn/snoRNA and other short Pol II transcripts termination [reviewed in (26)], is a potential candidate. However mutation of each of these activities failed to show any involvement in Pol I co-transcriptional cleavage using our Pol I minigene system (data not shown).
The identification of this new RNA cleavage site at the Pol I terminator adds a further parallel with the Pol II system. As well as the established torpedo mechanism (7,8,17,27), Pol I and Pol II termination rely on the presence of two distinct sites for transcript cleavage: for Pol I Rnt1 and T1 while for Pol II poly(A) site and CoTC cleavage sites (17). For Pol II the distance between poly(A) and CoTC sequences can be thousands of nucleotides apart and the process of termination is coupled with polyadenylation; in the rDNA terminator a simpler system may exist where the same elements are concentrated in a much shorter space. Understanding parallels between Pol I and Pol II termination mechanism may provide interesting insight into how the process of transcription termination has evolved in different eukaryotes and between different RNA polymerases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust grant (to N.J.P.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank J.R. Warner for the pGAL-REB1 yeast strain. The authors are grateful to members of the NJP lab for insightful discussion.
REFERENCES
- 1.Lang WH, Morrow BE, Ju Q, Warner JR, Reeder RH. A model for transcription termination by RNA polymerase I. Cell. 1994;79:527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 2.Grummt I, Rosenbauer H, Niedermeyer I, Maier U, Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986;45:837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- 3.Lang WH, Reeder RH. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol. Cell Biol. 1993;13:649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeder RH, Guevara P, Roan JG. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell Biol. 1999;19:7369–7376. doi: 10.1128/mcb.19.11.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansa P, Grummt I. Mechanism of transcription termination: PTRF interacts with the largest subunit of RNA polymerase I and dissociates paused transcription complexes from yeast and mouse. Mol. Gen. Genet. 1999;262:508–514. doi: 10.1007/s004380051112. [DOI] [PubMed] [Google Scholar]
- 6.Prescott EM, Osheim YN, Jones HS, Alen CM, Roan JG, Reeder RH, Beyer AL, Proudfoot NJ. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl Acad. Sci. USA. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allmang C, Tollervey D. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 1998;278:67–78. doi: 10.1006/jmbi.1998.1693. [DOI] [PubMed] [Google Scholar]
- 11.Henras AK, Bertrand E, Chanfreau G. A cotranscriptional model for 3′-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor. RNA. 2004;10:1572–1585. doi: 10.1261/rna.7750804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashe MP, Griffin P, James W, Proudfoot NJ. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 13.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 14.West S, Gromak N, Norbury CJ, Proudfoot NJ. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell. 2006;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Elela SA, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 16.Zanchin NI, Goldfarb DS. The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 1999;27:1283–1288. doi: 10.1093/nar/27.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West S, Gromak N, Proudfoot NJ. Human 5′ –> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 18.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wai H, Johzuka K, Vu L, Eliason K, Kobayashi T, Horiuchi T, Nomura M. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell Biol. 2001;21:5541–5553. doi: 10.1128/MCB.21.16.5541-5553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samarsky DA, Ferbeyre G, Bertrand E, Singer RH, Cedergren R, Fournier MJ. A small nucleolar RNA: ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl Acad. Sci. USA. 1999;96:6609–6614. doi: 10.1073/pnas.96.12.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens A, Poole TL. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 22.Morrow BE, Ju Q, Warner JR. A bipartite DNA-binding domain in yeast Reb1p. Mol. Cell Biol. 1993;13:1173–1182. doi: 10.1128/mcb.13.2.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong SW, Lang WH, Reeder RH. The release element of the yeast polymerase I transcription terminator can function independently of Reb1p. Mol. Cell Biol. 1995;15:5929–5936. doi: 10.1128/mcb.15.11.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 25.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 26.Rondon AG, Mischo HE, Proudfoot NJ. Terminating transcription in yeast: whether to be a ‘nerd' or a ‘rat'. Nat. Struct. Mol. Biol. 2008;15:775–776. doi: 10.1038/nsmb0808-775. [DOI] [PubMed] [Google Scholar]
- 27.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.