Abstract
Nearly all cellular and disease related functions of the transcriptional co-activator lens epithelium-derived growth factor (LEDGF/p75) involve tethering of interaction partners to chromatin via its conserved integrase binding domain (IBD), but little is known about the mechanism of in vivo chromatin binding and tethering. In this work we studied LEDGF/p75 in real-time in living HeLa cells combining different quantitative fluorescence techniques: spot fluorescence recovery after photobleaching (sFRAP) and half-nucleus fluorescence recovery after photobleaching (hnFRAP), continuous photobleaching, fluorescence correlation spectroscopy (FCS) and an improved FCS method to study diffusion dependence of chromatin binding, tunable focus FCS. LEDGF/p75 moves about in nuclei of living cells in a chromatin hopping/scanning mode typical for transcription factors. The PWWP domain of LEDGF/p75 is necessary, but not sufficient for in vivo chromatin binding. After interaction with HIV-1 integrase via its IBD, a general protein–protein interaction motif, kinetics of LEDGF/p75 shift to 75-fold larger affinity for chromatin. The PWWP is crucial for locking the complex on chromatin. We propose a scan-and-lock model for LEDGF/p75, unifying paradoxical notions of transcriptional co-activation and lentiviral integration targeting.
INTRODUCTION
Gene expression is regulated by transcriptional cofactors that fine-tune the interaction of the general transcription machinery with gene-specific transcription factors. Lens epithelium-derived growth factor (LEDGF/p75) was originally identified as a 75-kDa transcriptional co-activator interacting with the VP16 activation domain and with components of the general transcription machinery (1). LEDGF/p75 (530 amino acids) shares the first 325 amino acids with p52, an alternative splice variant from the same PSIP1 gene (1,2) (Figure 1A) and plays an important role in cell survival (3), oncogenesis (4–7), autoimmunity (8,9) and integration and replication of the human immunodeficiency virus type 1 (HIV-1) (10,11).
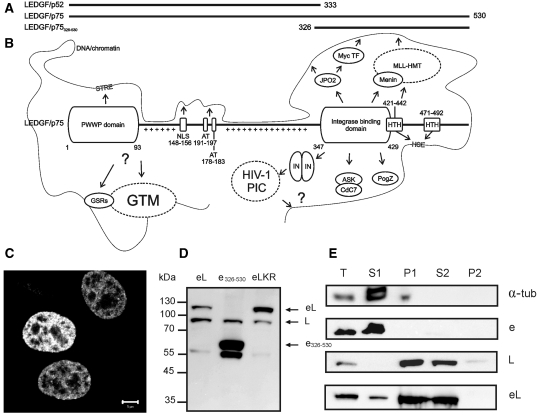
Figure 1.
The primary structure and interactome of LEDGF/p75. (A) Schematic representation of LEDGF/p75, its alternative splice variant LEDGF/p52 (13) and a deletion mutant, LEDGF/p75326–530 (61). (B) Primary structure and interactome of LEDGF/p75. Important domains of LEDGF/p75, interacting proteins or DNA-sequences are indicated. NLS, nuclear localization signal; AT, AT-hook domains; HTH, predicted Helix-Turn-Helix motifs; GSRs, gene specific regulators; GTM, general transcription machinery; STRE, stress-related regulatory element; HIV-1 PIC, pre-integration complex; IN, HIV-1 integrase; Plusses, positively charged regions in the p52 part of LEDGF/p75. (C) Confocal fluorescence image of HeLa cells expressing eGFP-LEDGF/p75. Scale bar = 5 µm. (D) Western blot with an anti-LEDGF/p75 antibody of HeLa cells transiently expressing eGFP-fusions. eL, eGFP-LEDGF/p75; e326-530, eGFP-LEDGF/p75326-530; eLKR, K56D-R74D; L, endogenous LEDGF/p75. (E) Cellular fractionation assay of HeLa cells transiently expressing eGFP-fusions. Western blot of different fractions is shown using antibodies to indicated proteins. T, total cell lysate; S1, Triton-soluble cellular fraction; P1, Triton-insoluble cellular fraction; S2, DNase/(NH4)2SO4-soluble cellular fraction; P2, DNase/(NH4)2SO4-insoluble cellular fraction.
LEDGF/p75 has an extensive interactome (Figure 1B). The protein contains multiple DNA/chromatin binding domains and a conserved protein–protein interaction domain. The N-terminal PWWP domain of LEDGF/p75 (amino acids 1–93) holds a conserved (though not invariant) Pro-Trp-Trp-Pro motif and belongs to the Tudor domain ‘Royal Family’ of protein domains regulating the chromatin function (12,13). This domain is generally involved in chromatin structure regulation through protein–protein interactions (14). A tripartite element in LEDGF/p75 consisting of the two AT-hooks (amino acids 191–197 and amino acids 178–183) and the nuclear localization signal (NLS) (amino acids 148–156) cooperates with the PWWP domain for interaction with DNA/chromatin, as has been shown in vitro (15) and in vivo (16). LEDGF/p75 allegedly interacts with heat shock elements (HSE) through its helix-turn-helix (HTH) motifs (amino acids 421–442 and amino acids 471–492) and with stress-related regulatory elements (STRE) through the PWWP domain to specifically promote expression of stress-related genes (17–21). LEDGF/p75 contains a high percentage of charged residues (39.4%) and contains two defined regions with positively charged residues (amino acids 94–142 and amino acids 208–325), that are involved in electrostatic interactions with DNA/chromatin (15). A conserved superhelical domain in LEDGF/p75 was originally identified as the domain for binding to HIV-1 integrase (IN), the enzyme that catalyzes the integration of HIV in the genome of an infected cell (22,23). By virtue of this interaction LEDGF/p75 biases lentiviral integration towards actively transcribed regions in the genome (24–26). The ‘integrase binding domain’ (IBD) of LEDGF/p75 (amino acids 347–429) shows structural homology to known protein–protein interaction motifs (27) and has been shown to interact with multiple other proteins in cells: (i) JPO2, a Myc transcription factor interacting protein (28), (ii) the menin tumor suppressor, implicated in cancer and transcriptional regulation as a component of the MLL-HMT (mixed lineage leukemia histone methyltransferase) complex (7), (iii) PogZ, pogo transposable element derived protein with a Zinc finger, a domesticated transposase (29) and (iv) heterodimeric Cdc7/ASK (30). Cdc7 is a Ser/Thr kinase essential for the initiation of DNA replication throughout the S-phase and its activity is controlled via interaction with a regulatory subunit, activator of S-phase kinase (ASK).
In this work we study the intracellular mobility of LEDGF/p75, in particular chromatin interactions, quantitatively and in vivo with a combination of fluorescence techniques. We label LEDGF/p75 with eGFP and perform a detailed characterization of the hybrid protein. We provide new insight in the transcription factor chromatin binding kinetics in vivo. We investigate interaction partner mediated chromatin tethering of LEDGF/p75 and demonstrate the role of the PWWP domain for this.
MATERIALS AND METHODS
Cell lines
HeLa cells were obtained from the NIH Reagent program and were grown in ‘Complete Medium’, high-glucose Dulbecco's Modified Eagle Medium (Gibco BRL, Belgium), supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich gmbh, Taufkirchen, Germany) and 50 μg/ml Gentamycin (Gibco BRL, Belgium), at 5% CO2 and 37°C in a humidified atmosphere. The generation of monoclonal HeLa cells stably suppressing endogenous LEDGF/p75 (HeLa-p75KD) has been described earlier (31). An average HeLa cell nuclei volume 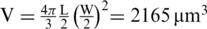 was calculated by measuring the length (L) and width (W) of seven nuclei by optical transmission microscopy and assuming an ellipsoidal nuclear shape. Assuming 10–100 000 genes per cell, this roughly corresponds to a gene concentration of ∼5–100 nM.
was calculated by measuring the length (L) and width (W) of seven nuclei by optical transmission microscopy and assuming an ellipsoidal nuclear shape. Assuming 10–100 000 genes per cell, this roughly corresponds to a gene concentration of ∼5–100 nM.
Western blotting, cellular fractionation assays
Western blotting was performed on whole cell lysates as described before (32) with a specific anti-LEDGF/p75 antibody (A300-847A, Bethyl Laboratories, Montgomery, TX, USA). Cellular fractionation assays were performed as described earlier (15), with the specific LEDGF/p75 antibody, an anti-RFP antibody for mRFP-IN (AB3216, Millipore N.V., Brussels, Belgium), an in-house polyclonal anti-GFP antibody and anti-α-tubulin (T5168, Sigma-Aldrich, Bornem, Belgium). Briefly, 24 h post-transfection cells were harvested and either lysed for checking overall expression (T) or extracted with Triton X-100 to separate the soluble (S1) and insoluble (P1) cellular proteins. DNaseI-treatment allowed subsequent separation into a soluble, chromatin binding fraction (S2) and an insoluble, non-chromatin binding fraction (P2).
Transfections
For the transient transfections 0.5–1 × 105 cells were seeded per well in a Lab-TekTM Chambered Coverglass (VWR International, Leuven, Belgium) to obtain 50–70% cell confluency after overnight incubation. Transfections were performed with a Mirus TransIT®-HeLaMONSTER® transfection kit (VWR International, Leuven, Belgium), with 1 μl TransIT reagent, a maximum of 0.5 μg plasmid DNA per well and 1 μl MONSTER reagent per μg of DNA. After the transfection mixture was prepared in 50 μl fresh OptiMEM per well (Gibco BRL, Belgium), 450 μl prewarmed Complete Medium was added and this mixture was slowly added to the cells and the cells were incubated for at least 12 h.
Theory: quantifying interactions with immobile structures
Consider a binding equilibrium of a protein L that can interact with an immobile binding site S on the chromatin:
| (1) |
| (2) |
| (3) |
| (4) |
with [L] the concentration of free protein L, in this case, LEDGF/p75, [S] the concentration of free immobile binding sites, [LS] the concentration of protein L bound to a binding site, Kass the association binding constant, kon the rate constant for association and koff the rate constant for dissociation. When the concentration of binding sites is high, k*on = konS0 is the pseudo first order rate constant for association. The tfree is the average time a protein spends between two successive binding events. The tbound is the residence time on the chromatin binding site. Intracellular binding reactions with immobile structures (nuclear envelope, chromatin, membrane) can be studied with fluorescence recovery after photobleaching (FRAP)(33), continuous photobleaching (CP) (34,35) and fluorescence correlation spectroscopy (FCS) (36). Generally, four dynamic regimes can be thought of:
Fast association (tfree ≤ τdiff,free) and slow dissociation (tbound > τdiff,free)
The protein binds with high affinity. It will be photobleached in the laser focus when FCS is performed. In this case, FRAP is a good method to analyse the binding kinetics: the dynamics will be slow and a model for reaction and diffusion is necessary because of the high probability of association (33).
Slow association (tfree > τdiff,free) and very slow dissociation (tbound > 100 ms)
The chance of binding is low but once bound the fluorochrome will be photobleached when measured with FCS. The fraction of the signal (from CP or FCS) that photobleaches is a measure for the K*ass = Kass[sites] (36):
| (5) |
When Equation 1 is solved to [LS] in function of total concentrations L0 and S0, K*ass (Kdiss) and the concentration of binding sites can be determined by non-linear least-squares fitting of the percentage photobleaching versus the total concentration of protein L0:
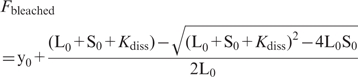 |
(6) |
The total concentration L0 can be measured with FCS. The parameter y0 is a constant accounting for the constant fraction of photobleaching at high concentrations. Alternatively, diffusion independent FRAP can be used to study the binding reaction (33).
Fast association (tfree < τdiff,free) and fast dissociation (tbound < τdiff,free)
The binding reaction is observed as a diffusion component in FRAP and FCS. The ‘effective’ slow diffusion coefficient defines the affinity (33,36,37):
| (7) |
In this regime, only the ratio of rate constants can be determined from the ACF. The diffusion coefficient of free cellular LEDGF/p75 was calculated with (assuming globular proteins):
| (8) |
Mr being the relative molecular mass and DeGFP = 33.0 µm2/s (as measured with FCS).
Slow association (tfree >> τdiff), moderate dissociation (τdiff,free < tbound < 100 ms)
The binding reaction is observed as an exponential component in FRAP and FCS (33,36). The fraction of free and ‘apparent slow’ diffusion components in FCS is a measure for the affinity:
| (9) |
In conclusion, in the first two regimes the binding reaction is not and in the last two regimes the binding reaction is observed during an FCS experiment.
Spot fluorescence recovery after photobleaching and half-nuclear fluorescence recovery after photobleaching
Diffusion rates and binding reactions of cellular proteins with slow/immobile cellular structures can be quantified with spot fluorescence recovery after photobleaching (sFRAP) (33,38). By illuminating a defined region-of-interest in the sample with a brief high-intensity laser pulse, the fluorescence is rapidly photobleached. It will recover due to the exchange of bleached with unbleached molecules. FRAP measurements were performed on a LSM510/ConfoCor2 (Carl Zeiss, Jena, Germany) at 488-nm excitation. Since bleaching-while-acquisitioning (39) and recovery-while-bleaching (40) influence the shape of the FRAP curve, careful optimization of laser power and imaging parameters was necessary.
sFRAP measurements were performed in a 1-µm radius (ω) circular spot. For eGFP and eGFP-LEDGF/p75, acquisitioning was every 65 ms at 5 µW excitation power (front objective aperture) and bleaching was performed with a single 65-ms scan at 1.6 mW. For eGFP-LEDGF/p75 in the presence of mRFP-IN, acquisitioning was every 5 s and bleaching was performed with a single 170-ms scan. Spot FRAP data represent averages of at least 10 cells. Analysis was performed with a standard diffusion model (38), a reaction-dominant model (33) or a full diffusion-reaction model (33), as described in detail in the Supplementary Information.
Half-nuclear FRAP (hnFRAP) measurements (41,42) were performed by photobleaching half of the nucleus with a single scan (1.4 s, 1.6 mW) and monitoring the subsequent fluorescence redistribution (every 1.4 s, 5 µW). Average nuclear pixel intensities per horizontal image line were calculated for each image with an ImageJ plug-in. Next, each post-bleach data point was normalized by the total average cell intensity, to account for whole-cell photobleaching. Next, each normalized data point was normalized by the corresponding pre-bleach data point, to account for cellular fluorescence inhomogeneities. Finally, vertical pixel distance was converted to an absolute distance scale (1 pixel = 90 nm) and data were plotted in a graph and fitted with (43):
| (10) |
with Fluonorm(x) the normalized fluorescence intensity as a function of the position x (µm), Fbleach the bleach depth, erf an error function describing the shape of the bleach profile, with b (µm) the position of the bleach border and σ(t) the displacement of the proteins as a function of time t (s). HnFRAP data for a single cell were fitted globally with Fbleach as a global fit parameter. From the slope of the mean squared displacement σ2(t) the diffusion coefficient D (µm2/s) was calculated (44):
| (11) |
Time zero was approximated with the moment during acquisition where half a bleaching iteration had been performed. HnFRAP data represent averages of at least five cells.
FCS and fluorescence cross-correlation spectroscopy
With FCS, diffusion and binding reactions of fluorescently labeled molecules can be studied in solution and in cells (45,46). On a confocal microscope the fluorescence in the sub-micrometer (<10−15 l) sized confocal volume is measured and then autocorrelated. The low time resolution decaying fluorescence trace contains information about diffusion and binding reactions, the high time resolution autocorrelation function (ACF) in addition provides the absolute protein concentration. For normal FCS/[fluorescence cross-correlation spectroscopy (FCCS)] measurements a commercial FCS/FCCS microscope (LSM510/ConfoCor2, Carl Zeiss, Jena, Germany) was used. Enhanced GFP (47) was excited at 488 nm (3 µW) and mRFP1 (48) was excited at 543 nm (9 µW). The excitation light was reflected by a dichroic mirror (HFT 488/543) and focused through a C-Apochromat 40×/1.2 W Korr/0.13–0.17 objective. The fluorescence emission light was split by a second dichroic mirror (NFT 570) into two separate beam paths and passed through a 505–530 nm bandpass filter and 70-µm pinhole for eGFP fluorescence and a 600–650 nm bandpass filter and 78-µm pinhole for mRFP1 fluorescence. The radial radius of the confocal volume at 1/e2 times its maximal intensity, ω1, was determined by measuring the τdiff of rhodamine 6G in water (D = 426 µm2/s)(49). In each cell cytoplasm or nucleus, 10 consecutive fluorescence intensity (detected photons per second) and correlation measurements of 20 s were performed. Correlation functions were fitted with a one- or two-component model, a normal or anomalous diffusion model (50), a reaction-dominant model (36) or a confined diffusion model (43), as described in detail in the Supplementary Information.
CP
CP traces were also recorded on the FCS set-up. Individual CP measurements were normalized to the first datapoint, averaged and fitted either to a biexponential decay (51) or to a model for a binding reaction with medium dissociation rate (35), as described in detail in the Supplementary Information.
Tunable focus FCS
Tunable focus FCS (TFFCS) was performed on a homebuilt set-up (52). The 30-mW 488-nm line of a Ar–Kr laser (Newport Spectra-Physics, Utrecht, The Netherlands) was split 50/50. One beam was maximally focused at the back-aperture of the objective (Olympus UPLFLN 100×/NA1.3/Oil, Olympus Belgium N.V., Aartselaar, Belgium) to create wide-field excitation for observing the fluorescence of the cells through the eyepiece of the microscope (Olympus IX). The other beam was carefully expanded, collimated and directed centrally through an adjustable diaphragm, to allow for TFFCS measurements. A fixed 50-µm pinhole was used for confocal detection on an avalanche photodiode (SPCM-AQR-15, Perkin-Elmer, Wiesbaden, Germany). The excitation power at each diaphragm setting was attenuated to give a constant power/area in the focal spot suitable for cellular FCS, avoiding potential intensity-dependent artefacts. Reproducible switching was achieved by measuring the residual power behind the diaphragm. TFFCS measurements were analysed with a 2-component model for normal diffusion.
Structure based selection of PWWP residues important for DNA binding
To select residues important for the interaction of the PWWP domain of LEDGF/p75 with DNA we used biomolecular modeling. First, we modeled the structure of the PWWP domain of LEDGF/p75 using the latest available version of Modeller (53), with the NMR structure (PDB 2B8A) of the HDGF-PWWP as a template (54). Second, we predicted putative DNA binding residues using the HotPatch algorithm (55). This analysis indicated residues K56 and R74 to be solvent exposed and not required for the stabilization of the tertiary structure of the protein, minimizing the chance of perturbing the PWWP folding. Of concern, these residues in the LEDGF/p75-PWWP are conserved in the HDGF-PWWP sequence and were already shown to be important for DNA binding of the HDGF-PWWP (54).
RESULTS
Fluorescent protein labeling of LEDGF/p75 preserves its function
To study the dynamics of LEDGF/p75 in living HeLa cells with fluorescence techniques, we first expressed and characterized the fusion of LEDGF/p75 and the enhanced green fluorescent protein (eGFP). This protein, eGFP-LEDGF/p75, displayed a heterogeneous nuclear distribution characteristic of endogenous LEDGF/p75 (Figure 1C) (22). Western blotting revealed no protein degradation of eGFP-LEDGF/p75 (Figure 1D). Chromatin binding properties of the fusion were similar to that of endogenous LEDGF/p75 as demonstrated by in vitro cell fractionation (Figure 1E): following extraction of the cells with Triton X-100, eGFP-LEDGF/p75 was present in the insoluble pellet (P1), whereas a DNaseI treatment of the P1 pellet released the protein into the supernatant (S2), as demonstrated earlier for endogenous LEDGF/p75 (15). In summary, an N-terminal fluorescent tag does not affect the functional properties of LEDGF/p75 analyzed in this study. In order to minimize the effects of endogenously expressed LEDGF/p75, we made use of HeLa cells stably depleted of endogenous LEDGF/p75 (HeLa-p75KD) (31). Complementation of these cells with wild-type LEDGF/p75 rescues HIV replication and lentiviral vector transduction (31). Likewise, we complemented LEDGF-depleted cells with eGFP-LEDGF/p75 and rescued viral replication and vector transduction to wild-type levels (data not shown), demonstrating that the eGFP label does not affect LEDGF/p75 functionality.
In vivo eGFP-LEDGF/p75 exists in a chromatin bound state, a slow state and a fast state
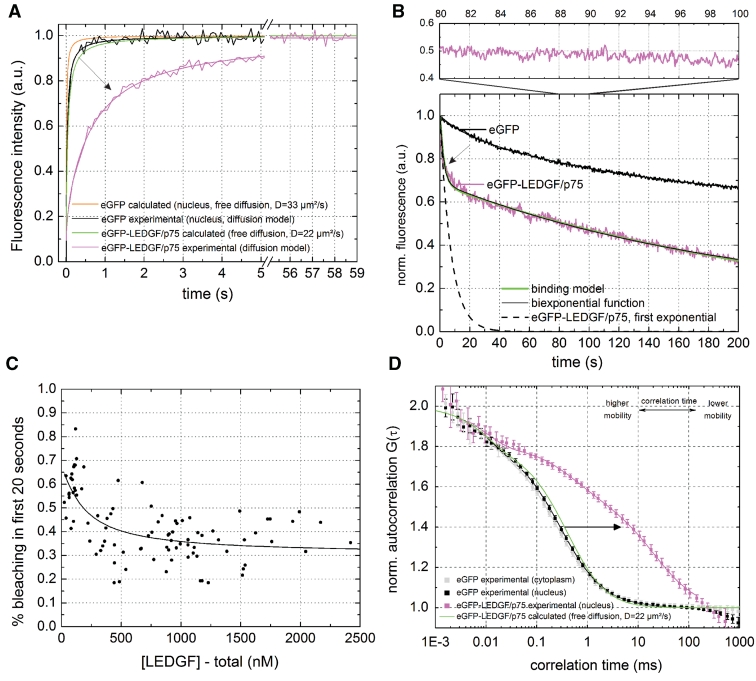
We performed sFRAP experiments in HeLa cells. The fluorescence of the eGFP control exhibited fast and complete recovery, as expected for a free diffusing, inert protein (Figure 2A black). The fluorescence recovery of eGFP-LEDGF/p75 was also complete but much slower (Figure 2A, magenta). A pure diffusion model provided a good fit of the experimental data and a diffusion coefficient D = 0.41 ± 0.06 µm2/s was calculated (Table 1), which is 53-fold lower than expected for free diffusion (Figure 2A, green, with D from Equation 8). Of concern, while diffusion time of eGFP (tdiff = 97 ms) could not be determined correctly due to recovery-while-photobleaching (40), the dynamics of eGFP-LEDGF/p75 (tdiff = 2.13 s) were slow enough to be measured accurately. A reaction-dominant model did not fit the data well and a full diffusion-reaction model did not provide a better fit than a pure diffusion model (Supplementary Figure S1A–C).
Figure 2.
Dynamic subpopulations of LEDGF/p75 revealed by sFRAP, CP and FCS. (A) sFRAP experiment of eGFP and eGFP-LEDGF/p75 in living HeLa cells. The diffusion coefficient we calculated for eGFP after fitting with a pure diffusion model was only 10.28 µm2/s (Table 1). For eGFP the observed recovery was lower than expected (solid orange curve) because of unavoidable recovery-while-photobleaching, which occurred because the diffusion time of eGFP (tdiff = 97 ms) was too small with respect to the acquisitioning time of the microscope (65 ms per iteration on our setup) (40). For eGFP-LEDGF/p75, the arrow indicates the decrease in mobility of eGFP-LEDGF/p75 due to chromatin interactions. DeGFP = 33 µm2/s is the D we measured with FCS, DeGFP-LEDGF/p75 = 22 µm2/s is calculated from DeGFP with Equation 8. See also Supplementary Figure S1. (B) CP experiment of eGFP and eGFP-LEDGF/p75. Upper panel is a 80–100-s zoom. (C) Concentration dependence of the amount of photobleaching of eGFP-LEDGF/p75 during the first 20 s. Solid line = fit with Equation 6. (D) FCS experiment of eGFP and eGFP-LEDGF/p75. Solid lines = two-component normal diffusion model. See also Supplementary Figure S2. Error bars = SD.
Table 1.
Dynamics of eGFP and eGFP-LEDGF/p75 measured with sFRAP, FCS, CP, TFFCS and hnFRAP
| eGFP |
eGFP-LEDGF/p75 |
||||||
|---|---|---|---|---|---|---|---|
| sFRAP | FCS/CP | TFFCS | sFRAP | FCS/CP | hnFRAP | TFFCS | |
| Fimm (%) | 0.1 ± 0.1 | – | – | −1.2 | – | – | – |
| Dfast or DFRAP (µm2/s) | 10.28 ± 0.63a | 33.0 ± 3.5 | 36.4 ± 3.1 | – | 22.3 ± 4.1b | – | 35.3 ± 10.7 |
| Ffast (%) | – | 97.3 ± 0.7 | – | – | 35.9 ± 3.0c | – | 44.1 ± 5.7d |
| Dslow or DFRAP (µm2/s) | – | 0.8 ± 0.7 | – | 0.41 ± 0.01 | 0.5 ± 0.1 | 1.02 ± 0.01 | 0.95 ± 0.01 |
| αe | – | 0.98 ± 0.02 | – | – | f | – | |
| Ffree (%)g | – | – | – | 1.8 ± 3.1 | 2.2 ± 0.4 | 4.6 ± 0.94 | 4.3 ± 1.0 |
| Kdiss (nM) | – | 103 ± 66 | – | ||||
| [sites]high (nM) | – | 61 ± 31 | |||||
Values represent averages of at least 10 (sFRAP), 20 (FCS/CP), 10 (TFFCS) or 5 (hnFRAP) measurements and are ±SD.
aThis value is an underestimation, because the shortest bleach iteration (65 ms) was still too long to avoid recovery-while-photobleaching.
bThis parameter likely does not represent a diffusion process, see Figure 3F and ‘Discussion’ section.
cThe contribution of each component did not depend significantly on the concentration.
dThe fractional contribution of each component did not depend on the focus spot size.
eThis parameter was obtained by fitting with a one-component anomalous diffusion fit.
fFit not satisfying, see Supplementary Figure S2B.
gFfree is calculated from the apparent diffusion law, see ‘Materials and Methods’ section, Equation 7, with DeGFP-LEDGF/p75 = 22.3 µm2/s.
Fimm, immobile fraction; Dfast and Dslow, diffusion coefficient from fitting the FCS curves; DFRAP, diffusion coefficient obtained from Supplementary Equations S1 and S2; Ffast, fractional contribution of the fast component observed with FCS; α, anomaly parameter; Ffree, fraction of the protein population observed with FCS that is freely diffusing.
Next, we applied CP (39). The CP signal from eGFP in HeLa cells decayed slowly and biexponentially (Figure 2B, black), as has been described before for eGFP (35). For eGFP-LEDGF/p75 (Figure 2B, magenta) a biphasic decay was also observed (Figure 2B, black). Resuming the CP measurement after a pause resulted in a similar decay (data not shown), arguing for the absence of an immobile fraction. It has been shown that slow diffusion in a finite compartment can indeed lead to a biphasic fluorescence decay (34). Settling of diffusion of eGFP-LEDGF/p75 (D = 0.41 µm2/s) in a HeLa nucleus (R = 8.74 µm) should take ∼47 s (Supplementary Information), which agrees quite well with the completion of the fast exponential decay we observed (Figure 2B, dashed line). Of concern, fitting the same data with a binding model with medium dissociation rates (Figure 2B, green) did not result in a better description of the experimental data than a simple biexponential decay (35), proving that diffusion and binding are necessary to describe the observed photobleaching.
Interestingly, in the <500 nM concentration range the fraction of the fast exponential was concentration dependent (Figure 2C), which suggests the presence of a low concentration of higher affinity binding sites. In line with this, a full reaction-diffusion model for analyzing the sFRAP data also suggested a low fraction of higher affinity binding sites (Supplementary Table S1). With Equation 6 the data from Figure 2C could be fitted to determine an average concentration of binding sites S0 = 61 ± 31 nM and a steady state dissociation constant Kdiss = 103 ± 66 nM.
The fluorescence signal after the initial photobleaching was divided in 20-s intervals which were autocorrelated individually (Figure 2B-top) and an average ACF was calculated with FCS (56). The eGFP exhibited free diffusion in the nucleus (Figure 2D, black and Table 1) with D = 33.0 ± 3.5 µm2/s (Table 1), which is 2.9 times slower than measured in solution (49) and in good agreement with previous findings in cells if the correct D of the calibration probe (426 µm2/s versus 280 µm2/s) is taken into account (57). The eGFP-LEDGF/p75 ACF was again strongly shifted into a slower time scale (Figure 2D, magenta) with respect to the calculated ACF for free diffusion (Figure 2D, green). A one-component (normal or anomalous) diffusion model did not describe the ACF well (Supplementary Figure S2A and B). A two-component normal diffusion model described the ACF very well (Supplementary Figure S2C): a fast component (35 ± 3%) with apparent D = 22.3 ± 4.1 µm2/s and a slow component (65 ± 3%) with D = 0.5 ± 0.1 µm2/s was determined (Table 1). The slow D is in good accordance with the D obtained from FRAP. A reaction-dominant fit model did not describe the FCS data well, implying that the slow component in the ACF cannot be described solely with a binding reaction (Supplementary Figure S2D). More complex two-component diffusion models that described confined diffusion resulted only in a modestly better fit (Supplementary Figure S2E and F) than a normal 2-component diffusion model, suggesting the latter is necessary and sufficient to describe the observed dynamics.
Finally, we performed control measurements to verify that the dynamics we observed for LEDGF/p75 can indeed be related to chromatin interactions. First, we performed similar experiments on known chromatin binding proteins to verify that chromatin interaction can indeed be observed in the experimental setup (Supplementary Figure S3A). Next, we deleted the N-terminal p52 region of LEDGF/p75 and performed similar experiments on this protein, eGFP-LEDGF/p75326–530. The CP curve of this protein resembled that of free eGFP and the ACF shifted almost completely back to free diffusion (Supplementary Figure S3B). This proves that chromatin interactions of LEDGF/p75 via its N-terminal p52 domain contribute significantly and protein–protein interactions of the IBD do not contribute significantly to the observed overall dynamics of LEDGF/p75.
In summary, we combined sFRAP, CP and FCS to study the mobility of eGFP-LEDGF/p75 in the nucleus of living HeLa cells. Overall, LEDGF/p75 does not interact strongly with chromatin, as implied from the absence of a permanent immobile fraction in sFRAP. Rather, we observed different dynamic states: (i) a slow state with D = 0.4–0.5 µm2/s, observed with sFRAP and FCS, (ii) a presumed bound state with ∼60 nM binding sites and a dissociation constant Kdiss ∼100 nM, suggested from sFRAP and CP measurements and (iii) a fast state, observed only with FCS.
Slow eGFP-LEDGF/p75 corresponds to hopping on chromatin
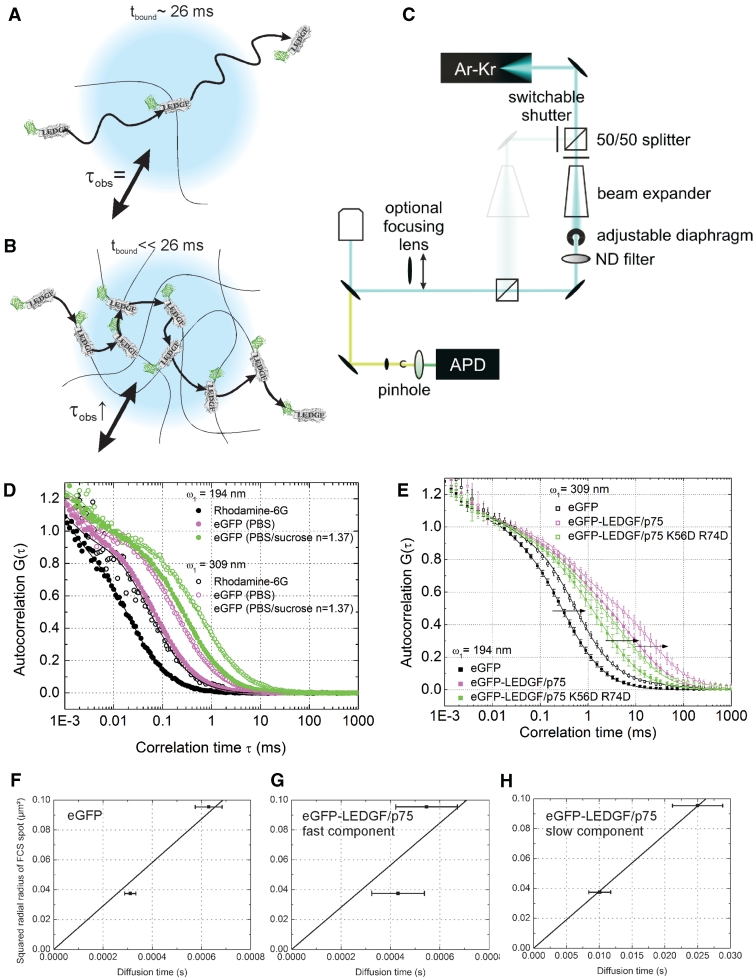
The slow moving fraction of LEDGF/p75 (D = 0.5 µm2/s, τdiff = 26 ms) is too fast to represent chromatin controlled movement of associated LEDGF/p75, which is reported to be 2–3 orders-of-magnitude slower (58). We performed TFFCS, where the excitation volume can be tuned in size, to verify the scaling of the observed dynamics with the radius of the measurement spot (33,59,60): if the observed diffusion time (τobs) does not scale with the size of the excitation volume, then τobs is solely determined by the residence time tbound (=1/koff) on the immobile structure (Figure 3A). If however τobs does scale with the size of the excitation volume, then the binding process is much faster than the diffusion time scale (Figure 3B). Put differently, the relaxation time of the binding process [=1/(k*on + koff)] will be smaller than τobs if τobs varies with the focal spot size. We first performed detailed in vitro control experiments to characterize this set-up (Figure 3C). In aqueous solution, both the apparent particle number (Supplementary Figure S4A) and the average diffusion time (Supplementary Figure S4B) of the standard probe rhodamine 6G increased when tuning the laser beam to a smaller diameter (and hence the focal spot to a larger diameter) (Figure 3D black). In a next step, we used eGFP as a probe. Both in phosphate buffered saline (PBS) (Figure 3D, magenta) and in a buffer with an index of refraction matching the intracellular environment (PBS, 23.5% w/w sucrose, n = 1.37) (Figure 3D, green) increasing the laser focus again shifted the experimental ACF to a slower time scale, as expected. The TFFCS set-up thus performs well for determining the dependence of the ACF on the size of the focus. We next performed similar measurements in living HeLa cells (Figure 3E). The diffusion time of eGFP was clearly dependent on the size of the focal spot (Figure 3E, black and Figure 3F), consistent with the free intracellular movement of the protein. Overall, the ACF of eGFP-LEDGF/p75 also shifted in a larger focal spot (Figure 3E, magenta). More specifically, while the apparent diffusion time (τdiff) of the fast component did not significantly shift to a slower time scale if the focal spot was increased, the τdiff of the slow component scaled perfectly with the focal spot size. In other words, the fast component likely does not represent a diffusion process, while the slow component clearly does. Moreover, the D we calculated from the slope of the linear fit was D = 36.4 ± 3.1 µm2/s for eGFP and 0.95 ± 0.01 µm2/s for eGFP-LEDGF/p75, in good accordance with the corresponding values obtained with sFRAP and normal FCS.
Figure 3.
TFFCS shows fast chromatin binding of LEDGF/p75. (A) The observed slow diffusion time (τobs = 25.9 ms, D = 0.5 µm2/s) is the binding time of LEDGF/p75 with chromatin, tbound = 1/koff. In this case τobs does not scale with the laser focus diameter, indicated in blue. (B) LEDGF/p75 continuously associates with and dissociates from the chromatin while diffusing through the measurement spot. The τobs scales with the laser focus diameter. (C) Schematic representation of the home built TFFCS set-up. The beam expander and diaphragm are used to vary the diameter of the collimated laser beam. ND, neutral density attenuation filter; APD, avalanche photodiode. The diameter of the diaphragm is controlled by measuring the power of the excitation light passing through it. Higher power through the diaphragm means a broader beam will enter the objective back aperture. The objective focuses a broader beam in a smaller focal point, since the beam diameter determines the effective numerical aperture of the objective (NA = n × sinα with n the refractive index and α the angular aperture). For further details, see the ‘Materials and Methods’ section. (D) In vitro TFFCS measurements of rhodamine 6G in water (black), eGFP in PBS (magenta) and PBS/sucrose with a viscosity and refractive index similar to that of the intracellular environment (n = 1.37) (green). ω1: radial radius of the excitation spot. (E) Intracellular TFFCS measurements of eGFP (black), eGFP-LEDGF/p75 (magenta) and eGFP-LEDGF/p75 K56D-R74D (green). Error bars = SD. (F–H) Plot of the diffusion time from the experimental ACF versus the squared radial radius of the confocal excitation spot for (F) eGFP, (G) the fast and (H) the slow component of eGFP-LEDGF/p75.
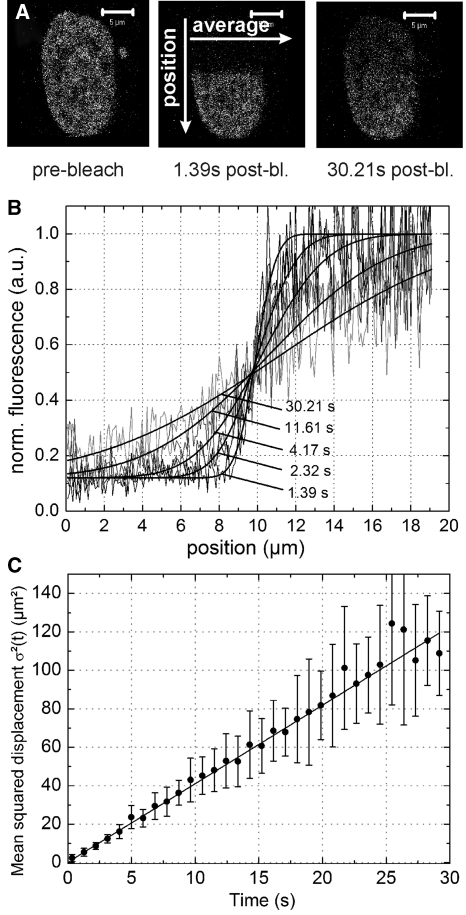
To further corroborate that the dynamics of the slow FCS component are indeed diffusion controlled, we performed hnFRAP. After photobleaching half a nucleus, we monitored the nuclear fluorescence equilibration over time, perpendicular to the bleach border, as illustrated in Figure 4A. The bleach profile changed slowly, but gradually over time, typical for a diffusion controlled binding reaction (Figure 4B), as has been described before for other known chromatin binding proteins. When we plotted the mean squared displacement, obtained by fitting the hnFRAP curves, versus time, we again obtained a nearly perfect linear relation and the D that was calculated from the slope was D = 1.02 ± 0.01 µm2/s, in perfect agreement with our TFFCS data. Furthermore, the linear MSD-time relationship corroborates our Brownian-like diffusion FCS fit model. In conclusion, LEDGF/p75 likely exhibits a hopping mechanism in the nucleus, characteristic of non-specific, frequent chromatin interactions (Figure 3B).
Figure 4.
LEDGF/p75 shows normal diffusion dependent dynamics. (A) Illustration of half-nucleus FRAP. Half of the nucleus is photobleached and recovery perpendicular to the bleach border is monitored versus time. (B) Nuclear intensity profile versus time along the arrow designated ‘position’ in (A) Solid lines = Equation 10. (C) Mean squared displacement – time plot. Error bar = SD. Solid line = Equation 11.
LEDGF/p75-PWWP contributes to chromatin binding of LEDGF/p75
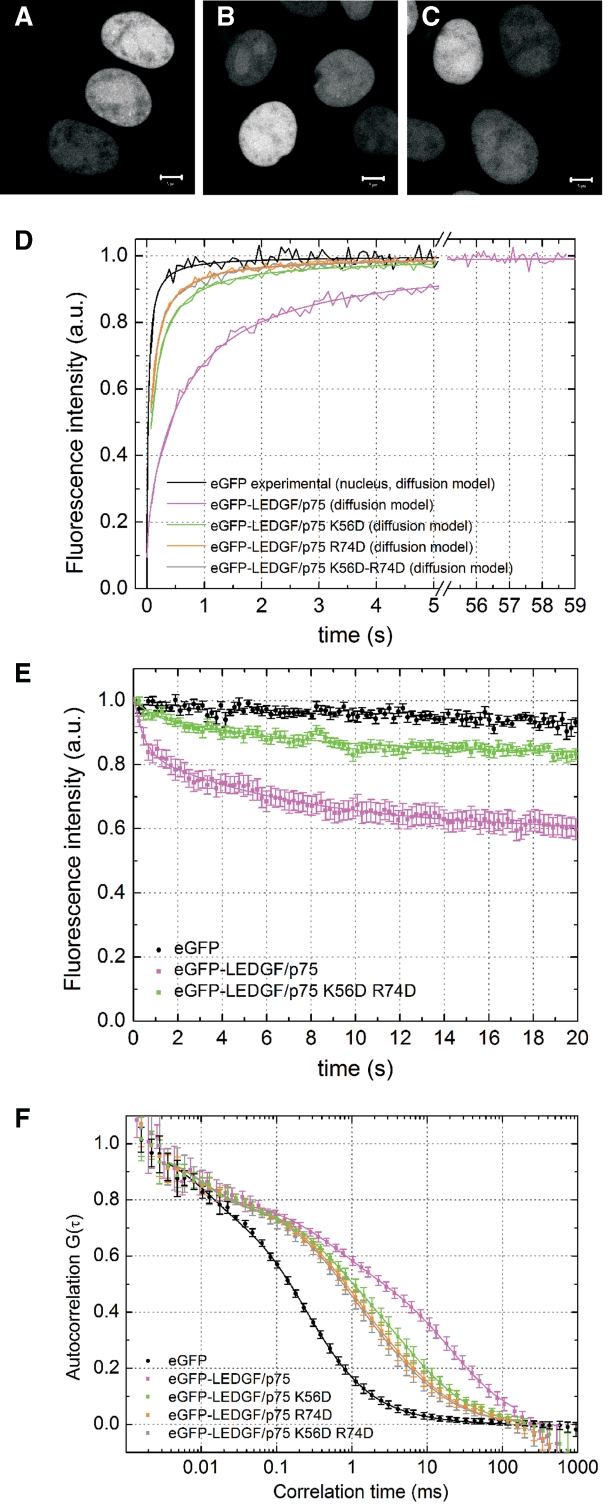
We set out to verify the previously documented contribution of the PWWP domain to the in vivo chromatin binding properties of LEDGF/p75 (15,16). To affect the overall protein structure as little as possible, we sought to specifically alter the affinity of this domain for chromatin. Based on the molecular model of the PWWP domain of HDGF in complex with DNA (54), we predicted that positively charged residues K56 and R74 in LEDGF/p75 are most likely interacting with the phosphates of the host DNA. Next, we constructed and purified two single mutant proteins of LEDGF/p75, K56D and R74D and the double mutant K56D-R74D. Correct overall folding was corroborated with circular dichroism spectroscopy (Supplementary Table S2). Next, eGFP-fusions of the same proteins were transiently expressed in HeLa cells. The proteins were characterized by a more diffuse nuclear localization (Figure 5A–C), compared with the typical heterogeneous nuclear distribution of the wild-type protein (22) (Figure 1C), suggesting that chromatin binding properties were affected. Next, we used sFRAP, CP and FCS to study the dynamics of the mutants. All mutants were considerably faster (4- to 8-fold) than wild-type LEDGF/p75 (Figure 5D and F and Table 2). The R74D mutation had a more pronounced effect than the K56D mutation (Figure 5D and F green and orange, respectively) while the double mutant did not show an additive effect compared with the R74D single mutation (Figure 5D and F, gray). All mutants were still considerably slower than expected for free diffusion, indicating they still have some residual interactions with chromatin. Importantly, the difference in cellular dynamics between the K56D and R74D mutants could not be inferred from a differential distribution of the proteins, but was only evidenced by our sFRAP and FCS measurements, demonstrating the sensitivity of these techniques. Finally, the ACF of eGFP-LEDGF/p75 K56D-R74D still varied with the focus size, implying that this protein also moves by diffusion (Figure 3E). In summary, by mutating a single amino acid residue in LEDGF/p75-PWWP, R74D, chromatin binding of the protein was severely impaired, as evidenced by an up to 8-fold increase in dynamics with respect to wild-type LEDGF/p75. The PWWP domain of LEDGF/p75 is thus important but not sufficient for in vivo chromatin binding.
Figure 5.
The PWWP domain of LEDGF/p75 contributes to high affinity chromatin binding and is crucial for chromatin tethering of HIV-1 integrase. (A–C) Confocal fluorescence images of HeLa cells expressing (A) eGFP-LEDGF/p75 K56D, (B) R74D and (C) K56D-R74D. (D) sFRAP experiment. (E) CP experiment of eGFP-LEDGF/p75 K56D-R74D. The CP curves of eGFP and eGFP-LEDGF/p75 are shown as a reference. (F) FCS experiment of eGFP-LEDGF/p75 mutants. The ACFs of eGFP and eGFP-LEDGF/p75 are shown as a reference. Error bars = SD.
Table 2.
Dynamics of LEDGF/p75 PWWP mutants measured with sFRAP and FCS
| eGFP-LEDGF/p75 K56D |
eGFP-LEDGF/p75 R74D |
eGFP-LEDGF/p75 K56D R74D |
||||
|---|---|---|---|---|---|---|
| sFRAP | FCS | sFRAP | FCS | sFRAP | FCS | |
| Fimm (%) | −0.1 ± 0.1 | 0.1 ± 0.1 | −0.1 ± 0.1 | |||
| Dfast (µm2/s) | – | 16.4 ± 3.5 | – | 13.3 ± 2.4 | – | 16.8 ± 5.1 |
| Ffast (%) | – | 44.5 ± 9.3 | – | 58.5 ± 8.6 | – | 62.8 ± 13.8 |
| Dslow (µm2/s) | 1.97 ± 0.09 | 1.0 ± 0.3 | 3.02 ± 0.18 | 1.4 ± 0.6 | 3.25 ± 0.20 | 1.5 ± 0.8 |
| α | – | 0.69 ± 0.05a | – | 0.78 ± 0.04 | – | 0.80 ± 0.05 |
Values represent averages of at least 10 (sFRAP) or 20 (FCS) measurements and are ±SD. Parameters Dfast, Ffast and Dslow are obtained from a two-component normal diffusion fit. Parameter α is obtained from a one-component anomalous fit.
aFit not satisfying. For more details on the parameters, see Table 1.
LEDGF/p75 locks the IN-LEDGF/p75 complex on chromatin through its PWWP domain
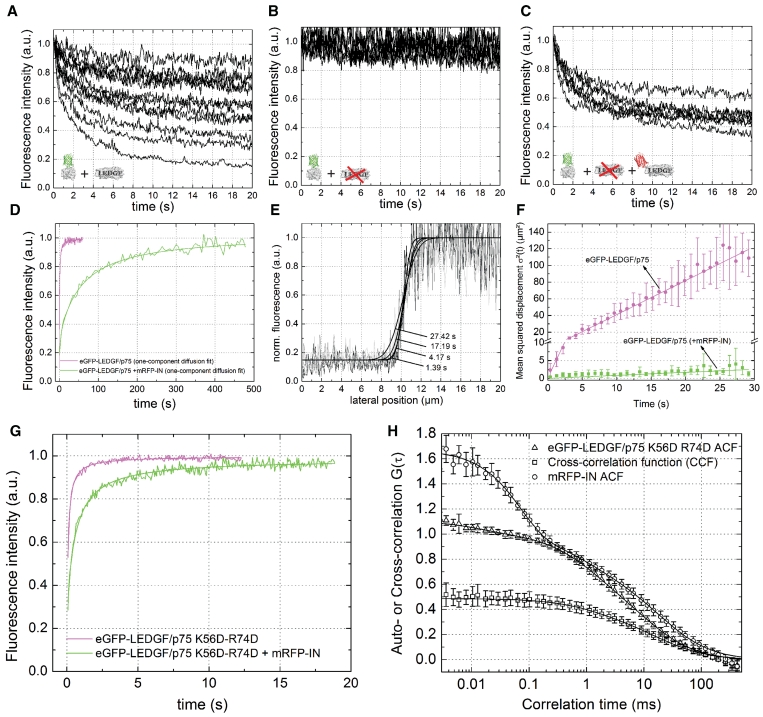
LEDGF/p75 has been shown to tether IN to chromatin by virtue of a direct protein–protein interaction (15,22,61–64). We performed CP measurements of eGFP-tagged IN in HeLa cells to show tethering in vivo: we observed variable photobleaching of eGFP-IN from cell to cell in wild-type HeLa cells (Figure 6A), but no significant photobleaching of eGFP-IN was observed in HeLa-p75KD cells (Figure 6B). When HeLa-p75KD cells were back-complemented with 500 nM of RNAi-resistant mRFP-LEDGF/p75, a large fraction of eGFP-IN was again photobleached (Figure 6C). The dynamics of IN are thus strongly influenced by LEDGF/p75 mediated chromatin tethering. We next quantified the effect of IN on in vivo chromatin binding kinetics of LEDGF/p75. Upon co-expression of mRFP-labeled IN the overall dynamics of LEDGF/p75 shifted to much slower time scale (Figure 6D). When fitting the sFRAP curve with a normal diffusion model (Supplementary Equation S1), a good fit was obtained (Figure 6D and Supplementary Figure S5A) and D = 0.0055 ± 0.001 µm2/s was calculated, about 75 times lower than in cells without co-expression of mRFP-IN (Tables 1 and 3). A reaction-dominant model (Supplementary Equation S3) did not give a good fit (Supplementary Figure S5B) and a full diffusion-reaction model (Supplementary Equation S4) did not result in a better fit than a normal diffusion model (Supplementary Figure S5C). With hnFRAP a similar decrease in dynamics was observed (D = 0.021 ± 0.004 µm2/s) (Figure 6E and F).
Figure 6.
LEDGF/p75-PWWP locks IN-LEDGF/p75 on chromatin. (A–C) CP experiment of eGFP-IN in (A) wild-type HeLa cells, (B) HeLa-p75KD cells and (C) HeLa-p75KD cells back-complemented with 500 nM mRFP-LEDGF/p75. (D) sFRAP experiment of eGFP-LEDGF/p75 in HeLa cells, without (magenta) and with (green) co-expression of mRFP-IN. Solid lines represent a fit to a normal diffusion model. Results from the fitting are presented in Table 3. See also Supplementary Figure S5. (E) HnFRAP experiment in nuclei expressing eGFP-LEDGF/p75 and mRFP-IN. Solid lines = Equation 10. (F) MSD-time plot. Solid line = Equation 11. (G) FRAP measurement of eGFP-LEDGF/p75 K56D-R74D in HeLa cells, without (magenta) and with (green) co-expression of mRFP-IN. Solid lines = Supplementary Equation S1. Results from the fitting are presented in Table 1. (H) FCCS measurements in cells expressing eGFP-LEDGF/p75 K56D-R74D and mRFP-IN. The high amplitude of the CCF means a strong protein–protein interaction. Error bars = SD.
Table 3.
Effect of HIV-1 integrase co-expression on the dynamics of LEDGF/p75 and LEDGF/p75 K56D-R74D measured with sFRAP and hnFRAP
| eGFP-LEDGF/ p75 + mRFP-IN) |
eGFP-LEDGF/ p75 K56D-R74D + mRFP-IN) | ||
|---|---|---|---|
| sFRAP | hnFRAP | sFRAP | |
| Fimm (%) | −4.5 ± 2.0 | – | 2.0 ± 1.0 |
| D (µm2/s) | 0.006 ± 0.001 | 0.021 ± 0.004 | 0.54 ± 0.02 |
| Ffree (%)g | 0.12 0.02 | – | 3.64 ± 0.13 |
Values represent averages of at least 10 (sFRAP) or 5 (hnFRAP) measurements and are ±SD. For more details on the parameters, see Table 1.
Next, we verified with sFRAP whether the mutations in the PWWP domain of LEDGF/p75 affected chromatin tethering of IN. Co-expression of mRFP-IN decreased the dynamics of the K56D-R74D mutant about six times, as calculated from the difference in D (Tables 2 and 3 and Figure 6G), which is much less than wild-type eGFP-LEDGF/p75 (note the difference in time scale in Figure 6D and G). To verify that the absence of chromatin tethering was not due to a loss of the IN-LEDGF/p75 interaction, we used cellular FCCS. A high relative cross-correlation amplitude, comparable with what we published before for IN-LEDGF/p75326–530 and IN-LEDGF/p75-K150A (61,62), was observed (Figure 6H), indicating that the interaction is not affected by the mutations. In conclusion, chromatin tethering of IN by LEDGF/p75 can be observed with FRAP. We showed for the first time in vivo that IN-LEDGF/p75 complexes have a 75-fold lower mobility as compared with LEDGF/p75. Since IN alone did not appear to interact strongly with chromatin, LEDGF/p75 thus tethers IN strongly to chromatin. Rational mutation of the PWWP domain resulted in a strongly decreased ability to tether IN, indicating that the PWWP domain in LEDGF/p75 constitutes a ‘protein lock’ that strongly tethers IN to chromatin.
DISCUSSION
Fluorescent LEDGF/p75 displays wild-type properties
Our experimental approach combines complementary fluorescence techniques to study fluorescently labeled LEDGF/p75 in living cells. It had been shown that the interactions with cellular partners through the IBD are not hampered by the presence of the eGFP label (28–30,63). Unlike a C-terminal eGFP (data not shown), an N-terminal fusion displayed a proper intracellular distribution. In addition, the overall chromatin binding characteristics of eGFP-LEDGF/p75 are not altered (Figure 1E); since LEDGF/p75 lost chromatin binding by mutating the PWWP domain, we are confident that chromatin binding of eGFP-LEDGF/p75 is conserved. Results are in agreement with Turlure et al. (16), who showed that eGFP-PWWP still functionally interacts with chromatin. All experiments in cells were performed at relevant concentrations of fluorescent LEDGF/p75. On average, the concentration of transiently expressed eGFP-LEDGF/p75 was similar to that of endogenous LEDGF/p75 (Figure 1D) and measurements were performed in a 1 nM–2.5 µM concentration range to study potential concentration dependent effects (Figure 1D). Furthermore, the ACF of eGFP-LEDGF/p75 was concentration independent (data not shown) and our investigation of chromatin tethering of IN (Figure 6A–C) led us to assume that the intracellular concentration of LEDGF/p75 lies in submicromolar range. In line with this, LEDGF/p75 has been shown to be a much more abundant cellular protein than its alternative splice variant p52 (22,65).
Dynamic in vivo chromatin interactions of LEDGF/p75
DNA binding and chromatin fractionation assays have been used in the past to study interactions of LEDGF/p75 with chromatin but these methods only provide a static and average view on interactions with DNA/chromatin (15,16,21). Also, irreconcilable notions of in vivo chromatin binding of LEDGF/p75 exist. On the one hand, LEDGF/p75 has been shown to be a ‘site-specific’ transcriptional co-activator, interacting with promoter elements of stress-related genes. On the other hand, LEDGF/p75 has been shown to be the factor that targets lentiviral integration to ‘random’ genomic regions of active transcription (21,24). We used sFRAP, CP, FCS, TFFCS and hnFRAP to monitor in vivo chromatin interactions of LEDGF/p75 for the first time. We showed that LEDGF/p75 does not permanently take part in immobile protein/chromatin complexes (Figure 2A).
The slow state was observed both with all techniques and represents low-affinity, likely non-specific chromatin scanning/hopping, a common phenomenon of transcription factors (66). By this mechanism, LEDGF/p75 likely targets lentiviral integration to random (actively transcribed) regions (24–26). On the other hand, the concentration dependent CP measurements strongly suggest the existence of low-concentration higher-affinity chromatin-bound states (Figure 2A–C and Table 1), which might result from association with promoters of stress-responsive genes (21,67) or more generally, as has recently been shown, from the association downstream of transcription start sites (TSS) of active transcription units (68). The concentration of binding sites we calculated (61 nM), indeed lies on the same order-of-magnitude as the concentration of TSS in a eukaryotic cell (∼5–100 nM, see ‘Materials and Methods’ section).
To our surprise, we only observed the fast state (Figure 2D and Table 1) with FCS. This state did not appear to represent translational motion of the protein (Figure 3G). The time scale and fraction of the fast process did not appear to be concentration or cell dependent (data not shown), ruling out free/bound protein fractions or a post-translationally modified fraction. This state might represent any process that changes the emissive state of the eGFP fluorophore, such as protonation (69), quenching or possibly rotational diffusion of chromatin bound eGFP-LEDGF/p75. In that respect, it has been shown that mRFPs can reside in dark states even in the milliseconds time scale under 1P excitation (70).
Quantitative information on in vivo chromatin hopping with TFFCS
We showed with hnFRAP that diffusion of eGFP-LEDGF/p75 in the slow (seconds) time scale seems normal in absence (Figure 4C) or presence (Figure 6F) of HIV-1 IN. By adapting a standard FCS microscope, we could also clearly distinguish the contribution of diffusion to the observed chromatin binding dynamics of LEDGF/p75 and showed that even in the fast (milliseconds) time scale diffusion still seems normal (Figure 3E and H). This implies that association with and dissociation from chromatin occurs faster than the transit time of the protein through the laser focus, as illustrated in Figure 3B. As such, the strength by which LEDGF/p75 is slowed down could be used to quantify the low-affinity chromatin binding of LEDGF/p75. Based on Dobserved = 0.5 µm2/s and the theoretical Dfree = 22 µm2/s, we calculated with Equation 8 that LEDGF/p75 remains 98% of the time bound to chromatin and moves during the remaining 2% of the time to a new binding site. The average tfree and tbound should thus be considerably shorter than the observed diffusion time (25.9 ms) and k*on ∼43 times larger than koff (Equation 7). In other words, koff should be bigger than 38.6/s and k*on > 1.7 × 103/s. This kinetic ‘effective diffusion’ regime of non-specific chromatin binding has been simulated before (36) and implies that transcription factor chromatin binding might be even more dynamic than currently appreciated (33,36,42,59).
Importantly, TFFCS as such would not be able to discern between slow movement of a (big) protein complex and frequent chromatin interactions of a (small) protein. Therefore, one has to verify that chromatin interactions are indeed contributing to the observed dynamics, which we did, both in vitro (Figure 1E) and in cells (Supplementary Figure S3). Furthermore, by rational mutation of the PWWP domain, a known chromatin interaction motif, we did prove that the PWWP contributes to the observed affinity of LEDGF/p75 for chromatin in vivo. Additionally, we could show cargo-mediated chromatin tethering by LEDGF/p75, which has been shown many times to be an important function of LEDGF/p75 for HIV replication. Finally, it must be noted that the ‘chromatin hopping’ we describe for LEDGF/p75 could also represent chromatin hopping of a larger complex containing LEDGF/p75 and other nuclear components and this complex would experience a larger effective viscosity in the chromatin matrix. Further research will provide more insight in this. In line with our kinetic regime of hopping, fast chromatin binding kinetics have recently also been experimentally observed for the Drosophila Hox gene Sex combs reduced transcription factor with DNA in vivo (60) and DNA-bound and slow apparent diffusion two-state chromatin binding has recently also been observed for Heterochromatin protein 1 (43), a protein we also studied as a control for chromatin binding (Supplementary Figure S3).
Extremely slow, yet diffusion controlled dynamics of LEDGF/p75-IN
It has been shown that LEDGF/p75 can act as a molecular tether coupling IN to the chromatin (15,22,61–64) and IN has been suggested to be present in a paracrystalline form in the viral particle (71). In the presence of its viral cargo, IN, LEDGF/p75 formed a 75-fold slower dynamical complex. Since interaction with IN likely does not decrease k*on of LEDGF/p75, the observed dynamics should still be diffusion controlled, which is what we observed (Supplementary Figure S5 and Figure 6E and F). The effective diffusion law (Equation 7) allowed us to calculate that while for LEDGF/p75, k*on/koff ∼43, for IN-LEDGF/p75, k*on/koff ∼2.7 × 103. The k*on is thus at least as large as for LEDGF/p75 alone and the koff likely decreases considerably after interaction with IN. The IN is known to be present at least in a dimeric form at nM–µM concentrations (72) and already a IN dimer contains two binding sites for LEDGF/p75 (73). As we have shown, IN does not bind strongly to chromatin (Figure 6A–C). Probably thus, IN-LEDGF/p75 complexes contain at least two LEDGF/p75 units that can both tether IN cooperatively to chromatin, resulting in the apparent diffusion controlled increase in affinity. An allosteric effect of IN on the intrinsic affinity of LEDGF/p75 for chromatin is unlikely, since the IBD and chromatin binding domains of LEDGF/p75 are located in distinct structural parts of the protein. It might be argued that the IN-LEDGF/p75 complex senses, due to its oligomeric size, a much larger viscosity in cells than does LEDGF/p75 alone and this might explain the observed 75-fold decrease in dynamics. As we have shown in Figure 6G and H, PWWP-mutated LEDGF/p75 still has wild-type affinity for IN, but its dynamics are only slowed down 6-fold by IN co-expression. Therefore, we are confident that the observed dynamical shift of IN-LEDGF/p75 does represent an increased affinity for chromatin.
The linker histone H1, which we also used as a control for our measurements (Supplementary Figure S3), has recently also been studied quantitatively and similar slow overall dynamics were observed (74). Because dynamics of H1 were described best with a full diffusion/reaction model while a simple diffusion model sufficed for IN-LEDGF/p75, this suggests that while for H1 very slow dissociation from chromatin is the cause of the slow overall dynamics, very fast association with chromatin is rather the cause of the slow dynamics for IN-LEDGF/p75.
The PWWP domain is the key to the chromatin lock
In recent years, LEDGF/p75 has become a protein of interest to understand various diseases such as leukemia, autoimmunity and AIDS (4,7,8,22). Nearly all normal and disease related functions of LEDGF/p75 involve tethering of interaction partners to chromatin through its conserved IBD. We investigated the role of the PWWP domain in the chromatin binding of LEDGF/p75 in vivo. The conserved PWWP domain of LEDGF/p75 contains a recently described general protein fold, present in proteins that carry chromatin-binding motifs and has been proposed to bind DNA in several independent structural biology studies (54,75). Other studies however suggested, albeit without experimental evidence so far, that the PWWP fold is implicated in non-DNA chromatin interactions (12,14). Although eGFP-PWWP did not show any DNA binding in vitro, it was shown to be localized on mitotic chromosomes, much alike full length LEDGF/p75 (16). Our rational mutagenesis of the PWWP domain of LEDGF/p75 resulted in a protein with up to 8-fold increase in overall mobility as compared with wild-type LEDGF/p75 (Figure 5D and F). The mutant protein exhibited diffusion-dependent anomalous diffusion (Figures 3E and 5F and Table 2), which could either result from residual interactions with the chromatin (76) or from interactions with other cellular cofactors, leading to obstructed diffusion (77). Nonetheless, we confirm that interactions of the PWWP domain with chromatin are necessary but not sufficient to convey wild-type affinity of LEDGF/p75 for chromatin.
Importantly, we are the first to show that upon mutation of the PWWP-domain, tight chromatin tethering of IN disappears, while the tight interaction of LEDGF/p75 with IN is conserved. This strongly argues for a sole role of LEDGF/p75, in particular the PWWP-domain, in associating an IBD-bound cargo to chromatin. The HDGF-PWWP has been shown to be able to dimerize (78) and this might be an important step in chromatin tethering. Currently, we are investigating whether cargo-induced locking is a general phenomenon for tethering cargos to chromatin. Being the key for locking cargos to chromatin, the PWWP-domain might be a promising novel target for blocking LEDGF/p75 malfunctioning in AIDS but also oncogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: University of Leuven (GOA/2006/02, IOF/KP/07/008); the Institute for the Promotion of Innovation through Science and Technology in Flanders (CellCoVir SBO grant 60813); the Research Foundation Flanders (FWO) (grant G.0530.08); the European Commission project Targeting HIV Integration Co-factors (grant HEALTH-F3-2008-201032); FWO postdoctoral fellowship (to J.Hendrix.); Mathilde-Krim postdoctoral fellowship (The Foundation for AIDS Research) (to J.D.R.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors acknowledge Davide Mazza (Laboratory for Receptor Biology and Gene Expression, National Cancer Institute, Bethesda, MD) for providing the Matlab code for analyzing the FCS data with the binding models, Antoine Delon (Université Joseph Fourier, France) for helpful discussions on CP and Werner Verbakel (Laboratory for Biomolecular Dynamics) for writing the ImageJ script for hnFRAP analysis.
REFERENCES
- 1.Ge H, Si YZ, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge H, Si YZ, Wolffe AP. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell. 1998;2:751–759. doi: 10.1016/s1097-2765(00)80290-7. [DOI] [PubMed] [Google Scholar]
- 3.Singh DP, Ohguro N, Kikuchi T, Chylack LT, Shinohara T. Gene sequence and functional studies of lens epithelial cell derived growth factor (LEDGF) Invest. Ophthalmol. Vis. Sci. 1998;33:S777. [Google Scholar]
- 4.Ahuja HG, Hong JM, Aplan PD, Tcheurekdjian L, Forman SJ, Slovak ML. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- 5.Daugaard M, Kirkegaard-Sorensen T, Ostenfeld MS, Aaboe M, Hoyer-Hansen M, Orntoft TF, Rohde M, Jaattela M. Lens epithelium-derived growth factor is an Hsp70-2 regulated guardian of lysosomal stability in human cancer. Cancer Res. 2007;67:2559–2567. doi: 10.1158/0008-5472.CAN-06-4121. [DOI] [PubMed] [Google Scholar]
- 6.Huang TS, Myklebust LM, Kjarland E, Gjertsen BT, Pendino F, Bruserud O, Doskeland SO, Lillehaug JR. LEDGF/p75 has increased expression in blasts from chemotherapy-resistant human acute myelogenic leukemia patients and protects leukemia cells from apoptosis in vitro. Mol. Cancer. 2007;6:31. doi: 10.1186/1476-4598-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels T, Zhang JY, Gutierrez I, Elliot ML, Yamada B, Heeb MJ, Sheets SM, Wu XW, Casiano CA. Antinuclear autoantibodies in prostate cancer: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate. 2005;62:14–26. doi: 10.1002/pros.20112. [DOI] [PubMed] [Google Scholar]
- 9.Ganapathy V, Casiano CA. Autoimmunity to the nuclear autoantigen DFS70 (LEDGF): what exactly are the autoantibodies trying to tell us? Arthritis Rheum. 2004;50:684–688. doi: 10.1002/art.20095. [DOI] [PubMed] [Google Scholar]
- 10.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Maele B, Debyser Z. HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev. 2005;7:26–43. [PubMed] [Google Scholar]
- 12.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 13.Singh DP, Kimura A, Chylack LT, Shinohara T. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene. 2000;242:265–273. doi: 10.1016/s0378-1119(99)00506-5. [DOI] [PubMed] [Google Scholar]
- 14.Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 15.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 16.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatma N, Singh DP, Shinohara T, Chylack LT. Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J. Biol. Chem. 2001;276:48899–48907. doi: 10.1074/jbc.M100733200. [DOI] [PubMed] [Google Scholar]
- 18.Fatma N, Kubo E, Chylack LT, Shinohara T, Akagi Y, Singh DP. LEDGF regulation of alcohol and aldehyde dehydrogenases in lens epithelial cells: stimulation of retinoic acid production and protection from ethanol toxicity. Am. J. Physiol. Cell Physiol. 2004;287:C508–C516. doi: 10.1152/ajpcell.00076.2004. [DOI] [PubMed] [Google Scholar]
- 19.Sharma P, Fatma N, Kubo E, Shinohara T, Chylack LT, Singh DP. Lens epithelium-derived growth factor relieves transforming growth factor-beta 1-induced transcription repression of heat shock proteins in human lens epithelial cells. J. Biol. Chem. 2003;278:20037–20046. doi: 10.1074/jbc.M212016200. [DOI] [PubMed] [Google Scholar]
- 20.Shinohara T, Singh DP, Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog. Retin. Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 21.Singh DP, Fatma N, Kimura A, Chylack LT, Jr, Shinohara T. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem. Biophys. Res. Commun. 2001;283:943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 22.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 23.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 24.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 25.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 28.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- 29.Bartholomeeusen K, Christ F, Hendrix J, Rain JC, Emiliani S, Benarous R, Debyser Z, Gijsbers R, De Rijck J. Lens epithelium derived growth factor/p75 interacts with the transposase derived DDE domain of pogZ. J. Biol. Chem. 2009;284:11467–11477. doi: 10.1074/jbc.M807781200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes S, Jenkins V, Dar MJ, Engelman A, Cherepanov P. Transcriptional co-activator LEDGF interacts with Cdc7-activator of S-phase kinase (ASK) and stimulates its enzymatic activity. J. Biol. Chem. 2010;285:541–554. doi: 10.1074/jbc.M109.036491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gijsbers R, Ronen K, Vets S, Malani N, De RJ, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandekerckhove L, Christ F, Van Maele B, De Rijck J, Gijsbers R, Van den Haute C, Witvrouw M, Debyser Z. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delon A, Usson Y, Derouard J, Biben T, Souchier C. Continuous photobleaching in vesicles and living cells: a measure of diffusion and compartmentation. Biophys. J. 2006;90:2548–2562. doi: 10.1529/biophysj.105.069815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wachsmuth M, Weidemann T, Muller G, Hoffmann-Rohrer UW, Knoch TA, Waldeck W, Langowski J. Analyzing intracellular binding and diffusion with continuous fluorescence photobleaching. Biophys. J. 2003;84:3353–3363. doi: 10.1016/S0006-3495(03)70059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michelman-Ribeiro A, Mazza D, Rosales T, Stasevich TJ, Boukari H, Rishi V, Vinson C, Knutson JR, McNally JG. Direct measurement of association and dissociation rates of DNA binding in live cells by fluorescence correlation spectroscopy. Biophys. J. 2009;97:337–346. doi: 10.1016/j.bpj.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crank J. The Mathematics of Diffusion. Oxford: Clarendon Press; 1975. [Google Scholar]
- 38.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters R, Brunger A, Schulten K. Continuous fluorescence microphotolysis: a sensitive method for study of diffusion processes in single cells. Proc. Natl Acad. Sci. USA. 1981;78:962–966. doi: 10.1073/pnas.78.2.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyvis TK, De Smedt SC, Van Oostveldt P, Demeester J. Fluorescence recovery after photobleaching: a versatile tool for mobility and interaction measurements in pharmaceutical research. Pharm. Res. 1999;16:1153–1162. doi: 10.1023/a:1011924909138. [DOI] [PubMed] [Google Scholar]
- 41.Beaudouin J, Mora-Bermudez F, Klee T, Daigle N, Ellenberg J. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys. J. 2006;90:1878–1894. doi: 10.1529/biophysj.105.071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller F, Wach P, McNally JG. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys. J. 2008;94:3323–3339. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller KP, Erdel F, Caudron-Herger M, Marth C, Fodor BD, Richter M, Scaranaro M, Beaudouin J, Wachsmuth M, Rippe K. Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys. J. 2009;97:2876–2885. doi: 10.1016/j.bpj.2009.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oancea E, Teruel MN, Quest AF, Meyer T. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J. Cell Biol. 1998;140:485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. Biopolymers. 1972;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 46.Malvezzi-Campeggi F, Jahnz M, Heinze KG, Dittrich P, Schwille P. Light-induced flickering of DsRed provides evidence for distinct and interconvertible fluorescent states. Biophys. J. 2001;81:1776–1785. doi: 10.1016/S0006-3495(01)75828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 48.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrasek Z, Schwille P. Precise measurement of diffusion coefficients using scanning fluorescence correlation spectroscopy. Biophys. J. 2008;94:1437–1448. doi: 10.1529/biophysj.107.108811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwille P, Korlach J, Webb WW. Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry. 1999;36:176–182. doi: 10.1002/(sici)1097-0320(19990701)36:3<176::aid-cyto5>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 51.Delon A, Usson Y, Derouard J, Biben T, Souchier C. Photobleaching, mobility, and compartmentalisation: inferences in fluorescence correlation spectroscopy. J. Fluoresc. 2004;14:255–267. doi: 10.1023/b:jofl.0000024557.73246.f9. [DOI] [PubMed] [Google Scholar]
- 52.Wawrezinieck L, Rigneault H, Marguet D, Lenne PF. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 54.Lukasik SM, Cierpicki T, Borloz M, Grembecka J, Everett A, Bushweller JH. High resolution structure of the HDGF PWWP domain: a potential DNA binding domain. Protein Sci. 2006;15:314–323. doi: 10.1110/ps.051751706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettit FK, Bare E, Tsai A, Bowie JU. HotPatch: a statistical approach to finding biologically relevant features on protein surfaces. J. Mol. Biol. 2007;369:863–879. doi: 10.1016/j.jmb.2007.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elson EL, Magde D. Fluorescence Correlation Spectroscopy. 1. Conceptual Basis and Theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 57.Maertens G, Vercammen J, Debyser Z, Engelborghs Y. Measuring protein-protein interactions inside living cells using single color fluorescence correlation spectroscopy. Application to human immunodeficiency virus type 1 integrase and LEDGF/p75. FASEB J. 2005;19:1039–1041. doi: 10.1096/fj.04-3373fje. [DOI] [PubMed] [Google Scholar]
- 58.Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 59.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vukojevic V, Papadopoulos DK, Terenius L, Gehring WJ, Rigler R. Quantitative study of synthetic Hox transcription factor-DNA interactions in live cells. Proc. Natl Acad. Sci. USA. 2010;107:4093–4098. doi: 10.1073/pnas.0914612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J. Virol. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, Witvrouw M, Engelborghs Y, Christ F, Gijsbers R, et al. Virus Evolution Reveals an Exclusive Role for LEDGF/p75 in Chromosomal Tethering of HIV. PLoS Pathog. 2007;3:e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 64.Shun MC, Botbol Y, Li X, Di NF, Daigle JE, Yan N, Lieberman J, Lavigne M, Engelman A. Identification and characterization of PWWP domain residues critical for LEDGF/p75 chromatin binding and human immunodeficiency virus type 1 infectivity. J. Virol. 2008;82:11555–11567. doi: 10.1128/JVI.01561-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mueller F, Mazza D, Stasevich TJ, McNally JG. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? Curr. Opin. Cell Biol. 2010;22:403–411. doi: 10.1016/j.ceb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh DP, Kubo E, Takamura Y, Shinohara T, Kumar A, Chylack LT, Jr, Fatma N. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J. Mol. Biol. 2006;355:379–394. doi: 10.1016/j.jmb.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 68.De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 2010;38:6135–6147. doi: 10.1093/nar/gkq410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haupts U, Maiti S, Schwille P, Webb WW. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc. Natl Acad. Sci. USA. 1998;95:13573–13578. doi: 10.1073/pnas.95.23.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hendrix J, Flors C, Dedecker P, Hofkens J, Engelborghs Y. Dark states in monomeric red fluorescent proteins studied by fluorescence correlation and single molecule spectroscopy. Biophys. J. 2008;94:4103–4113. doi: 10.1529/biophysj.107.123596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. http://nar.oxfordjournals.org/content/early/2010/05/19/nar.gkq410.full?sid=05793d0a-8913-4029-9c18-84ab8bb47d5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsiang M, Jones GS, Hung M, Mukund S, Han B, Liu X, Babaoglu K, Lansdon E, Chen X, Todd J, et al. Affinities between the binding partners of the HIV-1 integrase dimer-lens epithelium-derived growth factor (IN dimer-LEDGF) complex. J. Biol. Chem. 2009;284:33580–33599. doi: 10.1074/jbc.M109.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl Acad. Sci. USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stasevich TJ, Mueller F, Brown DT, McNally JG. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 2010;29:1225–1234. doi: 10.1038/emboj.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wedemeier A, Zhang T, Merlitz H, Wu CX, Langowski J. The role of chromatin conformations in diffusional transport of chromatin-binding proteins: cartesian lattice simulations. J. Chem. Phys. 2008;128:155101. doi: 10.1063/1.2895048. [DOI] [PubMed] [Google Scholar]
- 77.Weiss M, Elsner M, Kartberg F, Nilsson T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 2004;87:3518–3524. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sue SC, Lee WT, Tien SC, Lee SC, Yu JG, Wu WJ, Wu WG, Huang TH. PWWP module of human hepatoma-derived growth factor forms a domain-swapped dimer with much higher affinity for heparin. J. Mol. Biol. 2007;367:456–472. doi: 10.1016/j.jmb.2007.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.