Abstract
DNA cytosine methylation (5-meC) is a widespread epigenetic mark associated to gene silencing. In plants, DEMETER-LIKE (DML) proteins typified by Arabidopsis REPRESSOR OF SILENCING 1 (ROS1) initiate active DNA demethylation by catalyzing 5-meC excision. DML proteins belong to the HhH-GPD superfamily, the largest and most functionally diverse group of DNA glycosylases, but the molecular properties that underlie their capacity to specifically recognize and excise 5-meC are largely unknown. We have found that sequence similarity to HhH-GPD enzymes in DML proteins is actually distributed over two non-contiguous segments connected by a predicted disordered region. We used homology-based modeling to locate candidate residues important for ROS1 function in both segments, and tested our predictions by site-specific mutagenesis. We found that amino acids T606 and D611 are essential for ROS1 DNA glycosylase activity, whereas mutations in either of two aromatic residues (F589 and Y1028) reverse the characteristic ROS1 preference for 5-meC over T. We also found evidence suggesting that ROS1 uses Q607 to flip out 5-meC, while the contiguous N608 residue contributes to sequence-context specificity. In addition to providing novel insights into the molecular basis of 5-meC excision, our results reveal that ROS1 and its DML homologs possess a discontinuous catalytic domain that is unprecedented among known DNA glycosylases.
INTRODUCTION
DNA methylation at carbon 5 of cytosine (5-meC) is a reversible epigenetic mark for transcriptional gene silencing that plays critical roles in development and reproduction of most eukaryotic species. Animal DNA methylation is mostly confined to symmetrical CG sequences, but plants also have significant levels of methylated cytosines in CHG and CHH sequences (where H is A, C or T) (1,2). DNA methylation patterns are subject to dynamic regulation involving both methylation and demethylation processes (3,4) and dysfunction of methylation control is a key factor in several forms of human disease, including cancer (5,6). Active DNA demethylation in mammals occurs in early embryos and primordial germ cells but its molecular mechanisms are poorly understood (7). In plants, biochemical and genetic analyses have identified a family of DNA glycosylases that remove 5-meC as a free base and initiate a base excision repair demethylation pathway (8–12).
Plant 5-meC DNA glycosylases are typified by Arabidopsis ROS1 (REPRESSOR OF SILENCING 1) and DME (DEMETER) (8,9), which together with paralogs DML2 and DML3 (DEMETER-LIKE proteins 2 and 3) (13,14) are the founding members of the DML family. All four proteins remove 5-meC from DNA and cleave the phosphodiester backbone by successive β,δ-elimination, leaving a gap that has to be further processed to generate a 3′-OH terminus suitable for polymerization and ligation (10,11,13,14). In vivo, ROS1, DML2 and DML3 are needed to counteract the robust RNA-dependent DNA methylation pathway at hundreds of discrete regions across the plant genome (13–15), whereas DME contributes to genome-wide demethylation during endosperm development and is required for imprinting (11,16–18). Genes encoding putative DML proteins are only found in plant genomes, including mosses and unicellular green algae. Members of the DML family are large polypeptides containing a region that shows sequence similarity with members of the well-known HhH-GPD superfamily of DNA repair glycosylases (19). In addition, they also share a carboxy-terminal domain of unknown function (10), and a short amino-terminal domain significantly rich in lysine (20) (Supplementary Figure S1).
In an ongoing effort to elucidate the molecular basis of active DNA demethylation, we have chosen ROS1 as an archetypal 5-meC DNA glycosylase for detailed analysis. We have recently reported that the short lysine-rich amino-terminal domain is not required for catalytic activity, but mediates strong methylation-independent binding to DNA, and allows efficient excision of 5-meC in long substrates (20). However, it remains unknown how the enzymes of the DML family specifically recognize 5-meC in DNA and distinguish it from unmethylated C. The fact that ROS1 activity is strongly inhibited by replacement of the C5 methyl group by halogen derivatives, even if these substituents decrease the strength of the scissile C1′-N glycosidic bond, suggests an important role for selective steric recognition of the target base at the active site (21). After 5-meC excision, ROS1 remains bound to its reaction product (20,21). This binding leads to a highly distributive behavior of the enzyme on DNA substrates containing multiple 5-meC residues, and may help to avoid generation of double-strand breaks during processing of bimethylated CG dinucleotides or densely methylated DNA regions (21).
A comprehensive understanding of how plant 5-meC DNA glycosylases specifically recognize and excise their target base will require solving their crystal structure in complex with DNA. Nevertheless, some useful information may still be obtained by combining structural information available from DNA glycosylases of the HhH-GPD superfamily and the analysis of amino acids specifically conserved in the DML group. All HhH-GPD DNA glycosylases share a common core structure that consists of two helical domains whose interface contains the enzyme active site (22,23). One of these domains contains the signature HhH-GPD motif (a helix–hairpin–helix and Gly/Pro rich loop followed by a conserved catalytic aspartate) (19), which interacts with the DNA minor groove. An extensive body of evidence strongly suggests that all DNA glycosylases, including HhH-GPD enzymes, perform extrahelical base excision through a reaction path that involves (i) DNA distortion and base flipping, which gives enzyme access for a nucleophilic attack on the anomeric C1′carbon, and (ii) insertion of the base lesion into a substrate recognition pocket (22,23). In this base-flipping mechanism, the properties of the active site pocket rather than the HhH-GPD motif are a major component of the base specificity of each enzyme.
In the present study, we performed multiple sequence alignment and structural homology analysis to predict the location of several candidate ROS1 residues important for recognition and/or catalysis that were functionally-tested by site-directed mutagenesis. In addition to providing instructive clues on the molecular origins of 5-meC recognition and excision, our results reveal that proteins of the DML family are endowed with a discontinuous DNA glycosylase domain.
MATERIALS AND METHODS
Homology-based modeling
A multiple sequence alignment of DML proteins and several members of the HhH-GPD superfamily was performed using the program T-Coffee (24). The alignment was viewed, adjusted and refined manually with Jalview (25). A 3D model structure of the two aligned regions from Arabidopsis ROS1 (amino acids 567–625 and 883–1062) was built using Swiss-Model (26) and the 3D structure of Bacillus stearothermophilus Endonuclease III as a template [Protein Data Bank accession code: 1P59, (27)]. Nucleic acid coordinates extracted from 1P59 were used to superimpose a DNA structure with a flipped-out abasic (AP) site analog onto the ROS1 model. The structural figures were prepared with PyMOL (http://www.pymol.org). Protein structural disorder predictions were performed with VL3H [http://www.ist.temple.edu/disprot/Predictors.html; (28)].
DNA substrates
Oligonucleotides used as DNA substrates (Supplementary Table S1) were synthesized by Operon and purified by PAGE before use. Double-stranded DNA substrates were prepared by mixing a 5 μM solution of a 5′-fluorescein- or alexa-labeled oligonucleotide (upper-strand) with a 10 μM solution of an unlabeled oligomer (lower-strand), heating to 95°C for 5 min and slowly cooling to room temperature. Annealing reactions for the preparation of the 1-nt gapped duplex were carried out at 95°C for 5 min in the presence of a 2-fold molar excess of both unlabeled 5′-phosphorylated oligonucleotide (P30_51) and unlabeled oligonucleotide (CGR) with respect to the 5′-alexa-labeled 3′-phosphorylated oligonucleotide (Al-28P), followed by cooling to room temperature.
Production of ROS1 variants derivatives
Site-directed mutagenesis was performed using the Quick-Change II XL kit (Stratagene). The mutations were introduced into the expression vector pET28a (Novagen) containing the full-length wild-type (WT) ROS1 cDNA using specific oligonucleotides (Supplementary Table S2). Mutational changes were confirmed by DNA sequencing and the constructs were used to transform Escherichia coli BL21 (DE3) dcm− Codon Plus cells (Stratagene). WT and mutant versions were expressed and purified as N-terminal His-tagged proteins, as previously described (21) (Supplementary Figure S2). Protein stability was measured by limited proteolysis with thermolysin (29). WT and mutant proteins (160 µM) were preincubated (4 h) or not under DNA glycosylase assay conditions (see below) in the absence of DNA substrate, and then digested for 5 min with 5 µg/ml thermolysin. Samples were analyzed by SDS/PAGE and relative band intensities were used to estimate the percentage of stable protein remaining after pre-incubation (Supplementary Figure S2).
Enzyme activity assays
Double-stranded oligodeoxynucleotides (20 nM, unless otherwise stated) were incubated at 30°C for the indicated times in a reaction mixture containing 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 0.1 mg/ml BSA, and the indicated amounts of WT ROS1 or mutant variant in a total volume of 50 μl. When reactions included AP endonuclease 1 (APE 1, 5 U; New England BioLabs), EDTA was omitted and 5 mM mM MgCl2 was added. Reactions were stopped by adding 20 mM EDTA, 0.6% sodium dodecyl sulphate and 0.5 mg/ml proteinase K, and the mixtures were incubated at 37°C for 30 min. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol precipitated at −20°C in the presence of 0.3 mM NaCl and 16 μg/ml glycogen. Samples were resuspended in 10 μl of 90% formamide and heated at 95°C for 5 min. When measuring DNA glycosylase activity, samples were treated with NaOH 100 mM and immediately transferred to 90°C for 10 min. After adding an equal volume of 90% formamide, samples were heated at 95°C for 5 min. Reaction products were separated in a 12% denaturing polyacrylamide gel containing 7 M urea. Fluorescein-labeled DNA was visualized in a FLA-5100 imager and analyzed using Multigauge software (Fujifilm).
When measuring AP lyase activity, a fluorescein-labeled oligonucleotide duplex containing U opposite G (200 nM) was incubated at 30°C for the indicated times in a reaction mixture containing 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 0.1 mg/ml BSA, 2.5 U of E. coli Uracil DNA glycosylase (New England BioLabs), and the indicated amounts of WT ROS1 or mutant variant in a total volume of 5 μl. Reactions were stopped by adding 20 mM EDTA, 0.6% sodium dodecyl sulphate, and 0.5 mg/ml proteinase K. After adding 10 μl of 90% formamide, samples were heated at 95°C for 5 min. Products were resolved and analyzed as described above.
Kinetic analysis
As we have shown previously (20,21) ROS1 does not exhibit significant turnover in vitro due to strong product binding, and therefore a simple Michaelis–Menten model is inadequate for a correct kinetic analysis of this enzyme. Accordingly, we have used a previously described method (30) successfully employed to measure and compare single-turnover kinetics with different orthologs of thymine DNA glycosylase (TDG) (31). The standard reaction conditions were equimolar (20 nM) enzyme/substrate ratios and incubation at 30°C. Data were fitted to the equation [Product] = Pmax[1 – exp(–kt)] using non-linear regression analysis and the software Sigmaplot. For each mutant enzyme and substrate, we determined the parameters Pmax (maximum substrate processing within an unlimited period of time), T50 (the time required to reach 50% of the product plateau level, Pmax), and the relative processing efficiency (Erel = Pmax/T50). Representative examples of 5-meC DNA glycosylase assays and kinetic analysis are shown in Supplementary Figure S3.
Electrophoretic mobility shift assay
Standard band-shift reactions were performed with the indicated amounts of protein and 100 nM fluorescein- and/or alexa-labeled duplex oligonucleotides. Binding reactions were carried out at 25°C for 60 min, unless otherwise stated, in 10 nM Tris–HCl pH 8.0, 1 mM DTT, 10 μg/ml BSA, 1 mM EDTA, in a final volume of 10 μl. Complexes were electrophoresed through 0.2% agarose gels in 1× TAE. Electrophoresis was carried out in 1× TAE for 40 min at 80 V at room temperature. Fluorescein- and/or alexa-labeled DNA was visualized in a FLA-5100 imager and analyzed using Multigauge software (Fujifilm).
RESULTS
An unusual sequence insertion is present in the DNA glycosylase domain of DML proteins
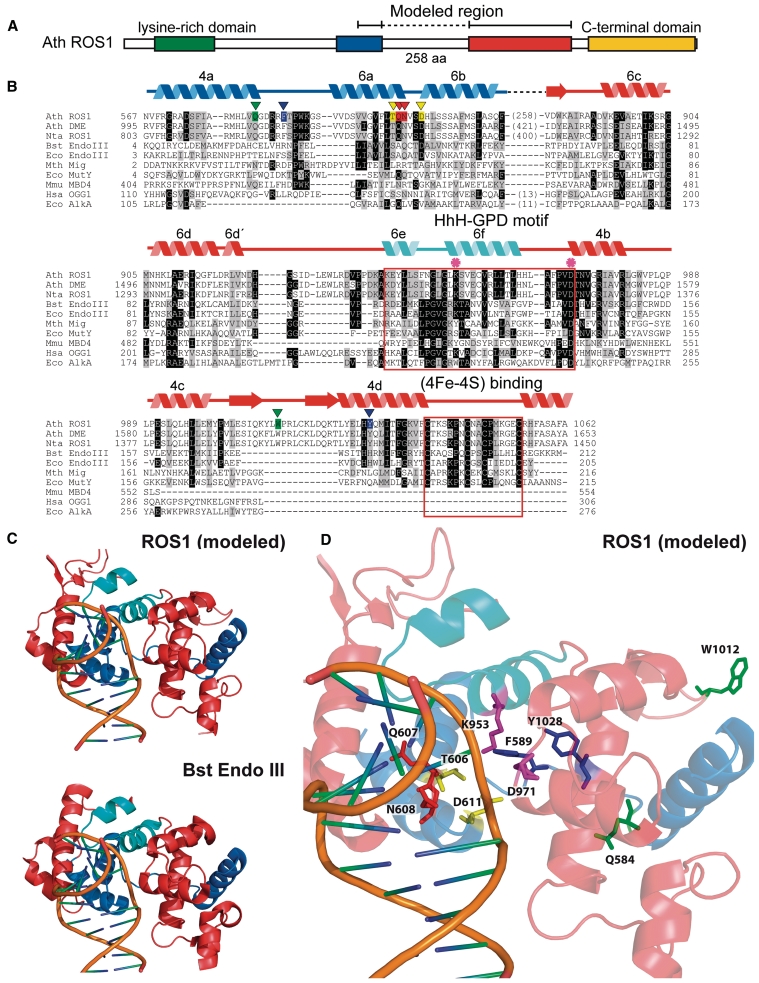
To gain insight into residues that comprise the DNA glycosylase domain of DML proteins, we performed a multiple sequence alignment that included Arabidopsis ROS1 and DME, Nicotiana tabacum ROS1, and several HhH-GPD proteins (Figure 1). The alignment revealed that sequence similarity to HhH-GPD enzymes in DML proteins is actually distributed over two non-contiguous segments. The first segment corresponds to a region that in HhH-GPD members contains the base-flipping wedge and its flanking alfa-helixes, as well as some of the residues that line the active site pocket (32). The second segment includes the HhH-GPD motif and its invariant aspartate, which is absolutely required for catalysis of 5-meC excision by DML proteins (10,11,13,14). This region also contains a lysine residue that is only present in the subset of HhH-GPD proteins with bifunctional DNA glycosylase/lyase activity (19) and a [4Fe–4S] cluster loop (FCL) motif that in some HHh-GPD proteins, such as E. coli Endo III and MutY, ligates a [4Fe–4S] cluster (19) (Figure 1B). The two separate segments with sequence similarity to HhH-GPD proteins are interconnected by a non-conserved linker region that is highly variable in sequence and length among members of the DML family (Figure 1B and Supplementary Figure S1). We applied a well-characterized disorder predictor [VL3H (28)] to analyze the location of ordered and disordered regions in ROS1 (Supplementary Figure S4). The results predict a high disorder content within this linker region, thus suggesting that it is intrinsically unstructured under native conditions.
Figure 1.
An unusual sequence insertion is present in the DNA glycosylase domain of members of the DML family. (A) Schematic diagram showing ROS1 regions conserved among DML proteins. (B) Multiple sequence alignment of DML proteins and several HhH-GPD superfamily members. Listed above the primary sequence are indicated secondary structure assignments from the ROS1 model prediction shown in (C), colored according to regions shown in (A). The helix–hairpin–helix of the HhH-GPD motif is shown in cyan. ROS1 amino acids mutated in this study are indicated by inverted triangles and highlighted in green (Q584 and W1012), blue (F589 and Y1028), yellow (T606 and D611) or red (Q607 and N608). The lysine residue that is diagnostic of bifunctional glycosylase/lyase activity, and the conserved aspartic acid residue in the active site are indicated by asterisks. The HhH-GPD and the [4Fe–4S] cluster loop (FCL) motifs are boxed. Names of organisms are abbreviated as follows: Ath, Arabidopsis thaliana; Nta, Nicotiana tabacum; Bst, Bacillus stearothermophilus; Eco, Escherichia coli; Mth, Methanobacterium thermoautotrophicum; Mmu, Mus musculus; Hsa, Homo sapiens. Genbank accession numbers are: Ath ROS1: AAP37178; Ath DME: ABC61677; Nta ROS1: BAF52855; Bst EndoIII: 1P59; Eco EndoIII: P20625; Mth Mig: NP_039762; Eco MutY: NP_417436; Mmu MBD4: 1NGN; Hsa OGG1: O15527; Eco AlkA: P04395. (C) Ribbon diagrams of the structural model for the DNA glycosylase domain of ROS1 and the crystallographic Bst EndoIII structure used as template. Structural elements are colored as in (A). The duplex DNA is shown in orange. Nucleic acid coordinates extracted from the Bst EndoIII-DNA trapped complex were used to superimpose a DNA structure with a flipped-out AP site analog onto the ROS1 model. (D) Close-up view of the ROS1 model. Mutated residues are shown as sticks and colored according to (B). The conserved lysine and aspartic acid residues are shown in magenta.
We used the crystal structure of Bacillus stearothermophilus Endonuclease III [Protein Data Bank accession code: 1P59, (27)] as a template to generate a 3D model structure of the two ROS1 polypeptide segments that show sequence similarity to HhH-GPD proteins (amino acids 567–625 and 883–1062) (‘Materials and Methods’ section). The model predicts a typical HhH-GPD core structure with two alpha domains: a six-helix barrel domain (6a–6f), and a four-helix domain formed by one N-terminal (4a) and three C-terminal (4b–4d) helixes (Figure 1). The non-conserved linker region of 258 amino acids is inserted between helixes 6b and 6c, which are part of the characteristically sequence-continuous six-helix barrel domain (Figure 1). Two other HhH-GPD proteins (AlkA and Ogg1) contain at this same location position a much shorter insertion (13 and 11 amino acids, respectively) (Figure 1). The model also predicts a second DML-specific insertion between helixes 4c and 4d. Thus, homology modeling predicts that proteins of the DML family have a discontinuous DNA glycosylase domain structure interrupted by an unusually long insertion.
T606 and D611 are critical residues for ROS1 DNA glycosylase activity
In order to identify residues specifically involved in 5-meC recognition and catalysis, we used this tentative model as a guide to design site-specific mutations of the ROS1 DNA glycosylase domain. Following a general approach that has been well detailed elsewhere (33), we searched not only for residues conserved among DML proteins and other HhH-GPD enzymes, but also for residues specifically conserved within the DML group. The former class may be important for the general catalytic mechanism, whereas the latter may contribute to specific recognition of 5-meC.
In E. coli Endonuclease III, amino acids S39 and D44 are both required for catalytic activity (34), and their homologous residues in ROS1 are T606 and D611, respectively (Figure 1B). The modeled structure of ROS1 predicts that T606 and D611 are positioned at the mouth of the groove that separates the six-helix barrel domain and the four-helix domain (Figure 1D). In HhH-GPD DNA glycosylases this groove lies between the DNA base stack from which the lesion is extruded and the base recognition pocket where it is inserted (35), and contains residues suitably disposed to access the N-glycosyl bond of the flipped base. To test the prediction that T606 and D611 have a role in catalysis, we mutated them to Leu (T606L) and Val (D611V), respectively.
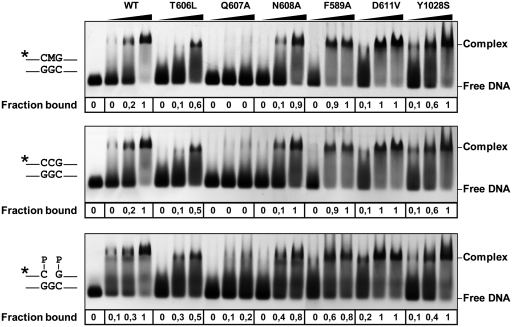
We examined the ability of WT and mutant proteins to process a 51-mer duplex oligo substrate that contained either 5-meC or T opposite G at position 29 in a CG context (Table 1). Consistent with our previously reported observations (10,21) we found that WT ROS1 processed 5-meC with higher relative processing efficiency than T (Table 1). We found that both the T606L and D611V mutations abolished the catalytic activity of ROS1 on both substrates (Table 1). For both ROS1 variants, neither 5-meC:G nor T:G processing was detected even after prolonged incubation times (data not shown). The DNA-binding capacity of WT and mutant proteins was assessed by electrophoretic mobility shift assay with different labeled substrates (Figure 2). As previously reported, ROS1 bound with similar efficiency to methylated and non-methylated DNA, and displayed a higher affinity for the 1-nt gapped reaction product. The T606L mutant enzyme exhibited a somewhat reduced binding activity compared to that of WT ROS1, whereas the D611V variant displayed higher binding capacity (Figure 2).
Table 1.
Relative substrate processing efficiencies of WT and mutant variants of ROS1a
| ROS1 variant | 5-meC:G |
T:G |
||||
|---|---|---|---|---|---|---|
| Pmax (nM) | T50 (h) | Erel | Pmax (nM) | T50 (h) | Erel | |
| WT | 10.24 ± 0.17 | 3.30 | 3.10 ± 0.05 | 7.64 ± 0.18 | 4.83 | 1.58 ± 0.04 |
| Q584L | 12.23 ± 0.45 | 5.07 | 2.41 ± 0.09 | 9.93 ± 0.51 | 5.42 | 1.83 ± 0.09 |
| F589A | 1.84 ± 0.11 | 5.10 | 0.36 ± 0.02 | 3.22 ± 0.21 | 4.76 | 0.68 ± 0.04 |
| T606L | n.d.b | n.a.c | n.a. | n.d. | n.a. | n.a. |
| Q607A | 1.19 ± 0.09 | 2.63 | 0.45 ± 0.04 | 0.29 ± 0.04 | 0.62 | 0.48 ± 0.06 |
| N608A | 13.64 ± 0.35 | 4.73 | 2.88 ± 0.07 | 12.07 ± 0.33 | 9.38 | 1.29 ± 0.04 |
| D611V | n.d. | n.a. | n.a. | n.d. | n.a. | n.a. |
| W1012A | 7.99 ± 0.27 | 5.01 | 1.60 ± 0.06 | 5.41 ± 0.45 | 6.62 | 0.82 ± 0.07 |
| Y1028S | 7.76 ± 0.22 | 5.11 | 1.50 ± 0.04 | 10.87 ± 0.62 | 5.66 | 1.92 ± 0.11 |
aPurified proteins (20 nM) were incubated at 30°C with 51-mer double-stranded oligonucleotide substrates (20 nM) containing either a single 5-meC:G pair or a T:G mispair. Reaction products were separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. Shown are the plateau levels of substrate nicking (Pmax) and the time required for processing of 50% of Pmax (T50). Relative processing efficiency was calculated as Erel = Pmax/T50. Values are mean ± SE from two independent experiments.
bn.d., none detected.
cn.a., not applicable.
Figure 2.
Binding of WT and mutant ROS1 proteins to substrate and product DNA. DNA-binding reactions were performed incubating increasing concentrations of WT ROS1 or mutant variants with 100 nM of fluorescein-labeled 5-meC:G substrate (upper panel), alexa-labeled homoduplex (center panel) or alexa-labeled 1-nt-gapped duplex product (lower panel). After nondenaturing gel electrophoresis, the gel was scanned to detect fluorescein- or alexa-labeled DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated. The fraction of bound DNA is indicated below each lane. The asterisk depicts 5′-end labeling of the upper strand. M: 5-meC.
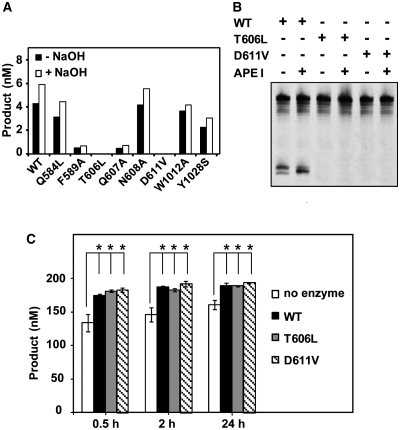
Since ROS1 is a bifunctional enzyme, we asked whether these mutant proteins lack DNA glycosylase activity, lyase activity or both. To differentiate 5-meC excision and strand incision, we analyzed the reaction products generated by different ROS1 variants with or without additional alkaline treatment with NaOH (Figure 3A). Incisions in the absence of NaOH reveal the combined DNA glycosylase/AP lyase action of the enzyme, whereas the alkaline treatment cleaves all AP sites generated by the enzyme and reflects DNA glycosylase activity. Consistent with our previously reported observations (21), we found that the amount of incision products generated by WT ROS1 was only slightly increased after NaOH treatment, thus suggesting that glycosyl bond scission is usually coupled to the AP lyase step. The same pattern was observed for all ROS1 mutant enzymes except T606L and D611V, which did not generate detectable incision products either in the absence or the presence of NaOH. The incapacity of both mutants to generate abasic sites was confirmed by performing reactions in the presence of human AP endonuclease APE1 (Figure 3B). We next tested whether T606L or D611V retained AP lyase activity by incubating both proteins with a 51-mer duplex oligo substrate that contained an AP site opposite G at position 29 in a CG context (Figure 3C). Although a significant level of spontaneous AP incision is observed in the absence of enzyme, the amounts of enzyme-dependent strand incision after 0.5, 2 and 24 h incubation were similar to those generated by WT ROS1 (Figure 3C; a representative gel is shown in Supplementary Figure S5). We also found that the incision product generated by both WT and mutant proteins migrates as a β-elimination product (Supplementary Figure S5). Altogether, these results indicate that both T606L and D611V mutants lack DNA glycosylase activity but retain AP lyase activity. In addition, they provide experimental evidence of the location of critical catalytic residues in the first segment of the discontinuous ROS1 DNA glycosylase domain.
Figure 3.
T606 and D611 are essential for ROS1 DNA glycosylase activity. (A) The generation of incision products was measured by incubating purified WT ROS1 or mutant variants (20 nM) at 30°C for 2 h with a double-stranded oligonucleotide substrate (20 nM) containing a single 5-meC:G pair. Samples were treated with or without NaOH 100 mM, and immediately transferred to 90°C for 10 min. Products were separated in a 12% denaturing polyacrylamide gel and the amounts of incised oligonucleotide were quantified by fluorescent scanning. (B) Purified WT ROS1 or mutant variants (20 nM) were incubated at 30°C for 2 h with a double-stranded oligonucleotide substrate (20 nM) containing a single 5-meC:G pair, either in the absence or the presence of human APE I (5 U), as indicated. Products were separated in a 12% denaturing polyacrylamide gel and the incised products were detected by fluorescent scanning. (C) A double-stranded oligonucleotide substrate containing an AP site opposite G (200 nM) was incubated at 30°C either in the absence of enzyme or in the presence of purified WT ROS1, T606L or D611V (100 nM). Reactions were stopped at the indicated times, products were separated in a 12% denaturing polyacrylamide gel and the amount of incised oligonucleotide was quantified by fluorescent scanning. Values are means ± SE (error bars) from two independent experiments. The asterisks indicate that the incision levels were significantly different (P < 0.05) from those observed in the absence of enzyme. The respective P-values were calculated using a Student’s unpaired t-test.
F589A and Y1028S mutations change ROS1 preference for 5-meC over T
To investigate residues possibly contributing to 5-meC specificity, we focused on two aromatic amino acids (F589 and Y1028) that are specifically conserved within the DML family (Figure 1B). Their positions correspond to residues that in other HhH-GPD enzymes interact with the lesion base (27,32,36), and the modeled ROS1 structure suggests that they are located in the base binding pocket of the enzyme (Figure 1D). To test the prediction that F589 and Y1028 are involved in the specific recognition of 5-meC, we substituted them with Ala (F589A) and Ser (Y1028S), respectively. Both F589A and Y1028S displayed a somewhat higher non-specific DNA-binding capacity than the WT protein (Figure 2).
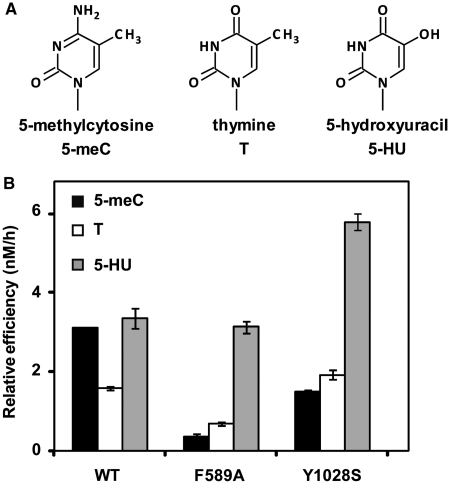
We found that mutation Y1028S produced a protein with a 2-fold reduced efficiency on 5-meC:G pairs, but with slightly increased activity on T:G mispairs (Table 1). On the other hand, the F589A mutation reduced enzyme efficiency on 5-meC:G ∼9-fold, but decreased activity on T:G only 2.3-fold (Table 1). As a result, both F589A and Y1028S exhibit a higher preference for T over 5-meC (Figure 4), which is just the opposite of the base specificity characteristic of WT ROS1 and its homologs (10,14). We also tested the effect of both mutations on excision of 5-HU and found that the mutant protein F589A exhibited a similar activity to that of WT ROS1, whereas Y1028S displayed ∼2-fold increased efficiency (Figure 4). ROS1 activity is strongly inhibited by replacement of the C5 methyl group by halogen derivatives (21), and we confirmed that all ROS1 variants, including F589A and Y1028S, retained a very low activity on 5-BrC and 5-BrU (Supplementary Figure S6). Altogether, these results indicate that F589 and Y1028 contribute to ROS1 preference for 5-meC over T, and suggest that they may be located in the base specificity pocket of the enzyme.
Figure 4.
F589 and Y1028 contribute to 5-meC specificity. (A) Chemical structures of substrate DNA bases tested. (B) Substrate processing ability of WT ROS1 and the mutant variants F589A and Y1028S. Relative processing efficiencies were determined in kinetic assays as described in ‘Materials and Methods’ section. Purified proteins (20 nM), were incubated at 30°C with 51-mer double-stranded oligonucleotide substrates (20 nM) containing at position 29 of the labeled upper-strand different target DNA bases paired with G. Reaction products were separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. Values are means ± SE (error bars) from two independent experiments.
Q607 is required for ROS1 base excision activity and stable DNA binding
HhH-GPD DNA glycosylases use a loop between two α-helixes to wedge into the minor groove of DNA, thus helping to extrude the lesion from the base stack (32). Despite structural conservation, the amino acid sequence of this loop, and in particular the identity of the residue that fills the vacant space left behind by the flipped base, varies widely among the HhH-GPD enzymes. Thus, the base-flipping residue is Q42 in Bst EndoIII (27), N149 in Hsa Ogg1 (37) and L125 in Eco AlkA (38) The alignment in Figure 1B shows that the homologous position in ROS1 corresponds to N608, and the modeled ROS1 structure suggests that N608 is located close to the DNA base stack (Figure 1D). However, we found that a N608A variant retained full catalytic activity, exhibiting the same processing efficiency than WT ROS1 on both 5-meC:G and T:G (Table 1), as well as a comparable DNA-binding capacity (Figure 2). Since mutation of the base flipping residue invariably causes a significant reduction in catalytic activity of DNA glycosylases (39–41), these results argue against the accuracy of our homology-based structure prediction in this portion of the ROS1 DNA glycosylase domain. This is not unexpected, given the low sequence conservation of the base flipping loop in HhH-GPD enzymes, and the high variability in the identity of the specific residue inserted into the DNA helix.
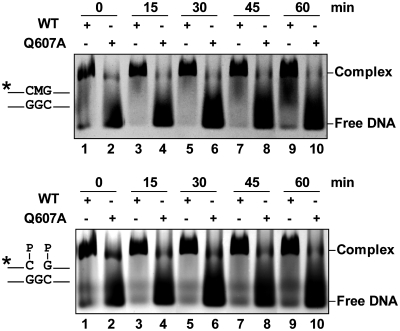
We searched for an alternative candidate residue conserved in DML proteins and, since most glycosylases use a bulky-side chain as nucleotide flipper, we decided to mutagenize the contiguous Q607. Mutation of Q607 to A resulted in an enzyme with strongly reduced activity on substrates containing either 5-meC or T (Table 1). The observed reduction in relative processing efficiency is higher for the 5-meC:G pair (6.9-fold) than for the T:G mispair (3.3-fold). Furthermore, and unlike all other ROS1 variants tested in this study, the Q607A mutant exhibited drastically reduced DNA binding to both methylated and non-methylated DNA, as well as to the 1-nt gapped reaction product (Figure 2). We reasoned that the Q607A mutation might compromise the stability of the protein–DNA complex. In order to investigate this possibility, we performed DNA-binding measurements at different time points to analyze the ability of Q607A to remain bound to DNA (Figure 5 and Supplementary Figure S7). We found that the mutant protein formed a detectable complex with either the methylated substrate or the gapped reaction product, but rapidly dissociated from both DNA probes. Altogether, these results indicate that Q607 is essential for base excision activity and stable DNA binding, and strongly support the hypothesis that it is a DNA-intercalating residue.
Figure 5.
Q607 is required for stable ROS1 binding to substrate and product DNA. Purified WT ROS1 or mutant variant Q607A (100 nM) were incubated with a mixture of 100 nM fluorescein-labeled 5-meC:G substrate and 100 nM alexa-labeled 1-nt gapped product, and the reactions were monitored for 60 min. After non-denaturing gel electrophoresis, the gel was scanned to detect fluorescein- (upper panel) or alexa-labeled (lower panel) DNA. Protein–DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated. The asterisk depicts 5′-end labeling of the upper strand. M: 5-meC.
N608 modulates 5-meC excision in different sequence contexts
Since plant DNA methylation is deposited in different sequence contexts, it is not surprising that DML proteins exhibit the capacity to excise 5-meC at CG, CHG and CHH sequences (10–14). The context specificity of DML family members has not been exhaustively characterized, but excision of 5-meC in vitro is apparently more efficient on those sequences more likely to be methylated in vivo. For example, DME and ROS1 remove 5-meC from a CHG context more efficiently when H = A than when H = C (10), in agreement with the fact that CCG is the sequence showing the lowest methylation level among CHG sites (42).
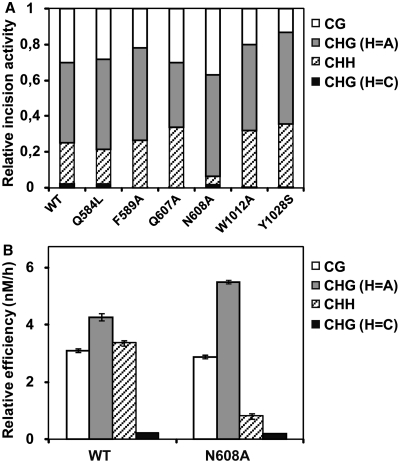
In order to determine whether any of the mutated residues contributes to sequence-context specificity, we tested all ROS1 variants for their relative capacity to excise 5-meC from CG, CHG and CHH sequences (Figure 6A). We found that all mutants except N608A exhibited a sequence-context specificity similar to that of WT ROS1. Unlike the WT enzyme and the rest of ROS1 variants, the N608A mutant showed a significantly reduced activity on the asymmetric CHH context (Figure 6A). To confirm this result, we performed a kinetic analysis to compare the relative processing efficiencies of WT ROS1 and N608A on 5-meC located in CG, CHG or CHH contexts (Figure 6B). We found that both enzymes displayed a similar efficiency on CG sites, and also showed a very low activity on the CHG context when H = C. However, the N608A mutation caused a higher efficiency than WT ROS1 on CAG sequences, and a strongly reduced activity on the asymmetric CHH context. These results indicate that N608 contributes to the sequence context specificity of ROS1, and suggest that this residue may contact the DNA bases surrounding the 5-meC.
Figure 6.
N608 contributes to sequence-context specificity. (A) Purified WT ROS1 or mutant variants (20 nM) were incubated at 30°C for 4 h with 51-mer double-stranded oligonucleotide substrates (20 nM) containing at position 29 of the labeled upper-strand a 5-meC residue in different sequence contexts. Products were separated in a 12% denaturing polyacrylamide gel and the amount of incised oligonucleotide was quantified by fluorescent scanning. For ease of comparison, the incision values for each substrate are normalized to the total incision detected in all four substrates for each individual enzyme. (B) Substrate processing ability of type ROS1 and the mutant variant N608 in different sequence contexts. Relative processing efficiencies were determined in kinetic assays as described in ‘Materials and Methods’ section. Purified proteins (20 nM), were incubated at 30°C with 51-mer double-stranded oligonucleotide substrates (20 nM) containing at position 29 of the labeled upper-strand a 5-meC residue in different sequence contexts. Reaction products were separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. Values are means ± SE (error bars) from two independent experiments.
Functional consequences of Q584L and W1012A mutations
We also tested the functionality of Q584 and W1012 residues. Q584 is located at an LVQ motif that is also present in mammalian MBD4 proteins (Figure 1B). The homologous Q423 in murine MBD4 is positioned in the active site, and modeling studies suggest it can make a hydrogen bond to the protonated N-3-H of thymine (43). W1012 is positioned in a short DML-specific sequence insertion between helixes 4c and 4d (Figure 1B). We substituted these two residues with Leu (Q584L) and Ala (W1012A), respectively. Both variants retained the same DNA-binding capacity than WT ROS1 (data not shown) and a similar base specificity (Supplementary Figure S6). The relative processing efficiency of Q584L was somewhat reduced on 5-meC:G pairs, and slightly increased on T:G mispairs, whereas W1012A exhibited ∼2-fold reduced activity on both 5-meC:G and T:G (Table 1). For both mutant enzymes, however, 5-meC remained as the preferred target.
DISCUSSION
A DNA glycosylase domain with a bipartite structural organization
A main finding of this study is that the DNA glycosylase domain of ROS1 and its homologs is composed of two non-contiguous segments connected together through a linker region that is highly variable in sequence and length across members of the DML family. It should be noted that a much shorter sequence insertion is present at an homologous position in two other DNA glycosylases (OGG1 and AlkA) (37,38), which suggests that this location has undergone a much more limited sequence expansion in other HhH-GPD enzymes. A remarkable feature of the linker region that connects the two DNA glycosylase segments in ROS1 is its predicted intrinsic disorder. Intrinsically disordered regions lack well defined conformation under native conditions and are common in a significant proportion of eukaryotic proteins (44). We can only speculate about the possible functions of such an unfolded region in the functionality of ROS1 and other DML proteins. A possibility is that the disordered link helps the protein to find its target. Thus, it has been proposed that a protein with disordered regions may sample the surrounding solution in search of a binding site with a higher capture radius than in the folded state, in a mechanism known as ‘fly-casting’ (45). In such a scenario, ROS1 would be partially unfolded before a productive encounter with DNA, and folding would be induced by DNA binding.
The first segment of the ROS1 discontinuous DNA glycosylase domain contains two essential residues for catalytic activity
We have found that both T606 and D611 residues are critical for ROS1 glycosylase catalysis, but they are dispensable for AP lyase activity and DNA binding. Their homologous residues in E. coli Endonuclease III, S39 and D44 respectively, are also required for catalytic activity (34). Thus, an S39L mutation abolishes the glycosylase activity of E. coli Endonuclease III but does not affect its AP lyase activity (34), which agrees with our data. However, a D44L mutant Endo III retains glycosylase activity but exhibits a greatly reduced lyase activity (34), which is the opposite result to what we found with a ROS1 D611L mutant. Such a discrepancy indicates that there must be specific differences in the precise catalytic mechanism followed by both enzymes. This is not unexpected, given the nature of their reaction products; whereas Endonuclease III and its orthologs only generate β-elimination products (46), in all DML proteins tested to date a significant amount of β-elimination incisions proceed to β,δ-elimination products (10–14). In any case, the fact that both T606 and D611 are required for catalytic activity strongly suggests that the first segment of what we propose as a bipartite DNA glycosylase domain truly contains residues located at the active site of ROS1.
The aromatic residues F589 and Y1028 are strong candidates for interaction with 5-meC in the base specificity pocket
In this work we have also aimed to identify molecular determinants of ROS1 substrate specificity. Similarly to other DNA glycosylases, such specificity is probably governed by direct contacts between the target base and residues in the active site pocket of the enzyme. Our homology modeling analysis allows a tentative identification of several residues that are likely located in the 5-meC-binding pocket of ROS1. Among these, we selected two amino acids (F589 and Y1028) that were specifically conserved in the DML family but not in the remaining HhH-GPD proteins. We have found that replacing either of these two residues (F589 to A or Y1028 to S) changes ROS1 substrate preference from 5-meC:G to T:G.
Our results are consistent with a role of F589 and Y1028 in base substrate specificity. Nevertheless, it could be also argued that, since melting a 5-meC:G base pair is less favorable than melting a T:G mispair, any shifting in preference in favor of T might be alternatively explained by a reduced base flipping efficiency irrespective of the substrate base. However, this hypothesis does not agree with the fact that the activity against a 5-HU:G mispair is unchanged in the F589A mutant, and is even higher than the WT in the Y1028S mutant (Figure 4). A reduced base flipping efficiency would be also difficult to reconcile with the fact that both F589A and Y1028S mutants do not display a reduced DNA-binding capacity (Figure 2). As discussed below, mutagenesis studies consistently report reduced DNA binding in base-flipping deficient mutants, both in DNA glycosylases and in other proteins that also rotate bases. Therefore, our results suggest that F589 and Y1028 play a role in base substrate specificity rather than base flipping.
Obviously, in the absence of detailed structural information it is not possible to identify the precise interactions providing the basis for specific base recognition However, it is conceivable that the aromatic side chains of F589 and/or Y1028 could help to stabilize the flipped-out 5-meC into the substrate-binding pocket of ROS1 through stacking interactions. Such stacking interactions have been suggested to be important in the recognition and binding of alkylated base lesions by 3-methyladenine DNA glycosylase MagIII (47). Interestingly, stacking interactions have also been reported as relevant for specific recognition of 5-meC by UHRF1, a mammalian protein that binds hemimethylated sites and is required for maintenance of DNA methylation (48–50). UHRF1 does not perform any catalytic reaction on 5-meC, but its SRA domain flips the methylated base out of the DNA helix and places it in a tight binding pocket, stacked between two aromatic residues (48–50).
N608 may contact bases adjacent to 5-meC
To our knowledge, no detailed information has been reported about the sequence context preference of any HhH-GPD DNA glycosylase. The only relevant data available pertain to human TDG, a DNA glycosylase belonging to another structural superfamily (51). TDG excises T from T:G mismatches with a preference for a CG context, and the crystal structure of the enzyme has revealed that part of this sequence-context specificity is due to contacts between base pairs neighboring the T:G mismatch and amino acids adjacent to the enzyme base-flipping residue (52).
ROS1 removes 5-meC from CG, CHG and CHH contexts (10–14), although shows a strongly reduced activity on a CHG sequence when H = C (10). We tested all ROS1 variants for altered context specificity and found that all of them retained a very low preference for CCG sequences. However, the N608A mutant exhibited a higher activity than WT ROS1 on a CAG context and a marked lower efficiency on an asymmetric CHH context. Our homology analysis initially predicted that N608 is the base-flipping residue of ROS1, but this hypothesis is unlikely since the N608A mutant retains both full catalytic activity and DNA-binding capacity. In fact, our results rather suggest that ROS1 uses the side chain of the contiguous amino acid (Q607) for base flipping (see below). However, given that these two residues are adjacent on the primary ROS1 structure, it is very likely that N608 forms part of the wedge used by ROS1 to contact the DNA minor groove. Altogether our results suggest that N608 is probably located in a position suitable disposed to make contacts with DNA bases surrounding the methylated cytosine.
Q607 is a putative base flipping residue required for stable DNA binding
Our results suggest that Q607 functions as a critical anchor to stabilize the protein–DNA complex, and support the hypothesis that ROS1 uses this residue to flip out 5-meC and compensates its extrusion by filling in the vacant space in the DNA base stack. Base flipping is a widespread mechanism used to gain access to the DNA helix by those proteins that need to interact with the bases rather than the phosphodiester backbone (53). Accumulating evidence point to the conclusion that such an extrusion process also plays a key role in stabilizing protein–DNA complexes. Thus, a consistent result repeatedly observed with structurally different DNA glycosylases is that mutating their base-flipping residue strongly reduces not only their base excision activity, but also their DNA-binding capacity (39–41), just as we observe with the ROS1 Q607A mutant. The relevance of base flipping for stable DNA binding has been also documented for cytosine-5 DNA methyltransferases (54), and there is evidence that also plays an important role in proteins that do not perform chemistry on bases. Thus, a mutation in the base-flipping residue of the SRA domain of UHRF1 results in a protein with significantly lower affinity for DNA (50). Recently, it has been reported that base flipping is also essential for stable DNA binding of MTERF1, a human mitochondrial transcriptional terminator (55).
Interestingly, we have found that the Q607A mutation is not only detrimental for stable ROS1 binding to methylated substrates but also for non-specific binding to unmethylated DNA. We have recently reported that methylation-independent DNA binding by ROS1 is largely mediated by a lysine-rich domain located at the amino terminus of the enzyme (20). The results reported here indicate that this domain is necessary but not sufficient for stable DNA binding. Furthermore, they strongly suggest that base flipping is a feature of both specific and non-specific DNA binding by ROS1, thus hinting at the possibility that the enzyme performs extrahelical interrogation of unmethylated base pairs.
It has been suggested that DNA glycosylases operating on modifications causing little or no disturbance of the DNA helix must extrude every base they encounter to recognize their target (56). Recent views, mostly gained through a combination of biophysical and structural approaches with the well-studied DNA glycosylases UNG, hOGG1 and MutM (57–59), propose that interrogation of normal bases is a transient phase of a general multi-step mechanism for base damage search and detection (60). The process would initially involve DNA sliding by the DNA glycosylase in a conformation designed as the ‘search complex’, followed by formation of a transient ‘interrogation complex’ that would extrude normal and damaged bases for inspection, and finally the conversion to a catalytically productive ‘excision complex’ upon encountering the cognate base modification (60). Unlike UNG, hOGG1 or MutM, the non-specific complexes of ROS1 with DNA do not dissociate rapidly and are fairly stable. The capacity to form stable non-specific complexes is also found in other DNA glycosylases such as TDG (51,61). It is therefore possible that each enzyme have evolved a different balance among the relative magnitudes of the search, interrogation, and excision stages of base repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science and Innovation and the European Regional Development Fund (grant number BFU2007-60956/BMC); Junta de Andalucía, Spain (grant number P07-CVI-02770); Junta de Andalucía, PhD Fellowship (to M.I.P.-M.); Fellowship for Initiation in Research from the University of Córdoba (to J.T.P.-D.). Funding for open access charge: Spanish Ministry of Science and Innovation and the European Regional Development Fund (grant number BFU2007-60956/BMC); Junta de Andalucía, Spain (grant number P07-CVI-02770).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank members of their laboratory for helpful discussions and advice.
REFERENCES
- 1.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 2.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roldan-Arjona T, Ariza RR. DNA demethylation. In: Grosjean H, editor. DNA and RNA modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution. Austin, TX: Landes Bioscience; 2009. pp. 149–161. [Google Scholar]
- 4.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson KD. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 7.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 8.Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 9.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 10.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl Acad. Sci. USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl Acad. Sci. USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl Acad. Sci. USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 19.Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 20.Ponferrada-Marin MI, Martinez-Macias MI, Morales-Ruiz T, Roldan-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J. Biol. Chem. 2010;285:23032–23039. doi: 10.1074/jbc.M110.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponferrada-Marin MI, Roldan-Arjona T, Ariza RR. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 2009;37:4264–4274. doi: 10.1093/nar/gkp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Dalhus B, Laerdahl JK, Backe PH, Bjoras M. DNA base repair–recognition and initiation of catalysis. FEMS Microbiol. Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 24.Poirot O, O’Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 26.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 29.Park C, Marqusee S. Pulse proteolysis: a simple method for quantitative determination of protein stability and ligand binding. Nat. Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 30.Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J. Biol. Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 31.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mol CD, Arvai AS, Begley TJ, Cunningham RP, Tainer JA. Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases. J. Mol. Biol. 2002;315:373–384. doi: 10.1006/jmbi.2001.5264. [DOI] [PubMed] [Google Scholar]
- 33.Zharkov DO, Grollman AP. Combining structural and bioinformatics methods for the analysis of functionally important residues in DNA glycosylases. Free Radic. Biol. Med. 2002;32:1254–1263. doi: 10.1016/s0891-5849(02)00828-6. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe T, Blaisdell JO, Wallace SS, Bond JP. Engineering functional changes in Escherichia coli endonuclease III based on phylogenetic and structural analyses. J. Biol. Chem. 2005;280:34378–34384. doi: 10.1074/jbc.M504916200. [DOI] [PubMed] [Google Scholar]
- 35.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fondufe-Mittendorf YN, Harer C, Kramer W, Fritz HJ. Two amino acid replacements change the substrate preference of DNA mismatch glycosylase Mig.MthI from T/G to A/G. Nucleic Acids Res. 2002;30:614–621. doi: 10.1093/nar/30.2.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 38.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallur AC, Feller JA, Abner CW, Tran RK, Bloom LB. Effects of hydrogen bonding within a damaged base pair on the activity of wild type and DNA-intercalating mutants of human alkyladenine DNA glycosylase. J. Biol. Chem. 2002;277:31673–31678. doi: 10.1074/jbc.M204475200. [DOI] [PubMed] [Google Scholar]
- 40.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 41.Maiti A, Morgan MT, Drohat AC. Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. J. Biol. Chem. 2009;284:36680–36688. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu P, Qiu C, Sohail A, Zhang X, Bhagwat AS, Cheng X. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J. Biol. Chem. 2003;278:5285–5291. doi: 10.1074/jbc.M210884200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 45.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl Acad. Sci. USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailly V, Verly WG. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem. J. 1987;242:565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eichman BF, O’Rourke EJ, Radicella JP, Ellenberger T. Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. EMBO J. 2003;22:4898–4909. doi: 10.1093/emboj/cdg505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 50.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 51.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair. 2006;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl Acad. Sci. USA. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts RJ, Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 54.Estabrook RA, Lipson R, Hopkins B, Reich N. The coupling of tight DNA binding and base flipping: identification of a conserved structural motif in base flipping enzymes. J. Biol. Chem. 2004;279:31419–31428. doi: 10.1074/jbc.M402950200. [DOI] [PubMed] [Google Scholar]
- 55.Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verdine GL, Bruner SD. How do DNA repair proteins locate damaged bases in the genome? Chem. Biol. 1997;4:329–334. doi: 10.1016/s1074-5521(97)90123-x. [DOI] [PubMed] [Google Scholar]
- 57.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 59.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 60.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.