Abstract
A biomimetic replacement for tooth enamel is urgently needed because dental caries is the most prevalent infectious disease to affect man. Here, design specifications for an enamel replacement material inspired by Nature are deployed for testing in an animal model. Using genetic engineering we created a simplified enamel protein matrix precursor where only one, rather than dozens of amelogenin isoforms, contributed to enamel formation. Enamel function and architecture were unaltered, but the balance between the competing materials properties of hardness and toughness was modulated. While the other amelogenin isoforms make a modest contribution to optimal biomechanical design, the enamel made with only one amelogenin isoform served as a functional substitute. Where enamel has been lost to caries or trauma a suitable biomimetic replacement material could be fabricated using only one amelogenin isoform, thereby simplifying the protein matrix parameters by one order of magnitude.
Keywords: Biomimetic material, Biomineralization, ECM (extracellular matrix), Fracture toughness: Genetic engineering, Mechanical properties, Molecular biology
1. Introduction
Tooth enamel can be destroyed by caries, the most prevalent infectious disease of mankind [1, 2]. Today, enamel lost to caries, trauma or developmental anomaly is replaced by a range of man-made materials, since enamel biomimetic materials are not presently available. Biomineralization uses a protein matrix precursor to guide the habit and orientation of mineral crystallite thereby producing a tissue with unique properties [3, 4]. Enamel is the biomineralized tissue covering teeth. Unlike bone, enamel is formed by cells derived from ectoderm and is based upon a collagen-free matrix that guides the formation of a ceramic composite containing substituted hydroxyapatite that is the hardest tissue in the vertebrate body. Enamel does not undergo repair, remodeling or regeneration and enamel once made must last the lifetime to allow the organism to feed, reach sexual maturity and reproduce.
The unique physical properties of enamel are seen to be dependent upon the properties of the protein matrix precursor that guides mineral formation, which balances, or optimizes, the competing mechanical properties of hardness and toughness in the mature tissue. Ectoderm-derived ameloblast cells synthesize, secrete and assemble an extracellular protein matrix, composed predominantly of amelogenin along with much smaller amounts of non-amelogenin proteins [5, 6]. In mouse and man, the X-chromosomal amelogenin gene can be alternatively spliced to give rise to more than a dozen mRNAs that encode a variety of protein isoforms, although a role for each isoform during enamel biomineralization is not known [7–10]. Unlike bone, a tissue that retains cells in the mineralized matrix, enamel is cell-free. The ameloblasts secrete the enamel matrix proteins into the extracellular space where they self-assemble into a matrix that guides the habit and organization of the mineral crystallites [11, 12]. Further unlike bone, the protein-rich enamel matrix precursor employed during formation is almost entirely reabsorbed with mineral replacement, leaving but small amounts of proteins behind that serve to modulate the material properties of the mineral phase, serving to plasticize or toughen it [13–17]. By the time the teeth erupt into positions for chewing, the ameloblast cells have died.
Amelogenin is the most abundant matrix protein serving to organize the hydroxyapatite mineral into parallel bundles of long, thin-crystallites that impart to the tissue the unique materials properties that allow enamel to serve a lifetime of chewing with little wear or fracture [16, 18, 19]. The primary transcript from the mouse amelogenin gene can be alternatively spliced to yield dozens of amelogenin proteins isoforms that participate in formation of the enamel matrix. Using genetic engineering we created a simplified protein matrix where only one, rather than dozens of amelogenin isoforms contribute to enamel formation.
2. Materials and Methods
2.1 Creation of the M180 Knock In Mouse
The methods used to create amelogenin knock in mice by targeted homologous recombination of a minigene encoding an engineered amelogenin have been published [20] and are largely used here. In brief, the targeting DNA construct was engineered using standard recombinant DNA techniques by inserting a mouse180 amino acid (M180) amelogenin minigene after the second exon of the endogenous gene followed by the SV-40 poly-A-signal sequence and the Neo resistant gene with flanking lox-sites. The length of the two homologous arms for the amelogenin genomic DNA, flanking the engineered amelogenin minigene, corresponded to about 6 Kbp at the 5’-end and 3.6 Kbp at the 3’-end. The nucleotide sequence across the homologous arms, amelogenin minigene and Neo resistance gene in the targeting construct were confirmed by DNA sequence determination. An internal or external DNA hybridization probe was used to discriminate homologous recombination from insertions. Two pairs of primers for PCR genotyping of cells and knock in mice were designed as follow: forward primer: 5’-GCC GCA CCT TCT TTT TGA TTA GC-3’, reverse primer: 5’-GAA TGC AGA GCA CAC AAT CTT GG; and forward primer: 5’-TCC ATC TGC ACG AGA CTA GTG AGA CG-3’, reverse primer: 5’-GAA TGC AGA GCA CAC AAT CTT GG-3’.
The mouse ES cell line 129/RW4 was grown in “knock-out” DMEM medium (Invitrogen) with 15% fetal bovine serum (Hyclone), 2 mM glutamine, 30 ug/ml of penicillin, 50 ug/ml of streptomycin, 0.1 mM β-mercaptoethanol, 1X non-essential amino acid and 1000 U/ml of mouse leukemia inhibiting factor (LIF) (Millipore Corp) on inactivated mouse primary fibroblast cells. The ES cell medium was changed every day and split every 2 days. Twenty-five micrograms of the targeting DNA was mixed with 1.1×107 ES cells in PBS and incubated at room temperature for 5 minutes before electroporation. The electroporation was performed in a 0.4 mm cuvette at 230 V/cm and 500 µF with a Bio-Rad Gene Pulser. After electroporation, 5 × 106 cells with 10 ml of ES cell medium were plated onto 100 mm dishes on top of a layer of feeder cells. Selection was performed 24 hours after electroporation by adding 300 ug/ml of G418 to the ES cell medium. The cells were kept under selection pressure for 7 days. Individual G418 resistant cell clones were physically isolated and dissociated into single cells by trypsin digestion. The single ES cells were cultured in 96 well plates with a feeder cell layer for 3 days.
Genomic DNA was recovered from ES cells digested with EcoRI and hybridized with the external and internal probes in Southern blot analysis to identify those cells that had undergone homologous recombination. Two positive knock in ES cell clones with the M180 minigene were chosen for blastocyst injection after confirming their normal karyotype. Blastocyst injections were performed at the UC Irvine Transgenic Mouse Facility.
Animals were created, housed and sacrificed according to a USC Institutional Animal Care and Use Committee in full compliance with federal guidelines.
2.2 Gene expression analysis
2.2.1 RT-PCR
Total RNA from the mandible of newborn knock in or wild type mice was extracted using Trizol reagent (Invitrogen). First strand cDNA was synthesized from 1ug of total RNA using a RT-PCR kit (Invitrogen) according the recommendation of the manufacturer and served as the template for amplification using standard reagents. Sets of oligodeoxynucleotide primers were synthesized that correspond to the amelogenin gene (NM_009666) and were used to amplify the first strand cDNA. Primers (Figure 1, panel C) correspond to the following nucleotide sequence: Primer 1), the first exon forward primer, 5’-ATC AGG CAT CCC TGA GCT TCA GAC; Primer 2), the sixth exon reverse primer, 5’-AGC TCA GGA AGA ATG GGG GAC A; Primer 3), the seventh exon reverse primer, 5’-CCA CTT CGG TTC TCT CAT TTT CTG. The sequences of primer pairs for amplifying the amelogenin and ameloblastin genes from the knock in or wild type mice are shown below. The forward primer is listed followed by the reverse primer. The primers for amplifying the amelogenin gene were: 5’-AGC TTC AGA CAG AAA CTC ACT GAG C and 5’-GGA GGC AGG CAA ACA AAA TCC. The primers for amplifying the ameloblastin gene were: 5’-TGG GAG CAC AGT GAA TGT CAG C and 5’-CCA GCT TGT TGA GGA AAT GCC.
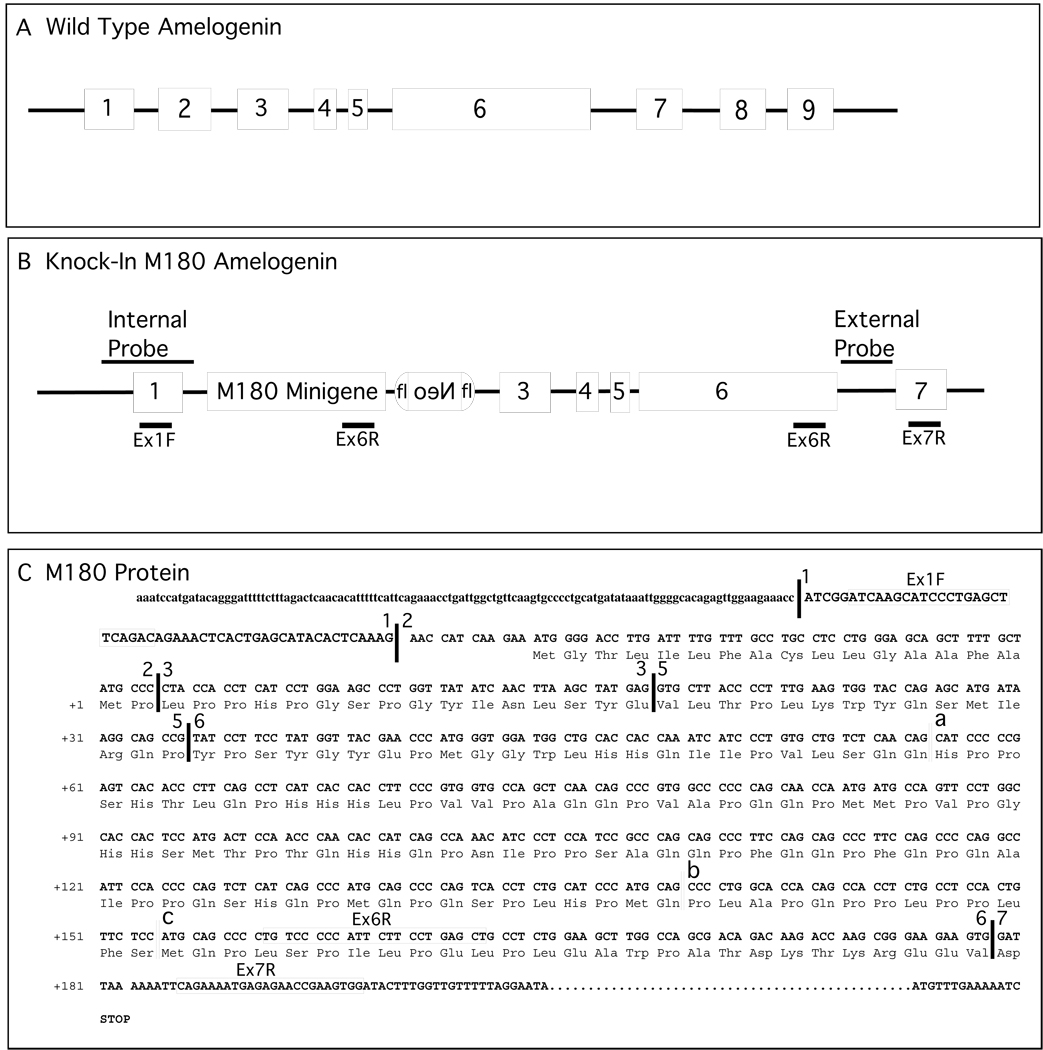
Figure 1. Physical map of the amelogenin locus, the knock in targeting construct and the M180 minigene.
Panel A. The map depicts the X-chromosomal amelogenin wild type parental state.
Panel B. Exon 2 is replaced by a minigene corresponding to M180. The minigene and the reverse oriented Neo cassette serves to restrict the alternative splice products produced. The physical positions for the “internal” or “external” DNA hybridization probes used to screen ES cells and founder animals are shown.
Panel C. Nucleotide and amino acid sequence for the M180 minigene. The nucleotide sequence of the DNA upstream of exon 1 and downstream of exon 7 are shown with non-coding nucleotides in lower case and coding nucleotides in upper case. The corresponding amino acid sequence for the mouse 180 amino acid amelogenin is shown using the three-letter amino acid code. The primers used for analysis of alternatively spliced RNA are labeled and enclosed by boxes.
Symbols: fl, floxed locus; Neo, reverse orientation neomycin gene used for selection; Ex1f, Ex6R, Ex7R, primer positions, with the sequence for each primer shown in panel C.
2.2.2 Western Blot
Analysis of total expressed proteins from the teeth of 3-day postnatal knock in or wild type mice were analyzed as previously described [20]. Protein was recovered by homogenizing the enamel organ epithelia in lysis buffer consisting of 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.02% sodium azide, 0.1% sodium dodecyl sulfate (SDS), 100 ug/ml phenylmethylsulfonyl fluoride (PMSF), 1 ug/ml aprotinin, 1% Nonidet P-40, and 0.5% sodium deoxycholate. Protein concentration was measured with a BioRad protein assay based on bovine serum albumin as the standard. Equal amounts of total enamel matrix proteins (20ug) were mixed with 2X loading buffer and size-resolved by electrophoresis in a 15% polyacrylamide gel. Size separated proteins were electrophoretically transferred to a PVDF membrane (Millipore Corp.) and immunoblotted with anti-amelogenin primary antibody. An enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia Biotech) was used to identify the antibody-antigen complex and a record produced by exposure to radiographic film followed by development in an automated film processor.
2.3 Enamel Morphology
The architecture of the enamel rods was analyzed as previously described [20]. The lower incisors from the 6 week-old knock in and wild type mice were carefully dissected from the mandible. The labial surface of the incisors was notched by sawing with a thin diamond wheel across the long axis at sites corresponding to the junction of the secretory and maturing zone of enamel and fractured. The resulting two pieces of the incisor were glued onto a metal stub and coated for 5 minutes with an intermittent pulse using gold-palladium alloy in Hummer V, set with the high voltage control knob at “9” to provide a coating thickness of approximately 100 Å. The enamel surface was visualized in a Cambridge 360 scanning electron microscope at the USC Center for Electron Microscopy.
2.4 Material Properties Testing
Microindentation techniques were used to measure enamel hardness, dentin hardness and bulk enamel fracture toughness [16, 21–23]. Ten freshly extracted lower left 9-week old wild type and 10 freshly extracted age matched lower left transgenic knock in M180 were kept moist at all times, embedded in epoxy resin, and sequentially ground to a 0.1 µm alumina finish using a semiautomatic polisher (Buehler, Lake Bluff, IL). Appropriate loads from 100 to 225 g were used with dwell times of 20 seconds using a customized manually-operated Vickers microhardness tester. For bulk enamel hardness and toughness, indentations were made approximately half way between the DEJ and the outer enamel surface. For bulk dentin hardness, indentations were made approximately half way from the DEJ to the site of the pulp chamber. All indentations were made on the incisal thirds of the teeth. Indentations were examined by light microscopy, using polarization, interference, light/dark field, and transillumination techniques to identify cracks and measurements were made using a digital micrometer. The enamel fracture toughness, Kz, was calculated as: Kz = γ P / e1.5, where “γ” is a fitting constant, “P” is the indenter load and “e” is half the length of the cracks. The solution represented by this equation only represents an approximation of a complex situation in a non-uniform anisotropic substrate, and its use is intended for comparative purposes rather than absolute values. Ten repetitions per tooth were averaged to describe each of the 10 teeth in each group; group means and standard deviations were then calculated. Data was analyzed by student t tests and p<0.05 (2-sided) was considered statistically significant.
3. Results
3.1 Knock In Mice Express only M180 Amelogenin
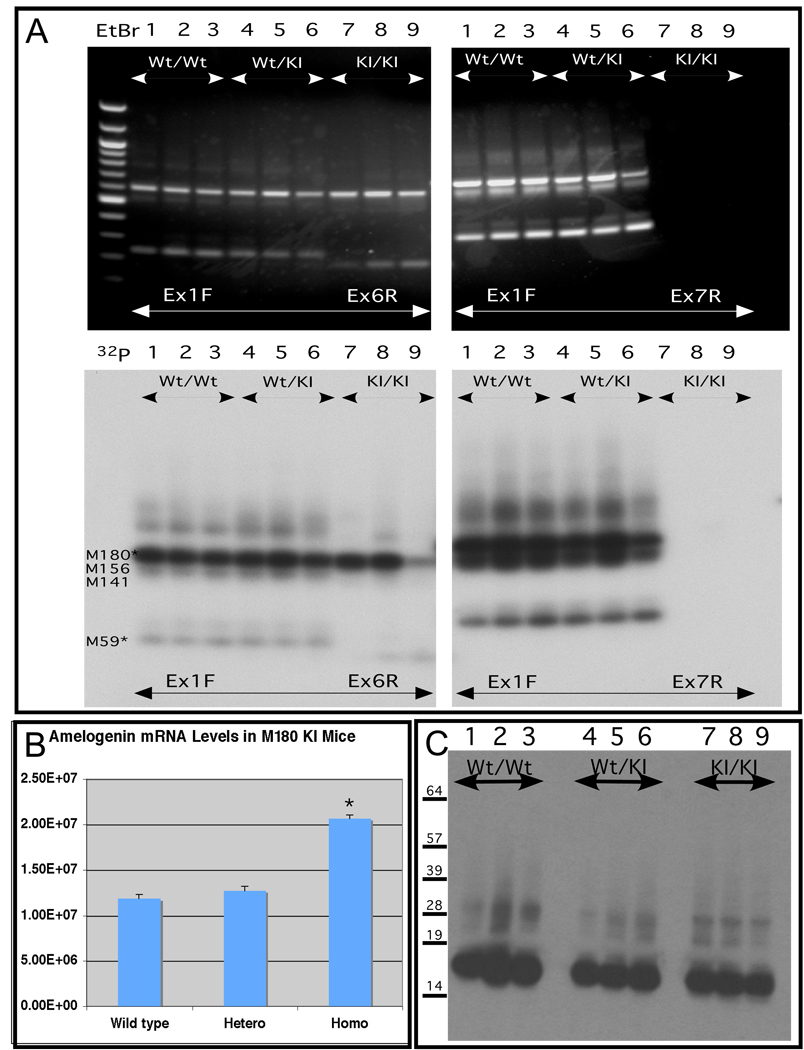
In this unique model of mouse enamel formation, by genetically replacing the endogenous amelogenin gene with the M180 amelogenin minigene (Figure 1), the ameloblasts should express only the mouse 180 amelogenin isoform and none of the other alternatively spliced products. However, if alternatively spliced cryptic products, generated between the engineered amelogenin minigene and the downstream endogenous amelogenin locus, were expressed by the ameloblasts of the knock in mice, it would interfere in our interpretation for the phenotype of the knock in amelogenin mice. Thus, we designed two pairs of primers that spanned across the amelogenin locus, from exon 1 to exon 6 and from exon 1 to exon 7 respectively. Multiple transcripts were detected using either pairs of primers in the wild type and heterozygous knock in mice (Figure 2, panel A, lanes 1 through 6 respectively). However, for the homozygous M180 knock in mice, only the inserted amelogenin minigene transcripts were detected with the primers spanning across exon 1 to exon 6 (Figure 2, panel A, lanes 7 through 9), and no transcripts were amplified with the primers spanning across exon 1 to exon 7 due to the lack of exon 7 in M180 minigene. These results indicated that no alternatively spliced products were expressed by ameloblasts from the M180 knock in mouse. To ensure that low abundance alternatively spliced mRNAs were not missed, we also performed these same assays in the presence of 32P-NTPs to improve sensitivity. The result from this additional sensitive technique corroborated the outcome from conventional RT-PCR (Figure 2).
Figure 2. Analysis of amelogenin RNA transcript and protein from animals bearing the knock in M180 amelogenin minigene.
Panel A. Identification of alternatively spliced transcript analysis performed by reverse transcription followed by polymerase chain amplification (RT-PCR). Total RNA was recovered from mouse mandibles, purified, subjected to reverse transcription with the resulting cDNAs characterized by polymerase chain amplification using specific amelogenin primers Ex1F, Ex6R, Ex7R, as described in the legend above. Homozygous wild type (Wt), heterozygous (Wt/KI) and homozygous knock in (KI) animals were analyzed. The upper two panels (black background) depict ethidium bromide stained amplicons. To increase sensitivity to detect low abundance transcripts, the same primers were used in the presence of 32P NTPs with the resulting radiolabeled amplicons shown in the lower two panels (white background). The predicted alternatively spliced product is marked to the left, using the nomenclature that corresponds to the number of amino acid residues encoded by the mRNA (e.g. M180, M156, M141 and etc). The presence of the wild type allele (lanes 1–6) supports the production of alternatively spliced products that are not observed for the homozygous knock in state (lanes 7–9). The failure to amplify a product with Ex1F and Ex7R confirms the absence of alternatively spliced products in the knock in since the M180 minigene lacks an exon 7 sequence (see Supplemental Figure S1).
Panel B. Quantitative RT-PCR analysis of transcript abundance based on threshold value. RNA was isolated, converted to first strand cDNA and then amplified. The resulting threshold cycle number was used to calculate the abundance of the starting transcript. RNA from 6 independent animals was used. Transcript abundance was normalized to ameloblastin. Amelogenin transcript abundance was slightly higher in ameloblasts from the M180 homozygous knock in teeth compared for the wild type amelogenin.
Panel C. Western blot analysis of amelogenin protein from teeth of knock in and wild type mice. Total protein from teeth from 3-day postnatal wild type (Wt) mice (lanes 1–3), heterozygous mice (lanes 4–6) or homozygous M180 knock in mice (KI) (lanes 7–9) mice was recovered and 20ug analyzed in each lane. No difference in the steady state amelogenin abundance between wild type and M180 knock in was identified.
To measure the relative abundance of the amelogenin transcripts produced from the knock in locus, we turned to quantitative real time PCR and primers from exon 1 and 6 (Figure 1, panel C). We found no difference in amelogenin transcript abundance between the wild type and the heterozygous mice, but noticed that amelogenin transcript abundance had increased in the homozygous M180 knock in animals (Figure 2, panel B). However, when amelogenin protein abundance was measured by Western blot of total protein from developing enamel matrix, we found there was no apparent change in amelogenin abundance between wild type, heterozygotes and M180 knock in mice (Figure 2, panel C). In addition to these molecular analyses, extensive histologic evaluations of developing tooth organs were performed (data not shown). These also revealed no abnormalities among the wild type, heterozygous and homozygous knock in mice. Cumulatively, we concluded that there were no overt alterations to enamel matrix generation during enamel formation in the M180 knock in mice.
3.2 Hierarchical Enamel Organization
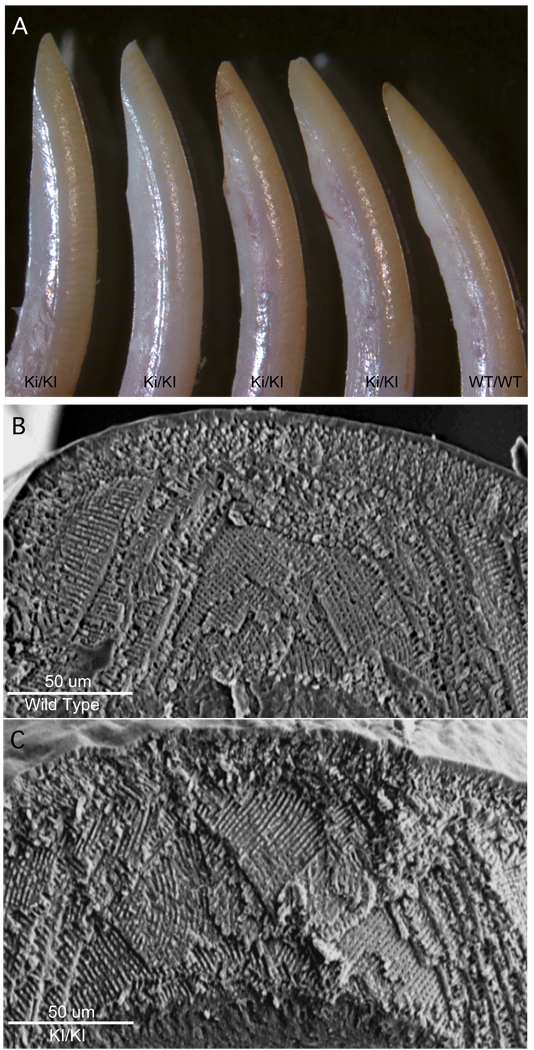
The clinical appearance of six-week old incisor M180 knock in (KI) incisors teeth revealed little or no apparent differences in their geometry or enamel wear patterns compared to their wild type (Wt) controls, as shown in Figure 3, panel A. Scanning electron microscopic examination of enamel rod architecture between wild type enamel (Figure 3, panel B) and enamel made from only the M180 amelogenin isoform (Figure 3, panel C) revealed no discernable differences. The rod-to-rod and interrod enamel organization appears to be unaltered in the M180 knock in compared to wild type enamel. An evolutionary adaption to enamel architecture of rodent incisor is that wear from chewing results in enamel self-sharpening and produces an edge of hydroxyapatite crystallites [24]. As shown by Figure 3, panels A through C, incisor self-sharpening is also unaffected by this simplified design of the biomineralized matrix
Figure 3. Appearance and enamel architecture of M180 knock in incisor teeth.
Panel A, incisors from either animals homozygous to the amelogenin M180 knock in allele (KI/KI), or wild type amelogenin allele (Wt/Wt), are shown at six weeks of age. No discernable alterations to the morphology of the incisors or their capacity to masticate food are identified.
Panel B and C, enamel was visualized by scanning electron microscopy from wild type (B) or M180 knock in (C) teeth and reveals no discernible alteration to enamel architecture.
3.3. Material Properties
We measured and compared the material properties of enamel from wild type and genetically simplified M180 knock in animals. These measurements serve as sensitive indicators for poorly formed enamel matrix precursor when it is converted to a mineral composite tissue, while also providing quantitative measures for such defects [15, 16, 31]. The results shown in Table 1 demonstrate a 22% reduction in fracture toughness in the knock in enamel accompanied by a 7% increase in hardness. These changes to the competing parameters of hardness and toughness suggest that simplifying the matrix precursor to a single amelogenin isoform may have allowed greater uniformity of crystallite orientation and packing, consistent with increased density and hardness, also consistent with decreased crack deflection and decreased toughness [21]. Hardness and toughness are considered to be surrogates for wear resistance and for fracture resistance.
Table 1. Material properties of enamel from wild type and M180 knock in animals; means and standard deviations.
Dentine and enamel hardness and enamel toughness were determined for incisors from M180 knock in and wild type animals. Statistically significant reduction (p<0.002) in toughness of the M180 knock in enamel was identified, suggesting that while it functioned normally in the animal for mastication, it was none the less 22% less tough than the wild type counterpart. Conversely, a statistically significant (p<0.001) increase in hardness of the M180 knock in animal was identified.
| ANIMAL TYPE |
ENAMEL HARDNESS GPa |
ENAMEL TOUGHNESS MPa·m1/2 |
DENTIN HARDNESS GPa |
|---|---|---|---|
| M180 KI | 2.6 (0.1) | 1.0 (0.1) | 0.66 (0.09) |
| Wild Type | 2.3 (0.1) | 1.3 (0.1) | 0.66 (0.07) |
| Change | + 7 % | − 22 % | none |
| P | 0.001 | 0.002 | 0.4 |
4. Discussion
Each ameloblast cell synthesizes and organizes a unique volume of enamel protein matrix that is known as the enamel rod (or prism) corresponding to the ameloblast cross-sectional diameter. The pattern between the cells and their secreted rods of enamel matrix serve to control enamel architecture at this hierarchical level of enamel organization. Neighboring rows of ameloblast cells cooperate with one another to contribute a shared portion of the matrix in a process that weaves the matrix into a protein continuum [16, 24]. It is within the enamel matrix that the biomineral is grown and eventually fully replaces the protein. Amelogenin is the dominant protein of the matrix precursor and is subject to self-assembly to form nanospheres [19, 25]. These spheres correspond to the nanoscale hierarchical level of organization and are believed to control protein-to-mineral and protein-to-protein interactions [7, 9, 19, 26, 27]. The wild type amelogenin locus is known to produce more than a dozen mRNAs and their encoded protein isoforms, although a role for each isoform is not yet recognized [8, 28–30].
Our genetic approach was to replace the wild type amelogenin locus with a knock in minigene that expresses only one amelogenin isoform resulting in more than one order of magnitude of reduced protein complexity during enamel formation. A combination of techniques was used to show that only the mouse 180 amino acid residue long amelogenin (M180) minigene is expressed, with no other amelogenin isoforms expressed [20]. We sought to identify all possible transcripts expressed from the amelogenin locus by using the sensitive method of reverse transcription followed by polymerase chain reaction (RT-PCR) amplification, incorporating radioactive nucleotides to provide enhanced sensitivity to detect transcripts of low abundance. We identified no alternatively spliced products between the knock in engineered amelogenin minigene with the endogenous amelogenin expressed by ameloblasts from the M180 knock in mouse (Figure 2).
To determine if a change in the abundance of amelogenin mRNA had occurred with the genetic knock in we used real time reverse transcription and amplification with the threshold cycle function used to establish transcript abundance, as previously described [20]. We chose to use the ameloblastin gene transcript as an internal standard since ameloblastin is another enamel specific product expressed during this developmental period by ameloblasts. The genetic approach used here serves to keep the endogenous promoter and upstream gene regulatory domains intact so that gene regulation is unaffected. The finding that the M180 amelogenin mRNA was more abundant in ameloblasts from the homozygous knock in mice than from wild type or heterozygous mice indicates that the expression of the M180 minigene was not diminished. It is interesting to speculate that the lack of alternative splicing amelogenin products may indirectly increase M180 mRNA abundance in homozygous knock in mice. However, the Western blot shows that the protein levels are not elevated among the knock in versus wild type enamel. We conclude that we have not adversely impacted amelogenin expression by this genetic recombination.
Since enamel does not remodel, errors in matrix assembly and mineral replacement with the simplified matrix would be recorded as defects in the final biomineralized product. Such defects are expected to contribute to altered patterns of wear or to altered materials properties of the tissue, thereby contributing to premature failure in a tissue designed to last the lifetime of the organism. It is noteworthy that the resulting enamel from the M180 knock in reflects essentially no change in appearance and the resulting enamel functions well allowing the animal to feed over its lifetime.
With increasing mineral content, mature enamel tissue retains only traces of enamel proteins due to their degradation and reabsorption [32–34]. Retained enamel matrix proteins that surround the mature enamel crystallites have been shown to favor increased toughness [16] and contribute to elastic behavior [35]. Multiple amelogenin isoforms, as occurs in the wild type enamel, contributes to the balance between hardness and toughness (Table 1). These properties could be altered during the matrix precursor phase where amelogenin isoforms containing exon 4, 8 and 9 are normally expressed in the wild type mouse [36] but are absent in the M180 knock in mouse. However, the only known protein to be retained with the mineral crystallites in mature enamel is the amelogenin degradation product known as tyrosine rich amelogenin peptide (TRAP) defined by residues 1–45 of the M180 amelogenin [37]. Since degradation of M180 must yield TRAP, it is unlikely that we have altered hardness and toughness by altering TRAP abundance since we have not reduced the abundance of M180, the TRAP precursor (Figure 2). Alternatively, a loss of non-amelogenin proteins in the final enamel could change enamel material properties. The most abundant non-amelogenin protein in forming enamel is ameloblastin, and its expression is not altered in the M180 knock in mice (Figure 2). We have also shown that there are no known protein-to-protein physical interaction between amelogenin and any of the other non-amelogenin proteins [7] suggesting that changes to protein stoichiometry in the matrix formation phase was not likely to contribute to the modulated material properties.
Genetic manipulation of the mouse permits a correlation between matrix precursors and the final biomineral product, allowing dissection of the role that specific enamel matrix proteins may play over the control of the mineral phase. Previous investigations have shown that despite knocking out the amelogenin gene, the remaining enamel matrix proteins were able to nucleate mineral, but produced only a thin and poorly organized enamel architecture resembling the human condition of amelogenesis imperfecta [38]. Attempting to rescue the amelogenin null matrix through the addition of a single 59 amino acid (M59/LRAP) amelogenin isoform expressed from an transgene promoter failed to restore enamel architecture and function [39]. In contrast, here we used a knock in approach [20] to replace all the other alternatively spliced amelogenin mRNAs and corresponding proteins with only the M180 amelogenin isoform. The knock in approach preserves all of the genetic regulatory machinery needed for expression while reducing amelogenin isoform complexity 12 fold. The knock in approach thereby avoids the pitfall of altered protein abundances as can occur with transgenic approaches.
The data contained in this manuscript supports the conclusion that the complex mix of amelogenin isoforms involved in enamel formation can be simplified to only the single M180 protein with only minor impact on the structure or material properties of the enamel bioceramic enabling it to provide satisfactory function over the life span of the mouse as it incessantly gnaws. This is in marked contrast to our previous efforts to decipher the role that highly conserved amelogenin domains contribute to proper enamel formation [20]. Deletion of either of these two amelogenin domains resulted in marked and domain specific disruption of cell to matrix and/or protein to mineral interactions with consequential disturbances in the hierarchical architecture of each of their resultant enamel bioceramic tissues.
While mice live for a substantially shorter period than human, we interpret the successful deployment of the simplified enamel to be a suitably intense, in vivo animal demonstration that such enamel performs adequately in a “real world” application. An enamel biomimetic filling material for human teeth could be fabricated in vitro using only the M180 amelogenin isoform, thereby simplifying manufacturing parameters by orders of magnitude with regard to protein complexity. Studies by Fowler and colleagues have shown that full-length amelogenin can guide the in vitro formation of organized mineralized structures through co-operative interactions between assembling protein and forming mineral [40]. Their insight is supported by the data from our in vivo animal study also deploying a simplified matrix. Thus, an enamel biomimetic, with acceptable, but imperfect, material properties can be created. Dental caries (e.g., loss of enamel) remains the most prevalent infectious disease of man [1, 2] and a replacement material based on biology for restoring lost tissue through an enamel biomimetic has a large application reservoir [41].
5. Conclusions
Genetic manipulation of the mouse permitted us to measure the role that a specific single enamel amelogenin protein isoform exerts in controlling biomineralization and biomechanical function. A reduction in enamel protein complexity by orders of magnitude produced an enamel tissue with essentially unaltered tissue architecture and with acceptable material properties.
Acknowledgements
We are grateful to our colleagues at the University of Southern California, the Center for Craniofacial Molecular Biology and elsewhere for their critical appraisal of this manuscript. We thank the anonymous reviewers and the editor for their helpful suggestions that have improved this manuscript. This work was supported by the USPHS, NIH, National Institute for Dental and Craniofacial Research DE 13045 and DE 06988 (MLS) and DE13404 (MLP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Caufield PW, Li Y, Dasanayake A. Dental caries: an infectious and transmissible disease. Compend Contin Educ Dent. 2005;26:10–16. [PubMed] [Google Scholar]
- 2.Taubman MA, Nash DA. The scientific and public-health imperative for a vaccine against dental caries. Nat Rev Immunol. 2006;6:555–563. doi: 10.1038/nri1857. [DOI] [PubMed] [Google Scholar]
- 3.Weiner S. Organization of extracellularly mineralized tissues: a comparative study of biological crystal growth. CRC Crit Rev Biochem. 1986;20:365–408. doi: 10.3109/10409238609081998. [DOI] [PubMed] [Google Scholar]
- 4.Lowenstam HA, Weiner S. On biomineralization. New York: Oxford University Press; 1989. [Google Scholar]
- 5.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 6.Hu JC, Yamakoshi Y, Yamakoshi F, Krebsbach PH, Simmer JP. Proteomics and genetics of dental enamel. Cells Tissues Organs. 2005;181:219–231. doi: 10.1159/000091383. [DOI] [PubMed] [Google Scholar]
- 7.Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20:273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- 8.Simmer JP, Snead ML. Molecular biology of the amelogenin gene. In: Robinson C, Kirkham J, Shore R, editors. Dental Enamel Formation to destruction. Boca Raton: CRC Press; 1995. pp. 59–85. [Google Scholar]
- 9.Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol. 2005;149:182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Gibson CW, Golub E, Ding WD, Shimokawa H, Young M, Termine J, et al. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochem Biophys Res Commun. 1991;174:1306–1312. doi: 10.1016/0006-291x(91)91564-s. [DOI] [PubMed] [Google Scholar]
- 11.Paine ML, Krebsbach PH, Chen LS, Paine CT, Yamada Y, Deutsch D, et al. Protein-to-protein interactions: criteria defining the assembly of the enamel organic matrix. J Dent Res. 1998;77:496–502. doi: 10.1177/00220345980770030901. [DOI] [PubMed] [Google Scholar]
- 12.Snead ML, Zhu DH, Lei YP, White SN, Snead CM, Luo W, et al. Protein self-assembly creates a nanoscale device for biomineralization. Materials Science & Engineering C-Biomimetic and Supramolecular Systems. 2006;26:1296–1300. [Google Scholar]
- 13.Fearnhead RW. Mineralization of rat enamel. Nature. 1960;188:509–510. doi: 10.1038/188509a0. [DOI] [PubMed] [Google Scholar]
- 14.Eastoe JE. Organic matrix of tooth enamel. Nature. 1960;187:411–412. doi: 10.1038/187411b0. [DOI] [PubMed] [Google Scholar]
- 15.Fong H, White SN, Paine ML, Luo W, Snead ML, Sarikaya M. Enamel structure properties controlled by engineered proteins in transgenic mice. J Bone Miner Res. 2003;18:2052–2059. doi: 10.1359/jbmr.2003.18.11.2052. [DOI] [PubMed] [Google Scholar]
- 16.White SN, Luo W, Paine ML, Fong H, Sarikaya M, Snead ML. Biological organization of hydroxyapatite crystallites into a fibrous continuum toughens and controls anisotropy in human enamel. J Dent Res. 2001;80:321–326. doi: 10.1177/00220345010800010501. [DOI] [PubMed] [Google Scholar]
- 17.Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Dental enamel matrix: sequences of two amelogenin polypeptides. Biosci Rep. 1981;1:771–778. doi: 10.1007/BF01114799. [DOI] [PubMed] [Google Scholar]
- 18.Imbeni V, Kruzic JJ, Marshall GW, Marshall SJ, Ritchie RO. The dentin-enamel junction and the fracture of human teeth. Nat Mater. 2005;4:229–232. doi: 10.1038/nmat1323. [DOI] [PubMed] [Google Scholar]
- 19.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 20.Zhu D, Paine ML, Luo W, Bringas P, Jr., Snead ML. Altering biomineralization by protein design. J Biol Chem. 2006;281:21173–21182. doi: 10.1074/jbc.M510757200. [DOI] [PubMed] [Google Scholar]
- 21.White SN, Paine ML, Ngan AY, Miklus VG, Luo W, Wang H, et al. Ectopic expression of dentin sialoprotein during amelogenesis hardens bulk enamel. J Biol Chem. 2007;282:5340–5345. doi: 10.1074/jbc.M604814200. [DOI] [PubMed] [Google Scholar]
- 22.White SN, Paine ML, Luo W, Sarikaya M, Fong H, Yu Z, et al. The dentino-enamel junction is a transitional zone uniting dissimilar composite bioceramics. J Am Ceram Soc. 2000;83:238–240. [Google Scholar]
- 23.White SN, Miklus VG, Chang PP, Caputo AA, Fong H, Sarikaya M, et al. Controlled failure mechanisms toughen the dentino-enamel junction zone. J Prosthet Dent. 2005;94:330–335. doi: 10.1016/j.prosdent.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Boyde A. Microstructure of enamel. Ciba Found Symp. 1997;205:18–27. doi: 10.1002/9780470515303.ch3. discussion -31. [DOI] [PubMed] [Google Scholar]
- 25.Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P, Jr., et al. Evidence for amelogenin "nanospheres" as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 26.Bartlett JD, Ganss B, Goldberg M, Moradian-Oldak J, Paine ML, Snead ML, et al. 3. Protein-protein interactions of the developing enamel matrix. Curr Top Dev Biol. 2006;74:57–115. doi: 10.1016/S0070-2153(06)74003-0. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Guan X, Du C, Moradian-Oldak J, Nancollas GH. Amelogenin promotes the formation of elongated microstructures in a controlled crystallization system. J Phys Chem C. 2007;111:6398–6404. doi: 10.1021/jp0675429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau EC, Simmer JP, Bringas P, Jr., Hsu DD, Hu CC, Zeichner-David M, et al. Alternative splicing of the mouse amelogenin primary RNA transcript contributes to amelogenin heterogeneity. Biochem Biophys Res Commun. 1992;188:1253–1260. doi: 10.1016/0006-291x(92)91366-x. [DOI] [PubMed] [Google Scholar]
- 29.Bartlett JD, Ball RL, Kawai T, Tye CE, Tsuchiya M, Simmer JP. Origin, splicing, and expression of rodent amelogenin exon 8. J Dent Res. 2006;85:894–899. doi: 10.1177/154405910608501004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papagerakis P, Ibarra JM, Inozentseva N, DenBesten P, MacDougall M. Mouse amelogenin exons 8 and 9: sequence analysis and protein distribution. J Dent Res. 2005;84:613–617. doi: 10.1177/154405910508400706. [DOI] [PubMed] [Google Scholar]
- 31.Bechtle S, Habelitz S, Klocke A, Fett T, Schneider GA. The fracture behavior of dental enamel. Biomaterials. 2009;31:375–384. doi: 10.1016/j.biomaterials.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 32.Smith CE, Chong DL, Bartlett JD, Margolis HC. Mineral acquisition rates in developing enamel on maxillary and mandibular incisors of rats and mice: implications to extracellular acid loading as apatite crystals mature. J Bone Miner Res. 2005;20:240–249. doi: 10.1359/JBMR.041002. [DOI] [PubMed] [Google Scholar]
- 33.Robinson C, Briggs HD, Atkinson PJ, Weatherell JA. Matrix and mineral changes in developing enamel. J Dent Res. 1979;58:871–882. doi: 10.1177/00220345790580024101. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro JL, Wen X, Okamoto CT, Wang HJ, Lyngstadaas SP, Goldberg M, et al. Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cell Mol Life Sci. 2007;64:244–256. doi: 10.1007/s00018-006-6429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie ZH, Swain MV, Swadener G, Munroe P, Hoffman M. Effect of microstructure upon elastic behavior of human tooth enamel. J Biomech. 2009;42:1075–1080. doi: 10.1016/j.jbiomech.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Iacob S, Veis A. Identification of the functional activity of the [A-4] amelogenin gene splice product in newborn mouse ameloblasts. Bone. 2008;42:1072–1079. doi: 10.1016/j.bone.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fincham AG, Hu Y, Lau EC, Slavkin HC, Snead ML. Amelogenin post-secretory processing during biomineralization in the postnatal mouse molar tooth. Arch Oral Biol. 1991;36:305–317. doi: 10.1016/0003-9969(91)90101-y. [DOI] [PubMed] [Google Scholar]
- 38.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 39.Chen E, Yuan ZA, Wright JT, Hong SP, Li Y, Collier PM, et al. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tissue Int. 2003;73:487–495. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- 40.Fowler CE, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. Co-operative mineralization and protein self-assembly in amelogenesis: silica mineralization and assembly of recombinant amelogenins in vitro. Eur J Oral Sci. 2006;114 Suppl 1:297–303. doi: 10.1111/j.1600-0722.2006.00288.x. discussion 27-9, 82. [DOI] [PubMed] [Google Scholar]
- 41.Evans CA, Kleinman DV. The Surgeon General's report on America's oral health: opportunities for the dental profession. J Am Dent Assoc. 2000;131:1721–1728. doi: 10.14219/jada.archive.2000.0118. [DOI] [PubMed] [Google Scholar]