Abstract
Meiotic crossover (CO) recombination establishes physical linkages between homologous chromosomes that are required for their proper segregation into developing gametes and promotes genetic diversity by shuffling genetic material between parental chromosomes. COs require the formation of double strand breaks (DSBs) to create the substrate for strand exchange. DSBs occur in small intervals called hotspots1-3 and significant variation in hotspot usage exists between and among individuals4. This variation is thought to reflect differences in sequence identity and chromatin structure, DNA topology and/ or chromosome domain organization1, 5-9. Chromosomes show different frequencies of nondisjunction (NDJ)10, reflecting inherent differences in meiotic crossover control, yet the underlying basis of these differences remains elusive. Here we show that a novel chromatin factor, X non-disjunction factor 1 (xnd-1), is responsible for the global distribution of COs in C. elegans. xnd-1 is also required for formation of double-strand breaks (DSBs) on the X, but surprisingly XND-1 protein is autosomally-enriched. We show that xnd-1 functions independently of genes required for X chromosome-specific gene silencing, revealing a novel pathway that distinguishes the X from autosomes in the germ line, and further show that xnd-1 exerts its effects on COs, at least in part, by modulating levels of H2A lysine 5 acetylation.
The C. elegans genome has an unusual organization with regard to recombination and gene organization. Recombination preferentially occurs towards the chromosome ends, which contain fewer and less highly expressed genes than the central gene cluster11 (Baillie, unpublished). The X chromosome shows a similar distribution of crossovers12, but genes are more evenly distributed along its length. The X is also transcriptionally silent in the germline except for a brief period during late oogenesis13. Thus, during the critical period of CO formation, the X chromosome is replete with histone post-translational modifications (HPTMs) indicative of heterochromatin. Although recombination can occur in heterochromatin, closed chromatin generally presents an unfavorable environment for DSB formation. This unique configuration of an entire chromosome provides an advantage of the nematode system for understanding the relationship between higher order DNA structure and CO regulation9.
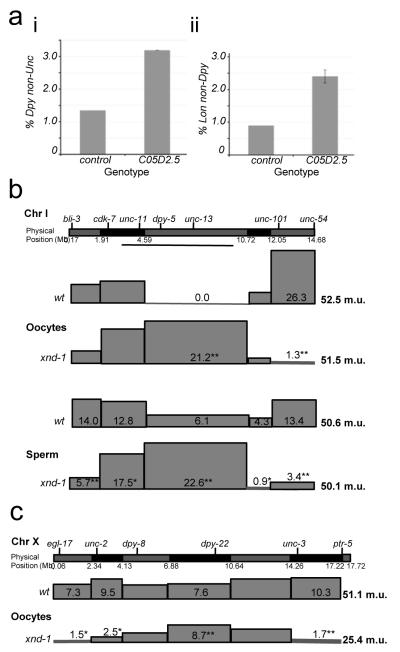
To identify meiotic recombination regulatory proteins, we screened over 350 known and putative chromatin binding proteins by RNA interference (RNAi) for segregation of triply marked chromosomes (see Methods). RNAi against C05D2.5 resulted in a dramatic increase in recombinant Dpy progeny (Figure 1a, i). A second triply marked chromosome confirmed these results (Figure 1a, ii), indicating that C05D2.5 affected recombination rather than expression of the genetic markers. C05D2.5 was renamed xnd-1 (X chromosome nondisjunction-1) based on its primary phenotypes. Further analysis was performed on two deletion mutations, ok708 and ok709 (Supplementary Figure 1).
Figure 1.
xnd-1 is needed for the normal recombination landscape in C. elegans. (a) C05D2.5 was identified by its increase in the number of recombinant progeny with the phenotype of the middle of three genetic markers, either unc-45 dpy-18 unc-64 (wt, n=8750, std. dev= 0.06; xnd-1, n=3980, std. dev= 0.15) (i) or dpy-1 lon-1 dpy-18 (wt, n=2229, std. dev= 0; xnd-1, n=1863, std dev= 0.21) (ii) Error bars represent the standard deviation from three or two independent experiments, respectively. (b) Chromosome I recombination maps for wt and xnd-1 oocytes (top) and sperm (bottom). (c) X chromosome recombination maps for wt and xnd-1 oocytes (data for b,c are found in Supplementary Tables 1-3). Genetic and physical markers are shown above and below the graphic representation of each chromosome, respectively. Boxes represent the relative map size for each interval as determined by SNP analysis (see Methods). Significant differences between wt and xnd-1 are marked (*p<0.05; **p<0.001).
Single nucleotide polymorphism (SNP) analysis was used to confirm a role for xnd-1 in CO regulation. In wild-type (wt), COs occurred in more gene-poor regions towards the autosome ends11(Figure 1b and Supplementary Table 1). In xnd-1 mutants, the global crossover landscape on autosomes was inverted; most COs occurred abnormally in the gene-rich region of chromosome I (Figure 1b and Supplementary Table 1). This dramatic shift in CO placement occurred without a change in the overall map size (i.e. total number of crossovers per chromosome) (Figure 1b and Supplementary Table 1). These data therefore indicate that xnd-1 is required for the normal distribution of meiotic crossovers on autosomes in C. elegans.
Because the X has a different chromatin state than autosomes, we also mapped COs on X. Like on autosomes, CO distribution is altered and occurs preferentially in the central region in xnd-1 mutants (Figure 1c). In contrast to autosomes, loss of xnd-1 reduced the genetic map of the X to half that observed in wt (24.4 m.u. versus 51.1 m.u. respectively; Figure 1c and Supplementary Table 2), indicating a two-fold decrease in recombination frequency. Thus, xnd-1 is required for CO distribution genome-wide, but is selectively required for the normal CO frequency on the X.
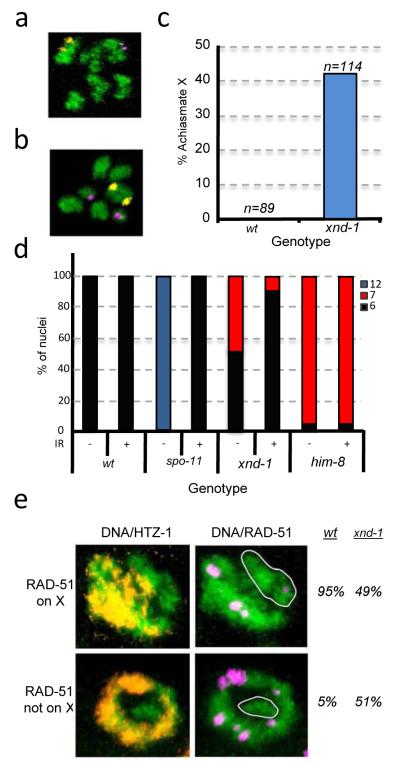
The increase in non-recombinant X’s was accompanied by an increase in the percentage of male progeny (XO genotype) arising from nondisjunction in XX hermaphrodites (~20% males versus <0.2% in wt; Supplementary Table 4), consistent with defects in meiotic CO formation14. Autosomal aneuploidy manifests as embryonic lethality, and on average, 35% of xnd-1 embryos do not hatch (Supplementary Table 4). To determine if both the X and autosomes were defective in CO formation, we analyzed diakinesis oocytes when homologues are held together at the CO and revealed as 6 DAPI staining bivalents in wt (Figure 2a). In xnd-1, 7 DAPI-staining bodies were evident in ~ 50% of oocytes: 5 bivalents and 2 smaller univalents are observed (Figure 2b), revealing that a single chromosome pair had frequently not received a CO. FISH confirmed that the achiasmate chromosome was the X in all instances (Figures 2a-c) demonstrating that xnd-1 is required for wt levels of X chromosome COs. Since no achiasmate autosomes were observed, the embryonic lethality associated with xnd-1 appears to be independent of its role in CO formation (see Methods).
Figure 2.
xnd-1 is required for efficient DSB formation on the X chromosome. (a-c) Achiasmate chromosomes are observed at diakinesis in xnd-1. FISH probes mark Chromosomes V (yellow) and X (magenta); DNA is stained with DAPI, (green). (a) Six DAPI-staining bodies indicate that all chromosomes have recombined, (b) seven reveal the achiasmate X’s in xnd-1 oocytes. (c) Quantification of achiasmate X frequency. Chromosome V was never achiasmate in wt or xnd-1. (d) Ionizing radiation (IR) rescues the CO defects of xnd-1. Quantification of IR rescue as assessed by the number of DAPI staining bodies at diakinesis 24 hrs post-irradiation. (wt −/+ IR, N=91, 82; spo-11 −/+ IR, N=73, 100; xnd-1 −/+ IR, N= 190, 157; him-8 −/+ IR, N=83, 65). (e) Fewer RAD-51 foci are observed on the X chromosome (white circles) in xnd-1. rad-54(RNAi) treated animals9 were dissected and germlines co-stained for DNA (green), HTZ-1 (which marks autosomes17, yellow), and the DNA repair protein, RAD-51 (magenta), which acts as a marker of DSBs9). The percentage of nuclei in which RAD-51 foci were observed on the X was quantified (wt, N=369; xnd-1, N=391).
The ability of xnd-1 to affect CO formation and distribution raised the possibility that xnd-1 might be involved in regulating DSBs. Meiotic DSBs are catalyzed by SPO-1115 and the failure to form bivalents in spo-11 mutants can be rescued by exogenous breaks induced by ionizing radiation16 (IR, Figure 2d). IR significantly reduces the number of nuclei with 7 DAPI staining bodies at diakinesis in xnd-1 (Figure 2d) indicating that the deficit in X COs is either the result of a failure to make DSBs or a failure to make CO-competent DSBs.
To determine whether xnd-1 is required for DSB formation, we assayed recruitment of the DSB repair protein RAD-51 to the X chromosome9. By co-labeling for the autosomal-associated HTZ-1 protein17 (Supplementary Figure 2), we assessed the frequency that RAD-51 associated with the (unlabeled) X. RAD-51 foci were detected on X in ~95% of wt nuclei (Figure 2e and Supplementary Movie 1). In contrast, only ~50% of xnd-1 mutant nuclei had detecTable RAD-51 foci on X (Figure 2e, and Supplementary Movie 2) indicating that xnd-1 has a crucial role in recruiting DSBs to the X.
In C. elegans, pairing and synapsis are independent of DSB formation16. Therefore we reasoned these processes should be unaffected by xnd-1 loss. FISH and immunofluorescence revealed that the kinetics and extent of pairing and synapsis did not differ between xnd-1and wt (Supplementary Figure 3a, b). Thus, the SC was fully polymerized between all homologues upon entry into pachytene (Supplementary Figure 3b and data not shown). Nevertheless, by late pachytene, SYP-1 and SYP-2 were no longer visible on the X in half of the nuclei (Supplementary Figure 3b, c) and a subset of X’s prematurely separated (Supplementary Figure 3a). Irradiation prevented desynapsis and separation (Supplementary Figure 3d, e), indicating that these phenotypes are a consequence of insufficient DSB formation. These observations suggest that the formation of DSBs is communicated to the SC, allowing for its stabilization. These results compliment our understanding that DSBs influence SC polymerization in C. elegans18, revealing an additional role for DSBs in SC maintenance.
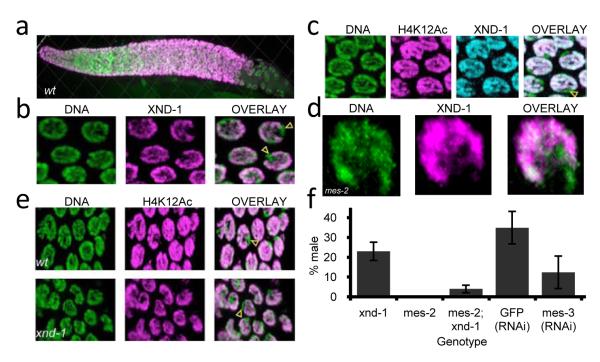
To further understand xnd-1 function, we investigated its subcellular localization. Labeling with anti-XND-1 antibodies revealed nuclear staining from the most distal region of the gonad through mid- to late pachytene (Figure 3a). XND-1 was not detected in somatic tissues or in xnd-1 mutant gonads (Supplementary Figure 4). Close examination of germline nuclei revealed that a single chromosome pair lacked XND-1 labeling (Figure 3b and Supplementary Figure 5). Remarkably, co-staining germline nuclei with autosome-specific HPTMs indicated that the unstained chromosome was the X (Figure 3c, Supplementary Movie 3). Thus, XND-1 is an autosomally-enriched protein required for efficient DSB formation on the X.
Figure 3.
XND-1 is an autosomal protein that regulates X chromosome crossing over. (a) Anti-XND-1 antibody staining of a wt hermaphrodite germline. (b) Close-up of wt nuclei reveals the absence of staining on one chromosome (yellow arrowheads). (c) Co-staining of wt pachytene nuclei with anti-XND-1 and anti-H4K12Ac reveals that these proteins are coincident, indicating that XND-1 is enriched on autosomes. A yellow arrowhead indicates the unstained X. (d) Localization of XND-1 is independent of the X chromosome silencing gene mes-2. XND-1 antibody staining in mes-2 (M-Z-) mutants with rare pachytene nuclei reveals normal XND-1 localization (X marked by white arrowhead). (e) Activating HPTMs remain excluded from the X in xnd-1mutants (yellow arrowheads). Histone H4K12Ac (magenta) is enriched on autosomes in wt (top) and xnd-1 (bottom). (f) Suppression of xnd-1 HIM phenotype by mes-2(bn11) and mes-3(RNAi ) (xnd-1, n = >8000; mes-2= >2000; mes-2; xnd-1, n=733; GFP(RNAi), n= 192; mes-3RNAi), n= 283). Error bars represent standard deviation from at least three experiments.
The localization of XND-1 is strikingly similar to MES-4 and MRG-1, which together with the X-enriched proteins, MES-2, -3, -6, regulate X chromosome gene silencing19. These factors could function to exclude XND-1 from the X. However, in mes-2 (M−Z−) germlines, XND-1 protein did not relocalize to the X (Figure 3d and Methods). XND-1 localization was also not altered in other mutants affecting gene silencing, meiotic DNA structure or X-specific behaviors, nor in mutants that affect pairing, axis and SC formation, DSB formation and repair (Supplementary Table 6). Thus, XND-1 localization appears to be independent of factors required for X chromosome silencing and vice versa: HPTMs associated with transcriptional repression remain on the X and those associated with activation are not recruited to the X (Figure 3e and data not shown). Furthermore, xnd-1 is not required for the similar process of transgene silencing20 (Supplementary Table 7). Thus, while XND-1 enrichment on autosomes is similar to MES-4 and MRG-1, it confers a novel and independent level of sex chromosome-specific behavior.
One possibility for the deficit in COs on the X is the establishment of a more closed, inaccessible chromatin environment. If this were the case, mutations that lead to more open chromatin on the X, such as mes-2, -3, -4, and -6, might counteract the effect of xnd-1 mutation. We observed that mes-2 or mes-3 partially suppressed the HIM phenotype of xnd-1 (Figure 3f) suggesting that xnd-1 and the mes genes have opposing effects on X chromosome COs: xnd-1 ensuring that it receives COs and the mes genes inhibiting CO formation through the imposition of heterochromatic gene silencing. These results suggest that xnd-1 acts at the level of chromatin structure to control recombination.
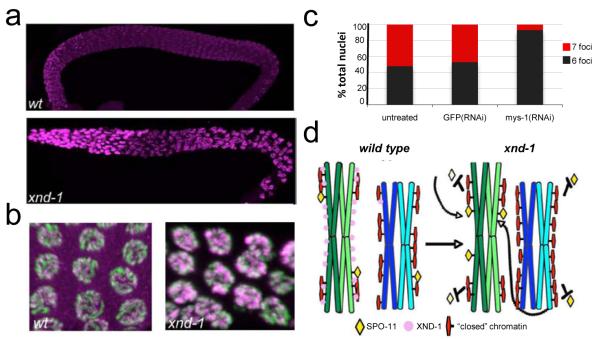
To determine whether xnd-1 has a direct effect on germline chromatin, we examined HPTMs in mutant gonads. In xnd-1 mutants, a striking increase in H2A Lysine5 acetylation (H2AK5Ac) was observed (Figures 4a, 4b). In wt, H2AK5Ac appeared faint and punctate whereas in xnd-1, H2AK5Ac labeling was more intense and uniform (Figure 4b). No differences were observed for related HPTMs, including H4K8Ac, H4K12Ac, H2BK5Ac and H3K4me2 or me3. Our data suggest that the H2AK5Ac mark may be critical for the regulation of meiotic crossovers in C. elegans.
Figure 4.
Germline chromatin is altered in xnd-1 mutants. (a) Histone H2A lysine 5 acetylation is increased in the xnd-1 mutant. wt (upper) and xnd-1 (lower) mutant gonads were stained on one slide and imaged identically revealing the more intense anti-histone H2A K5Ac staining in the xnd-1 gonad. (b) Mid-pachytene nuclei from wt and xnd-1 gonads (DNA, green; anti-H2A K5Ac, magenta) showing the more intense and uniformly distributed staining in the mutant. The gain in the wt image has been increased to reveal the modification in these nuclei. (c) mys-1(RNAi) suppresses the X chromosome CO defect of xnd-1 mutants. Quantification of DAPI staining bodies at diakinesis: 6 foci, gray; 7 foci, red (untreated, n= 94 nuclei; GFP(RNAi), n=124; mys-1(RNAi), n=288). (d) Proposed model for XND-1 function. The presence of XND-1 on autosomes allows for SPO-11 to gain access to the transcriptionally silent, heterochromatin-like X chromosomes. In the absence of XND-1, SPO-11 targets the gene-rich clusters (more transcriptionally active regions) on autosomes and titrates DSBs away from the silent X chromosomes and the less transcriptionally active autosomal arms.
In vitro, the human Tip60 protein can acetylate H2AK521. We reasoned that if H2AK5Ac is responsible for the CO defects in xnd-1, then mutating the C. elegans homologue of Tip60, mys-122 might suppress the mutant. No meiotic defects were observed in mys-1(RNAi) treated or wt controls as assessed by analyzing diakinesis Figures. As compared to xnd-1; GFP(RNAi) or untreated xnd-1 animals, which each reveals ~50% of nuclei with 7 foci, the xnd-1; mys-1(RNAi) treated worms showed only ~12% of oocytes in this class (Figure 4c). mys-1(RNAi) also decreased the amount of the H2AK5Ac mark observed in these animals (Supplementary Figure 6). These data strongly support the conclusion that xnd-1 is influencing meiotic crossover formation in C. elegans through modulation of germline chromatin.
Characterization of xnd-1 suggests that chromatin structure is a major determinant of chromosome-specific meiotic behaviors and may explain why chromosomes have different propensities to missegregate10. We identified XND-1 as an autosome-associated factor required for regulation of X chromosome recombination in C. elegans. Since xnd-1 and mes appear to act antagonistically with regards to X COs, we suggest xnd-1 may have evolved to ensure the proper disjunction of the nearly heterochromatic X, and in so doing, altered the distribution of COs genome-wide. xnd-1 may limit association (or action) of the DSB machinery with the most permissive sites in the genome, thereby increasing the pool of DSB machinery available to interact with the X chromosome and the gene-sparse regions on autosome arms (Figure 4i).
xnd-1 expands our understanding of genes involved in CO control. In C. elegans, the (as yet uncloned) rec-1 mutant shows a similar redistribution of CO into the gene cluster23, but does not affect the frequency of COs on the X. Thus, redistribution of COs to the gene clusters, per se, does not cause a deficit in COs on the X. Double mutant analysis with xnd-1 will likely inform how these gene products collaborate to regulate global chromatin organization and CO distribution. In humans, a significant fraction of hotspot usage can be explained by variants of Prdm9, a zinc-finger containing histone methyltransferase24-26, and divergent 13mer motifs to which it binds27. Although Prdm9 promotes H3K4 trimethylation28, it alone cannot explain hotspot choice since this modification localizes to a majority of transcriptionally active loci. Our work implicates another HPTM, H2AK5Ac, as a critical determinant of DSB distribution in C. elegans. Little is known about the in vivo role of H2AK5Ac; our study provides the first functional evidence that H2AK5Ac plays a deterministic role in the meiotic DSB landscape. Our data suggests that Tip60/MYS-1 modulates H2AK5Ac levels and thereby functions in CO control. In mammals, Tip60 is recruited to sites of DSB repair by phosphorylated H2AX and leads to acetylation of histone H4, a hallmark of recombinational repair7. In C. elegans, however, H2AX is not conserved. This raises the possibility that DSB initiation and repair may be coupled through the recruitment or regulation of MYS-1 and/or H2AK5 acetylation.
FULL METHODS
Worm Strains and Culture Conditions
All strains were grown and maintained under standard conditions at 20°C29. xnd-1(ok708)III and xnd-1(ok709)III were obtained from the C. elegans Gene Knockout Consortium, outcrossed at least 6 times, and maintained balanced over hT2::GFP. The mutations display maternal rescue (10% HIM in M+Z− animals (M, maternal load; Z, zygotic synthesis) compared to ~20% in M−Z−) and maternally deposited XND-1 protein can be seen in M+Z−germ cells. Therefore, our analyses of xnd-1 were restricted to M−Z− progeny.
xnd-1(ok708) and (ok709) gave equivalent percentages of males as compared to cosuppression lines, suggesting that they are loss of function alleles (Supplementary Table 1). We present analysis of ok709, but similar results have been obtained with ok708. Approximately 40% of the xnd-1 population is “clear” steriles, which are easily distinguished from fertile animals on adult day 1. These animals were separately analyzed for meiotic defects and show no differences in pairing, SC formation, or DSB formation as compared to wt. However, these animals have a cellularization defect in which multiple pachytene nuclei are aberrantly packaged into a single oocytes creating a disorganized diakinesis region (e.g. Supplementary Figure 7). These defects result in oocytes and eggs with an increased mass of DAPI staining material and massive aneuploidy and likely explains the increase in embryonic lethality.
Strains used for these analyses are listed in Supplemental Table 6 and below:
LGII: mes-2(bn11) unc-4(e120)/mnC1 dpy-10(e128)unc-52(e444).
LGIII CB185 lon-1(e185). CB364 dpy-18(e364). QP224 unc-45(e286), dpy-18(e364), unc-64(e246). QP231 dpy-1(e1), lon-1(e185), dpy-18(e364) III. RB868 xnd-1(ok708). RB869 xnd-1(ok709).
LGIV: AV106 spo-11(ok79) IV/nT1[unc-?(n754) let-?](IV;V). AV157 spo-11(me44)/nT1[unc-?(n754) let-? qIs50](IV;V). CA257 him-8(tm611)
LG V: JK2663 dpy-11(e224) mes-4(bn67) V/nT1[unc-?(n754) let-? qIs50](IV;V); zuIs178 [his-72(1kb)::HIS-72::GFP); unc-119(ed3)]
LG X: CB678 lon-2(e678).
Extrachromosomal array: let-868::GFP20
Recombination Screen
Triply marked mutant hermaphrodites, either unc-45, dpy-18, unc-64 III or dpy-1, lon-1, dpy-18 III, were crossed to N2 males to produce F1 progeny heterozygous for each of the 3 markers. Approximately five L4 hermaphrodite F1 progeny per plate fed dsRNA for gfp (control) or chromatin-associated genes according to established protocols31. Plates were scored 3-5 days later for F2 progeny that were either Dpy non-Unc or Lon non-Dpy, respectively, indicating they were homozygous for the middle marker and heterozygous or wild type for flanking markers. Because distinguishing the flanking Unc or Dpy mutant phenotypes from each other was not possible, the assessment of recombination rates was expressed as a fraction of the total progeny. Three to ten plates per genotype were scored, with a minimum of two trials per genotype.
Characterization of xnd-1 gene and transcripts
To confirm the endpoints of the deletion alleles ok708 and ok709, primers flanking the putative breakpoints were used to amplify the genomic region from homozygous animals. PCR products were sequenced using primers spanning the region. ok708 is a simple deletion and ok709 a more complex deletion/inversion. Breakpoints of both deletions are shown in Supplemental Figure 1. Additional sequence information is available from WormBase.
Seven cDNA clones were obtained from Y. Kohara, National Institute of Genetics, Japan and were sequenced using standard procedures. Five of the seven cDNAs matched the reported gene structure, according to WormBase. Two cDNAs revealed the presence of a novel splice variant (Supplemental Figure 1).
XND-1 Antibody Production
A GST fusion protein to the XND-1 N-terminus (amino acids 1-329) was expressed from pGex-6P-1 (GE Healthcare) in BL21 DE3 cells. Protein was induced with 100uM IPTG for 2 hrs at room temp prior to cell lysis and protein purification. The protein was run over a glutathione-agarose column and cleaved with PreScission protease (GE Healthcare) to isolate untagged protein for inoculation. A GST-tagged construct of the XND-1 C-terminus, amino acids 530-702, was also made and protein purification was performed and both sets of proteins were inoculated into animals for antibody production by the Proteintech Group Inc (Chicago, IL). The XND-1N protein was inoculated into guinea pigs; XND-1C protein into rabbits.
Immunostaining
Fixation and treatment of gonads for XND-1N staining was performed according to established procedures30 using 1% paraformaldehyde fix for five minutes prior to freeze cracking and ethanol treatment. For XND-1C, gonads were dissected in PBS, immediately freeze-cracked, and fixed in methanol (20 minutes) and acetone (10 minutes). Antibody concentrations are provided in Methods.
XND-1 antibodies were preabsorbed against xnd-1(ok708) animals32. XND-1N was used at a final concentration of 1:600; XND-1C at 1:200. Antibodies were generously provided to us by numerous labs and were used at the following concentrations: guinea pig (gp) anti-HTP-3(1:500) and gp anti-HIM-8 (1:500) from A. Dernburg; rabbit anti-HIM-3 (1:200) from A. Villeneuve; and gp anti-SYP-1N (1:200), rabbit anti-SYP-1C (1:200), and rabbit anti-SYP2 (1:200) antibodies from M. Colaiacovo; rabbit anti-RAD-51 from A. la Volpe; rat anti-HTZ-1 (1:2000) antibody from G. Csankovszki. Rabbit anti-RAD-51 (1:10000) and anti-REC-8 (1:10000) antibodies were purchased from SDI (Newark, DE); rabbit anti-Histone H4 (Ac12) (1:1000) from AbD Serotec (Oxford, UK); rabbit anti-dimethyl histone H3 (Lys4) (1:1000) from Upstate (Lake Placid, NY); rabbit anti-acetyl H2A (Lys 5) (1:500; 1:2500) from Cell Signaling/Millipore (Billerica, MA) and Serotec (Kidlington, UK). Secondary antibodies were Alexa-conjugated guinea pig 488 or 561; rabbit 561; or rat 488 (Invitrogen Molecular Probes, Eugene, OR).
For staining mes M−Z− germlines, the first F1 daughters of heterozygous mes mothers were individually plated and their first daughters were analyzed. By this method, we selected for granddaughters with the greatest deposition of (grand)maternal product which enhanced our ability to isolate F2 progeny with germ cells. We excluded from our analysis, animals in which germ cell morphology became indistinct.
FISH analysis
FISH XR, XL, and 5S probes were synthesized and labeled as previously described33. FISH gonad preparation, fixation, and hybridization of were performed according to published protocols16. FISH in combination with antibody staining (SYP1C, 1:200; XND-1, 1:600) was performed as described34.
Imaging
Except where noted, all studies were analyzed by confocal microscopy on either Leica TCS SP2 or SP5 microscope followed by 3-D analysis using Imaris software (Bitplane, St. Paul, MN).
Irradiation
L4 larvae aged for 20 hours and exposed to 2krad of ionizing radiation using a 137Cs source (MARK I irradiator, J.L. Shepard and Associates). Animals were further aged for 8-24 hours prior to fixation and staining. To determine if IR suppressed desynapsis, samples were analyzed 8-16hrs post-IR; for analysis of DAPI staining bodies, 24 hrs post-IR.
Analysis of Break Formation on the X chromosome
rad-54 or gfp RNAi were performed as described9. L4 larval progeny were dissected, fixed and stained with DAPI, rat anti-HTZ-1, and rabbit anti-RAD-51 antibodies. Quantification of for X chromosomal RAD-51 foci was performed using Imaris 3-D software and only mid- to late pachytene nuclei with 8 or more foci were evaluated.
Analysis of Pairing and Synapsis
xnd-1 and wild type germlines were dissected and co-labeled with 5S and XL FISH probes (described above) followed by labeling with anti-SYP-1 antibody and DAPI. Confocal images of full germlines were analyzed by dividing the transition zone to end of pachytene region into four approximately equal part and assessing the presence or absence of apposed FISH probes (indicative of pairing chromosomes) and the staining of SYP-1 along the length of a DNA fragment adjacent to the marked chromosome.
SNP mapping
xnd-1(ok708) was introgressed 6 times into the CB4658 Hawaiian background and the presence of Hawaiian-specific polymorphisms was confirmed by genotyping. Spontaneous males from this stock were crossed to lon-2(e678); xnd-1(ok709) hermaphrodites and non-Lon, xnd-1transheterozygous hermaphrodites were subsequently crossed to GFP+ males. GFP+ L4 hermaphrodites were individually plated and genotyped as described for Chr X35 and Chr I36. Chr I primers are:PM 1/2, PM 3/4, PM 7/8, PM 9/10, PM 11/12, PM 15/1636. To map sperm recombination, the Lon, transheterozygous males from the above cross were mated to dpy-18 hermaphrodites and non-Dpy cross progeny were individually plated and their progeny were genotyped as described above for chr I markers. Wild type controls for both sperm and egg were prepared simultaneously and X chromosome mapping was described in Lim et al35. Chr I mapping was performed as described above.
Hatching Rates
Individual L4 larvae were plated onto to freshly seeded NGM plates and transferred every 12 hours until the cessation of egg-laying. Eggs were counted immediately after transferring and adult animals were counted by picking worms off the plates 60-96 hours later. The number of wild type hermaphrodite (XX), male (XO), and DPY hermaphrodites (XXX) progeny were counted.
RNA interference
RNAi was performed according to established protocols9. For mys-1(RNAi), xnd-1 or wild type L4 larvae were placed on bacteria expressing the dsRNA for 48 hours and allowed to recover for 48 hours prior fixation and DAPI staining. Diakinesis stage oocytes were assayed as described above for the number and appearance of the DAPI-staining bodies.
Supplementary Material
Acknowledgements
We thank J. Bembenek, A. Bortvin, G. Deshpande, M. Halpern, V. Jantsch, A. MacQueen, and D. Mets for comments on the manuscript; the Koshland lab and Baltimore Area Worm Club for discussions; Mahmud Siddiqi for microscopy support. We are indebted to the Caenorhadbitis Genetics Center and the C. elegans Knockout Consortium for worm stocks and Yuji Kohara for cDNAs. This work was supported by the Carnegie Institution of Washington, NIH K01AG031296, and MWRI start-up funds to J.L.Y.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions
The authors declare no competing financial interests.
References
- 1.Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 5.Borde V, et al. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 6.Mieczkowski PA, Lemoine FJ, Petes TD. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:1010–1020. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 7.Buard J, Barthes P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16 Spec No. 2:R203–208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 11.Barnes TM, Kohara Y, Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly WG, et al. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 16.Dernburg AF, et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 17.Petty EL, Collette KS, Cohen AJ, Snyder MJ, Csankovszki G. Restricting dosage compensation complex binding to the X chromosomes by H2A.Z/HTZ-1. PLoS Genet. 2009;5:e1000699. doi: 10.1371/journal.pgen.1000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smolikov S, Schild-Prufert K, Colaiacovo MP. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet. 2008;4:e1000088. doi: 10.1371/journal.pgen.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong Y, Bender L, Wang W, Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 22.Ceol CJ, Horvitz HR. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- 23.Zetka MC, Rose AM. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics. 1995;141:1339–1349. doi: 10.1093/genetics/141.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 29.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan RC, Severson AF, Meyer BJ. Condensin restructures chromosomes in preparation for meiotic divisions. J Cell Biol. 2004;167:613–625. doi: 10.1083/jcb.200408061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 31.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 32.Colaiacovo MP, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 33.Phillips CM, et al. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123:1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai CJ, et al. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 2008;22:194–211. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim JG, Stine RR, Yanowitz JL. Domain-specific regulation of recombination in Caenorhabditis elegans in response to temperature, age and sex. Genetics. 2008;180:715–726. doi: 10.1534/genetics.108.090142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammarlund M, Davis MW, Nguyen H, Dayton D, Jorgensen EM. Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans. Genetics. 2005;171:1047–1056. doi: 10.1534/genetics.105.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.