Abstract
Basal-like breast cancers have several well-characterized distinguishing molecular features, but most of these are features of the cancer cells themselves. The unique stromal-epithelial interactions, and more generally, microenvironmental features of Basal-like breast cancers have not been well characterized. To identify characteristic microenvironment features of Basal-like breast cancer we performed cocultures of several Basal-like breast cancer cell lines with fibroblasts and compared these to cocultures of Luminal breast cancer cell lines with fibroblasts. Interactions between Basal-like cancer cells and fibroblasts induced expression of numerous interleukins and chemokines, including IL-6, IL-8, CXCL1, CXCL3, and TGFbeta. Under the influence of fibroblasts, Basal-like breast cancer cell lines also showed increased migration in vitro. Migration was less pronounced for Luminal lines, but these lines were more likely to have altered proliferation. These differences were relevant to tumor biology in vivo, as the gene set that distinguished Luminal and Basal-like stromal interactions in coculture also distinguishes Basal-like from Luminal tumors with 98% accuracy in 10-fold CV and 100% accuracy in an independent test set. However, comparisons between cocultures where cells were in direct contact and cocultures where interaction was solely through soluble factors suggest that there is an important impact of direct cell-to-cell contact. The phenotypes and gene expression changes invoked by cancer cell interactions with fibroblasts support the microenvironment and cell-cell interactions as intrinsic features of breast cancer subtypes.
INTRODUCTION
The heterogeneity of human breast tumors is well established (1), with Basal-like breast cancer (BBC) widely recognized as a particularly aggressive subtype of disease. Several molecular features that distinguish Basal-like breast cancers from other breast cancer subtypes have been identified including higher rates of p53 mutations (2), BRCA1 mutations and methylation (3, 4), EGFR-RAS-MEK pathway defects (5), and retinoblastoma (RB1) LOH (6). These molecular defects are characteristics of the cancer cells themselves. However, higher local recurrence rates for BBC (7) relative to other breast cancer subtypes suggests that these tumors may also be characterized by field defects arising either from epithelium, stroma, or from altered stroma-epithelium interactions (8).
The importance of stroma-epithelial interactions is well established [reviewed in Bissell and Radisky (9)]. Stroma plays a critical role in epithelial proliferation and organization through production of extracellular matrix, paracrine signaling and direct cell contact (9). Fibroblasts are a major component of the stroma and their numbers are greatly enriched in tumors (10). These cancer-associated fibroblasts also are qualitatively different than normal fibroblasts and can facilitate the progression of cancer (11). Some studies have suggested that normal fibroblasts may inhibit breast cancer (12), but other work by Wadlow et al. demonstrated that the phenotypic effects of stroma-epithelial interaction vary widely across a panel of cancer cell lines and fibroblasts derived from different anatomic sites (13). Gene expression lists reflective of stromal-epithelial interactions in breast cancer have previously been obtained using a variety systems (i.e. fibroblasts responding to serum (14), fibroblast-epithelial cocultures (15), microdissected tumor stroma and adjacent normal stroma (16), desmoid-type fibromatosis and solitary fibrous tumors to identify stromal reaction patterns (17, 18), and endothelial-epithelial cocultures (19)). Many of these stromal signatures, when evaluated in tumor microarray datasets, show prognostic value and correlate with tumor subtype (17, 20). However, as recognized by a recent panel convened to identify gaps in breast cancer research (21), “the stroma-derived signals are poorly understood, as is the reciprocal communication between epithelium and stroma”.
We hypothesized that Basal-like and Luminal breast cancers have distinct and characteristic interactions with stroma. We therefore performed cocultures where different breast cancer cell lines were grown in contact with fibroblasts, the predominant stromal cell type in breast tissue. The relevance of the observed gene expression changes from coculture were tested in vivo by evaluating the expression of these genes in patient samples. These results establish a model system for studying the molecular dynamics that govern stroma-cancer cell interactions and document novel and important interactions between Basal-like and Luminal cancers and their microenvironments.
METHODS
Cell lines
SKBR3 and MDA-MB-468 were purchased from American Type Culture Collection (ATCC) and maintained in ATCC-recommended media and conditions. MDA-MB-231 was from ATCC and grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 50 units per mL penicillin and 50 units/mL streptomycin. SUM102, SUM149, SUM159 cells were a gift from Steve Ethier of the Karmanos Cancer Center at Wayne State University. SUM102 was maintained in HuMEC (Invitrogen). SUM149 and SUM159 were maintained in Ham’s F-12 (GIBCO) supplemented with 5% FBS (Sigma Chemical Co., St. Louis, MO), 5 μg/mL insulin, 1 μg/mL hydrocotisone, 5 μg/mL Gentamycin (GIBCO/Invitrogen) 0.5 μg/ml Amphotericin B (Sigma). MCF7, ZR-75-1, ME16C were procured and cultured as described previously (22). T47D, HCC1937, and RMF (htert-immortalized fibroblasts from reduction mammoplasty, (23)) were maintained in RPMI1640 with L-glutamine (GIBCO) supplemented with 10% fetal bovine serum (Sigma) and 50U/mL penicillin and 50 units/mL streptomycin (GIBCO). SKBR3 was maintained in McCoy’s 5 (Cellgro [http://www.cellgro.com/]) supplemented with 10% fetal bovine serum (Sigma) and 50U/mL penicillin and 50U/mL streptomycin (GIBCO). All cell lines were tested for mycoplasma by the University of North Carolina at Chapel Hill Tissue Culture Facility. Cells were maintained at 37°C and 5% carbon dioxide, except where ATCC protocols suggested ambient carbon dioxide (MDA-MB-231 only). Cultures were two-dimensional, grown on plastic.
Coculture Conditions
Cancer cell lines and fibroblasts cells were grown in the appropriate cancer cell media (e.g. MCF-7 in RPMI) for the duration of the 48 hour coculture studies, after performing growth curves to ascertain that the RMFs maintained doubling times in each media similar to the doubling times observed in RPMI1640. Two types of cocultures were performed. First, we refer to direct cocultures, defined as a coculture where the two cell types are grown in physical contact, in the same well. The following RMF:cancer cell ratios were plated for most direct cocultures: 0:1, 1:4, 1:2, 1:1, 2:1, 1:0, and cells were maintained in coculture for 48 hours prior to harvesting cells for RNA isolation. Second, we refer to interaction cultures, defined as a coculture where fibroblasts and cancer cells are grown separated by a membrane, but where they are in contact via soluble factors. Interaction cocultures were plated at a 1:1 ratio in 6 well transwell plates and were maintained in coculture for 48 hours prior to harvesting cells for RNA isolation. These studies were performed by seeding one of the two cell types in the insert layer of Corning Transwell plates with 0.4 μm pore polycarbonate membranes while the other cell type was grown in the bottom well.
Cell Based Assays
MTT cell proliferation assays (Promega) were performed according to the manufacturer’s protocol in the bottom well of transwell plates with an 0.4 μm pore polycarbonate membrane after 48 hours of exposure to fibroblasts grown in the upper insert. The following RMF:cancer cell ratios were plated for 48 hours in 96-well interaction cocultures: 0:1, 1:4, 1:2, 1:1, 2:1, 1:0. Sixteen wells were performed per condition and triplicates experiments were performed. Absorbance at 570 nm minus absorbance at 490 nm was measured on an EL800 BioTek plate reader. Percent change in absorbance relative to control was calculated as the sample absorbance divided by the average absorbance of the control without fibroblasts, multiplied by 100.
Migration assays were performed using a 6-well transwell plate with an 8 μm pore polycarbonate membrane. The bottom well contained 2×105 fibroblasts or media alone and cultures were maintained for 48 hours. The insert was then seeded with 1×105 cancer cells and cancer cells were allowed to migrate for 6 hours. The chamber was disassembled and membranes were fixed in 10% formalin for 5 minutes. The membranes were rinsed in distilled water and then stained in 0.2% crystal violet for 15 minutes, followed by two washing steps in distilled water. The cells on the upper side of the membranes were scraped off and pictures were taken of the cells on the bottom of the membrane with an Olympus IX81 at 10X magnification. Four fields were counted per well using ImageJ software (http://rsbweb.nih.gov/ij/). All migration assays were replicated 3–5 times.
Scratch Assays were performed using a 6-well transwell plate with a 4 μm pore polycarbonate membrane. In the bottom well, 2×105 Basal-like cells or 5×105 Luminal cells were plated and allowed to grow for 48 hours with either media or RMF cells plated at a 1:1 ratio in the insert. The chamber was disassembled and a wound was made with a pipette tip. Cells were cultured to allow wound closure for 6 hours, at which time pictures were taken with an Olympus IX81 at 4X and 10X magnifications. Scratch assays were performed in triplicate.
RNA and Expression Microarrays
Cells were harvested by scraping in RNA lysis buffer. Total RNA was isolated using the RNeasy mini kit (Qiagen) and RNA quality was analyzed on an Agilent 2100 Bioanalyzer using an RNA6000 nano chip. Quantification was performed on a ND-1000 Nanodrop spectrophotometer. Microarrays were performed according to Agilent protocol using two-color Agilent 4×44K (Agilent G4112F) human arrays and 244K (Agilent G4502A) custom human arrays. Only probes present on the 4X44K array were utilized and all 4X44K probes were present on the 244K custom array. We used the Agilent Quick Amp labeling kit and protocol to synthesize Cy3-labeled reference from Stratagene Universal Human Reference spiked at 1:1,000 with MCF-7 RNA and 1:1,000 with ME16C RNA to increase expression of breast cancer genes. The identical protocol was applied to total RNA from cocultured or monocultured cell lines to label these samples with Cy5. Labeled cDNAs were hybridized to arrays overnight and washed prior to scanning on an Agilent G2505C microarray scanner. All array data are available through the Gene Expression Omnibus (GSEXXXX, pending).
Flow Cytometry
Cell were grown to 80% confluence and dyed with 5 uM of Invitrogen Cell Tracker (Green CMFDA for RMF; Red CMTPX for MCF7 and SUM149 cells). Cells were washed with PBS and grown in media for two hours before plating as monocultures or direct cocultures which were grown for 48 hours as previously described. Cells were trypsinized and kept on ice and sorted on a MoFlo by Beckman. Cells with high green expression were separated as the fibroblast population. RNA was isolated from green and non-green populations as described above.
Coculture data normalization and analysis
We selected three Basal-like and three Luminal breast cancer cell lines as a training set for identifying Basal-like specific coculture interactions. The Basal-like and Luminal cell line representatives were selected such that they met both of the following: (1) they were identified as Basal-like or Luminal when all of our monoculture cell line microarray data was median centered and the PAM50 assay was applied (24), and (2) this classification by PAM50 matched the classification by Neve et al. (25). The Basal cell lines meeting these criteria were SUM149 and HCC1937 and the Luminal lines were MCF-7, ZR-75-1 and T47D. To select a third Basal-like line, we added SUM102 which is a CK5/6 positive cell line that is Basal-like according to the PAM50 assay but was not assayed by Neve et al. The PAM50 and Neve predictions for each of the 11 cell lines studied in this manuscript are listed in Table 1.
Table 1.
Breast Cancer Cell Lines Cocultured with Reduction Mammoplasty Fibroblasts and their Subtype Classifications Given Various Predictors
| Cell Line | Training or Test | PAM50 | Neve et al. | Coculture-14 Subtype |
|---|---|---|---|---|
| MCF-7 | Training | Luminal | Luminal | |
| ZR-75-1 | Training | Luminal | Luminal | |
| T47D | Training | Luminal | Luminal | |
| HCC1937 | Training | Basal-like | Basal A | |
| SUM102 | Training | Basal-like | N/A | |
| SUM149 | Training | Basal-like | Basal B | |
| MDA-MB-231 | Test | Basal-like | Basal B | Basal-like |
| MDA-MB-468 | Test | Basal-like | Basal A | Basal-like |
| SUM159 | Test | Basal-like | Basal B | Basal-like |
| ME16C | Test | Basal-like | N/A | Basal-like |
| SKBR3 | Test | HER2E | Luminal | Luminal |
Data from 99 microarrays (25 monocultures, 49 direct cocultures, and 25 interaction cocultures representing 11 different cell lines) were included in this study. Only those genes where more than 70% of microarrays had signal in both channels >10 dpi were included. Data were Lowess normalized and missing data were imputed using k-nearest neighbors’ imputation. For the direct coculture analyses, we excluded genes that did not have at least 2-fold deviation from the mean in at least one sample and the method of Buess et al. (15) was used to normalize cocultures to appropriate monocultures performed in the same media and under identical conditions. The Buess method is an example of an expression deconvolution approach applied to coculture data; these methods have been widely used to analyze microarray data, yielding important insights for a variety of problems (26, 27). This method estimates the percent of fibroblasts and cancer cells in each coculture, followed by calculating a coefficient of interaction for each gene. The percent of fibroblasts and the percent of cancer cells were estimated by computing the percentage of monoculture gene expression from each cell type required to achieve the observed coculture gene expression using equations as described in Buess et al. The most extreme 20% of values for fibroblast percentage were excluded and the remaining 80% were averaged to obtain an estimate of fibroblast content. The percentages of cell types were used to compute the expected gene expression for each gene. The Buess interaction coefficient, I, was then calculated as the ratio of observed to expected gene expression. The estimated I can be thought of as an indicator of the fold-change in a given gene’s expression level relative to the expected level based on the monocultures alone. For interaction cultures, we compared their expression data to the matching monocultures performed under the same conditions and computed the interaction fold change (FC) for all genes. The I-matrix as well as the interaction FC across all samples and genes was analyzed using Significance of Microarrays (SAM) (28) and Prediction Analysis of Microarrays (PAM) (29).
Buess interaction coefficients were computed and used to perform a two-class, unpaired SAM analysis comparing cocultures in which fibroblasts and Basal-like (SUM102, SUM149, HCC1937) cancer cells were grown together to those in which fibroblasts and Luminal (MCF7, ZR-75-1, T47D) cancer cells were grown together. The same analyses were performed comparing interaction FC for Basal-like to Luminal, examining the expression of the cancer cells and the fibroblasts separately for each interaction coculture. To identify predictive gene sets for distinguishing cell line subtypes based on coculture data, PAM (29) was applied 100 times in ten-fold cross validation to identify the median class-specific predictive accuracy for a given number of genes in the predictor. The number of genes that provided the maximum class-specific predictive accuracy in both classes (Basal-like and Luminal) was selected. This set of genes was then applied to the test set cell lines that had been preprocessed using identical methods, and these gene sets were also applied to tumor data from public datasets. Functional and pathway analyses were done using Ingenuity Pathway Analysis (IPA) (Redwood City, California), with Benjamini-Hochberg multiple testing correction to identify significant functions and pathways with p-values less than 0.05.
Patient microarrays and subtype prediction
Public datasets were analyzed for this study, specifically using data from 295 tumors samples from the Netherlands Cancer Institute (NKI295) (30) and 227 samples from the University of North Carolina (UNC227) (31). In both datasets the PAM50 algorithm (24) was applied as follows: First, to median center the data the largest possible subset of microarrays consisting of 50% ER positive and 50% ER negative tumors was sampled from each dataset. The median was calculated from this group and applied to median center the entire dataset. This median centering method improves comparability across datasets by median centering given similar subtype composition. Second, the PAM50 single sample predictor, with parameters as described in Parker et al. (24), was used to identify subtype. This PAM50 subtype was used for comparison against all culture-derived subtype predictors.
To identify a coculture-based subtype predictor, the probes that distinguished Basal-like and Luminal cocultures were filtered to identify genes that were (1) up-regulated in Basal-like cocultures and above the median for at least 60% of the Basal-like tumors or (2) up-regulated in Luminal cocultures and above the median for at least 60% of Luminal tumors. This criterion ensured that only those genes identified in coculture that share the same direction in tumors were included. This gene set was further refined to identify the minimum number of predictive genes that had maximum class-specific predictive accuracy in ten-fold cross validation using PAM. To identify a fibroblast-derived or cancer-cell derived interaction signature we performed the same filtering methods to the interaction culture data representing fibroblast or cancer-cell responses to coculture. Each of these PAM models was trained on a dataset from NKI consisting of 45 Basal and 217 Luminal breast cancers. Genes from the training dataset were applied to identify subtype on the independent test sets described above.
RESULTS
Basal-like vs. Luminal gene expression in fibroblast cocultures
Two different types of cocultures were employed: direct cocultures where cells are in physical contact and interaction cocultures using transwell plates. Using established methods for identifying gene expression interactions in direct cocultures (15), we performed two-class Significance Analysis of Microarrays (SAM) to identify interactions that were distinct between Basal-like and Luminal cocultures. A total of 1320 probes showed statistical evidence of distinct interaction given a false discovery rate less than 0.001. Among these 1320 probes, we eliminated genes that behaved idiosyncratically in one cell line (rejecting genes that were not uniformly up-regulated in Basal-like or Luminal cell lines) to yield 488 probes representing 369 genes. A hierarchical cluster of these genes, displaying their coculture interaction coefficients, is shown in Figure 1 and the complete list of genes and their interaction coefficients is provided in Supplemental Table 1. In Figure 1, 140 genes are upregulated specifically in Luminal lines compared to Basal-like, and 229 genes are upregulated in Basal-like lines relative to Luminals. This suggests that interactions with fibroblasts are distinct between Basal-like and Luminal cancer cells.
Figure 1. Basal-like and Luminal Cocultures Show Distinct Gene Expression Profiles.
Hierarchical clustering shows 369 genes that were significantly and consistently differentially expressed between Basal and Luminal breast cancer, when interacting with fibroblasts. The heatmap shows the log2(R/G) of the Buess interaction coefficient, so the color represents the fold change relative to monoculture. A complete list of genes and their interaction coefficients across these samples are available in Supplemental Table 1.
To confirm the gene expression changes identified by computational deconvolution in Figure 1, we performed direct cocultures using dye-labeled cells and then separated cells by fluorescence activated cell sorting (FACS) for expression analysis. Microarrays were performed using RNA from the sorted cells and Table 2 (and Supplemental File 1) illustrates that 94–95% of the expression changes identified by deconvolution were observed in either fibroblasts, cancer cells, or both.
Table 2.
Genes identified in computational deconvolution are differentially expressed in sorted cells.
| Computational Results Validated in Sorted Cellsa | Cellular Subpopulation Expressing Validated Genesb | ||||
|---|---|---|---|---|---|
| Yes | No | Cancer Cells | Fibroblasts | Both | |
| SUM149 (basal) | 199 (94%) | 13 (6%) | 24 (12%) | 67 (34%) | 108 (54%) |
| MCF7 (luminal) | 124 (95%) | 7 (5%) | 50 (40%) | 22 (18%) | 52 (42%) |
343 genes from the original 369 identified in Figure 1 passed filters for image quality. 212 of these were genes originally upregulated in basal (evaluated in SUM149) and 131 were originally upregulated in luminal (evaluated in MCF7).
Validated genes included 199 total for SUM149 cocultures and 124 for MCF7 cocultures.
Several pathways were enriched specifically in Basal-like or Luminal direct cocultures. By Ingenuity Pathway Analysis (IPA), Luminal cocultures showed signaling changes in amino acid metabolism, DNA replication, nucleic acid metabolism, and post-translational modification (Supplemental Table 2). Interestingly, many of these categories are those that are upregulated in cellular proliferation (32). The Basal-like cocultures were enriched for pathways in hepatic fibrosis, activation of IRF, TREM1 signaling, LXR/RXR Activation, glucocorticoid receptor signaling, IL-6 Signaling, p38 MAPK Signaling, LPS/IL-1 mediated inhibition of RXR Function, and Growth Hormone Signaling (Supplemental Table 3). Thus, enrichment for pathways involved in immune response was observed in Basal-like cocultures. We had previously observed an increased wound response signature in the normal tissue adjacent to breast tumors (20). Our analyses of cocultures show that this wound response signaling is up-regulated in this model system, with both Basal-like and Luminal cocultures showing altered expression of genes in the 200-probe/130 gene wound response signature. However, consistent with our previous observations in vivo (33), in coculture different genes appeared to be upregulated in each cancer subtype with CCL2, CSF3, CXCL1, LAMC2 TLR2, TNFRSF6B and TREM1 up-regulated in Basal-likes and CEACAM6, RECK, S100A8 and TMC5 up-regulated in Luminals. Relative to monocultures comparing Basal-like and Luminal cancer cells, cocultures produced a broader range of expression differences. Comparing the expression of the three mono-culture Basal-like cell lines against the three Luminal cell lines using the same methods and FDR thresholds, we found only 10 probes corresponding to 8 genes. These genes were TFF3, PDZK1, KRT19, FLJ12650, EFHD1, CD109, FLJ10134 and MGC34923. Thus fibroblast interactions appeared to enhance the differences between Basal-like and Luminal cancers.
Cell proliferation and migration phenotypes for cancer cells in fibroblast cocultures
Based on our observation from gene expression that proliferation-related processes were upregulated due to interactions between Luminal cells and fibroblasts, we performed MTT assays to evaluate change in growth rate for Luminal and Basal interaction cocultures. While there was no net difference considering all luminal cell lines as a group (vs. all basal-like cell lines as a group), all three Luminal cell lines showed statistically significant differences in proliferation in coculture, and were more likely to display altered proliferation (Figure 2A). MCF-7 and T47-D increased cell number by 22% (p=1.0×10−6) and 14.4% (p=5.0×10−7) relative to monocultures. Only the Luminal line ZR-75-1 showed a significant decrease in proliferation (−8.7%, p=0.02). Among the Basal-like lines, only one line showed a significant change in proliferation with coculture; the SUM149 cells appeared to increase growth by approximately 15% (p=5.0×10−16), while the SUM102 and HCC1937 had non-significant decreased growth rates of 6% and 0.5%, respectively.
Figure 2. Basal-like vs. Luminal Differences in Proliferation and Migration in Response to Coculture.
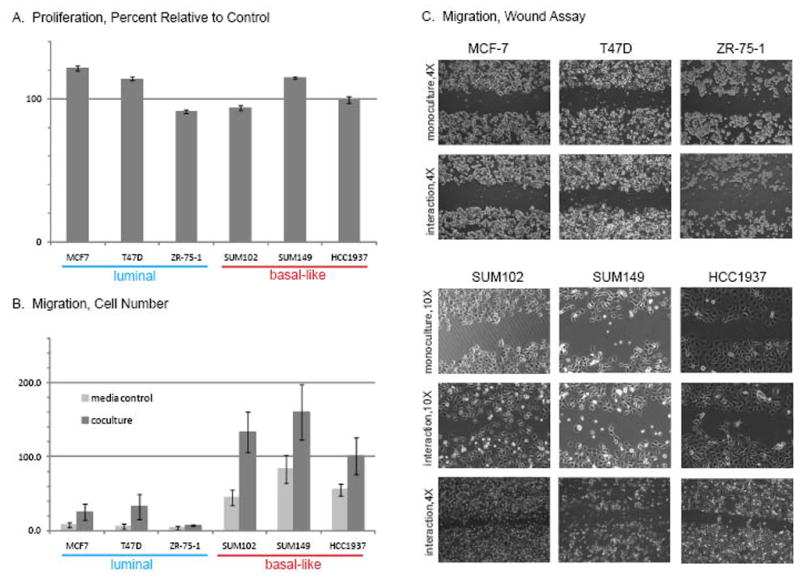
Interaction cocultures were used to measure proliferation using an MTT assay (A), migration using a Transwell migration assay (B) and wound healing using a scratch assay (C). Relative to monoculture, cocultures induced significant proliferation changes for all three Luminal lines and for one of the three Basal-like lines. Migration and wound healing were more pronounced both at baseline and following coculture for the Basal-like breast cancer cell lines.
In contrast, all three Basal-like lines showed increased cellular migration in interaction cocultures. Although these cell lines display migration capacity in monoculture, the number of migratory cells increased substantially when cocultured in a transwell migration assay (Figure 2B). In addition the ability of the Basal-like lines to close a wound was enhanced in the presence of RMF cells (Figure 2C). These results show a functional change in the cellular behavior in response to coculture. These results mirror observations based on gene expression data, where the Basal-like cells showed upregulation of epithelial-to-mesenchymal transition-related pathways such as HGF signaling (hepatic fibrosis in Supplemental Table 3), and also upregulated genes like TGFB1 and TWIST.
Direct vs. Interaction Coculture Differences
The direct cocultures suggested phenotypes that we were able to confirm using interaction cocultures phenotypic assays (MTT and Transwell migration assays), but we hypothesized that there may be differences in the character and strength of the response to coculture depending upon whether the cells were in direct contact or interacting only through soluble factors. Figure 3 shows that few of the genes expressed in direct coculture (Figure 3A) were similarly expressed in interaction cultures [The heatmap in Figure 3B is based on two Basal-like (SUM102, SUM149), and two Luminal breast cancer cell lines (MCF-7 and T47D)]. Despite the small percentage of genes that showed strong overlap (such as those in Figure 3C), it cannot be excluded that important interactions are mediated through soluble factors, because the interaction cocultures were able to induce the phenotypic effects of increased proliferation and migration. In fact, TWIST, IL6, IL8, CXCL3, and SOD2 are induced in both direct and interaction Basal-like cocultures.
Figure 3. Gene Expression Differences Observed in Direct Coculture are Not Fully Recapitulated in Interaction (Transwell) Cocultures.
This 1-dimensional (genes clustered only) cluster of the 369 genes from coculture included direct (A) and interaction (B) cocultures where either fibroblasts alone (but under the influence of Luminal or Basal-like breast cancer cell lines) or breast cancer cell lines alone (but under the influence of fibroblasts) were used to harvest RNA for microarrays. The color bars along the top indicate which cell type was represented in the microarray: orange indicates fibroblasts, blue indicates Luminal cells, and red indicates Basal-like cells. A small percentage of genes (C) show strong overlap between direct and interaction cocultures.
SAM analysis of direct cocultures to identify genes differentially expressed in the fibroblasts under the influence of Basal-like vs. Luminal cancer cell lines identified 233 probes that were differentially expressed (with FDR<0.001), and 204 of these probes (representing 152 genes) were consistently expressed across both Basal-like or both Luminal cocultures. Likewise, for cancer cell lines in interaction cultures with fibroblasts, 279 probes (with FDR<0.001) overall and 248 probes (189 genes) uniformly expressed by cell type were identified. Only three genes were common to the fibroblast and cancer cell lists and changed in the same direction in response to coculture: PGK1, MGC21688 and PTX3. Two genes were regulated in opposite directions in fibroblasts and cancer cells (TNFAIP2, ARID5A). The remaining genes were significant in either fibroblasts or cancer cell lines, but not in both. The direct cocultures appear to have a broader transcriptional response, enriched for more Gene Ontology (GO) functions and more signaling pathways than the transwell cultures. But both the direct and interaction coculture gene lists were enriched for common GO terms such as cellular movement, cellular growth and proliferation, and cell death. The coculture gene list is additionally enriched for cell morphology, cellular development, cell growth and proliferation, and drug metabolism (Supplemental Table 4) as well as signaling pathways such as hepatic fibrosis/hepatic stellate cell activation, role of macrophage, fibroblasts and endothelial cells in rheumatoid arthritis, role of pattern recognition receptors in recognition of bacteria and viruses, antigen presentation pathway, oncostatin M signaling and glucocorticoid receptor signaling (Supplemental Table 5).
Independent Validation In Vitro and Subtype Prediction In Vivo
To test whether these results were generalizable to other cell line models of breast cancer, we next employed direct cocultures with 5 additional cell lines as a test set. The test set included: SUM159, SKBR3, MDA-MB-231, MDA-MB-468, ME16C. Several fibroblast:epithelial ratios were performed for each cell line. Using our training samples and the 369 genes selected by SAM analyses as significantly associated with subtype, we performed 10-fold cross validation with PAM to identify a set of 14 genes that could reliably distinguish Basal-like from Luminal cocultures. We then applied this set of 14 genes, “the Coculture14” (Supplemental Table 6) to predict the subtype of the test set cocultures. The predicted subtype for each coculture is shown in Table 1. The enrichment for cytokines in the predictive signatures suggests that high expression of cytokines is a key feature in predicting Basal-like-fibroblast interactions.
To evaluate whether these genes were differentially expressed in vivo as a function of breast cancer subtype, we used the coculture derived lists to distinguish Basal-like and Luminal cancers. Supplemental Figure 1 shows the heat map of 140 genes from the coculture 369-gene list that passed an interquartile range (IQR) filter (IQR>1.0) across the 295 samples from the NKI. From this supervised cluster, it is evident that Basal-like and Luminal breast cancers differentially express the genes identified in Basal-like vs. Luminal cocultures. The fourteen genes in our Coculture14 predictor derived from the cell lines mapped to the NKI dataset and this gene set showed 91% training accuracy in identifying Basal and Luminal tumors (Table 3), but the class-specific accuracy was low for Basal-like breast cancers. We also rebuilt a second predictor using 105 genes that mapped to the dataset that were concordant between tumor and cell lines as described in Methods. Using this approach we identified a signature of 29 genes, “Coculture29”, (Supplemental Table 6) which had 98% cross-validation accuracy in predicting Basal vs. Luminal subtype, and good class-specific accuracy for both Basal-like and Luminals. Testing this signature on UNC227 [24] gave similar predictive accuracy (Table 3). Interestingly, the HER2 tumors were more likely to be classified as Luminal than Basal-like, suggesting that these tumors share some emergent features with Luminal cancers.
Table 3.
Predictor tumor subtype according to PAM50, 14 gene coculture, 29 gene coculture, 18-gene cancer cell, 30-gene fibroblast
| PAM50 | Coculture14, trained in vitro | Coculture29, trained in vivo | CancerCell18, trained in vivo | Fibroblast30, trained in vivo | |||||
|---|---|---|---|---|---|---|---|---|---|
| Basal-like | Luminal | Basal-like | Luminal | Basal-like | Luminal | Basal-like | Luminal | ||
| TRAINING NKI 295 | |||||||||
| Basal-like (n) | 45 | 21 | 24 | 45 | 0 | 40 | 5 | 38 | 7 |
| HER2E (n) | 25 | 2 | 23 | 4 | 21 | 7 | 18 | 3 | 22 |
| Luminal (n) | 217 | 0 | 217 | 4 | 213 | 2 | 215 | 0 | 217 |
| Normal (n) | 8 | 0 | 8 | 2 | 6 | 2 | 6 | 1 | 7 |
| Subtype-Specific Accuracy (%)a | 46.7% | 100.0% | 100.0% | 98.2% | 88.9% | 99.1% | 84.4% | 100.0% | |
| Overall Accuracy (%)a | 90.8% | 98.5% | 97.3% | 97.3% | |||||
| TEST UNC 227 | |||||||||
| Basal-like (n) | 66 | 24 | 42 | 66 | 0 | 48 | 18 | 26 | 40 |
| HER2E (n) | 26 | 0 | 26 | 5 | 21 | 0 | 26 | 1 | 25 |
| Luminal (n) | 118 | 0 | 118 | 0 | 118 | 0 | 118 | 0 | 118 |
| Normal (n) | 17 | 1 | 16 | 7 | 10 | 4 | 13 | 3 | 14 |
| Subtype-Specific Accuracy (%)b | 36.4% | 100.0% | 100.0% | 100.0% | 72.7% | 100.0% | 39.4% | 100.0% | |
| Overall Accuracy (%)b | 77.2% | 100.0% | 90.2% | 78.3% | |||||
Percent accuracy considering only 262 tumors, Basal-like (n=45) and Luminal (n=217).
Percent accuracy considering only 184 tumors, Basal-like (n=66) and Luminal (n=118).
Predictive Value of Fibroblast-Derived and Cancer-Cell Derived Gene Lists In Vivo
Having established the relevance of the direct coculture signatures in predicting subtype in vivo, gene expression in interaction cultures was used to evaluate the significance of fibroblast-derived and cancer-cell derived gene expression in characterizing tumor subtype. We found that signatures derived from the interaction cocultures had poor predictive accuracy in vivo relative to signatures derived from direct contact. Using 189 genes that were differentially expressed by Basal-like vs. Luminal cells when under the influence of fibroblast-derived soluble factors, a set of 18 epithelial-derived genes (CancerCell18) were identified that had maximum overall (97%) and class-specific accuracy (89–99%) in identifying Basal-like and Luminal cancers. However, in testing this gene set, overall accuracy dropped to 90% and Basal-like accuracy dropped to 72%. Likewise, using the 152 genes altered in fibroblasts under the influence of cancer cells, a 30-gene list (Fibroblast30) was identified that had 97% overall accuracy and 84% accuracy in identifying Basal-like tumors. However, testing this signature on a second dataset (UNC227) resulted in only 40% accuracy among Basal-like tumors. The interaction coculture gene sets (Supplemental Table 6) were not as relevant to in vivo subtype as the direct coculture signatures.
DISCUSSION
The microenvironment features that are unique to Basal-like vs. Luminal breast cancer have not been well characterized. Cancers develop and evolve through interaction with the microenvironment and thus the disease ultimately represents molecular features that accommodate both tumor cell and stromal cell characteristics. In other words, the tumor in vivo is more than the sum of its parts and represents the product of reciprocal interactions. The stromal-epithelial interactions may be targetable to slow growth or reduce metastases, so characterizing these interactions has translational relevance. To better understand the stromal cell interactions of breast cancer subtypes, we studied representative cell lines using fibroblast-cancer cell cocultures. Across 11 different cell lines, emphasizing six that were selected a priori as the representatives of Basal-like or Luminal cancers, we identified stromal interactions that distinguish Basal-like and Luminal breast cancers. This signature had a high degree of predictive accuracy (>98%) in distinguishing Basal from Luminal breast tumors in independent data. In fact, the direct coculture signatures (relative to interaction coculture signatures) resulted in predictors with the highest accuracy in predicting tumor subtype. The biological relevance of direct interaction is supported by studies in mouse xenograft models, where co-injection of cancer cells and fibroblasts or injection of cancer cells pre-treated with direct coculture-conditioned media results in tumor growth promotion (34, 35). This suggests that cell-cell contact in the microenvironment is a fundamental characteristic of breast cancer subtype.
Our observations are consistent with the idea that a tumor is more than the sum of its parts. The gene expression signatures we report represent an “emergent” property of breast cancer subtypes, defined as properties that cannot be identified through functional decomposition. By using a computational deconvolution process that preserves cell-cell contact and more closely approximates functional composition in vivo, we were able to identify biological signatures with implications for breast cancer. We confirmed that changes identified by computational deconvolution were also changed in the same direction in physically separated cells. As our result show, even if soluble interactions are preserved, such as in transwell interaction cultures, the gene expression differs markedly from that observed in direct cocultures where cell-cell contact is intact. However, the interaction cultures preserved some important signals involved in migratory phenotypes (e.g. IL6, IL8, etc.) and induced migratory behavior. Thus both systems have applicability for studying different aspects of the heterotypic interactions between cell types.
The role of fibroblast-cancer cell interactions in mediating many of the Basal-like pathways we identified had not previously been demonstrated. For example, oncostatin M dysregulation and alterations in hepatic fibrosis-associated genes were significantly altered in Basal-like coculture. Oncostatin M is a proinflammatory mediator that induces transmigration of human neutrophils through endothelium by stimulating expression of adhesion molecules and chemokines (36). Treatment of breast cancer cell lines with oncostatin M has previously been shown to reduce proliferation and induce cellular morphology changes and EGFR expression (37, 38), the latter of which is known to be a common occurrence in Basal-like breast cancer (5). Oncostatin M signaling also plays a critical role in differentiation and death of mammary epithelial cells (39). Thus, this pathway merits further investigation in relation to Basal-like breast cancer pathogenesis, emphasizing the role of cell-cell communication in modulating that pathway. STAT1 is a component of several of the significant pathways and this is noteworthy because this gene and interferon-gamma signaling were previously shown to be dysregulated in fibroblast-breast cancer coculture systems (15). Thus our results for STAT1 are consistent with previous literature, but also demonstrate that STAT1 alterations vary with subtype. The observed association between high STAT1 protein expression and poor disease specific survival (15) may be due to STAT1 expression acting as a surrogate biomarker for Basal-like breast cancer. Our objective was to identify how patterns of interactions differ between two important breast cancer subtypes. While some of these specific genes and pathways may be targetable, future studies should explore mechanism in greater detail for some of these pathways. However, the patterns we observed show that our coculture studies rediscovered some well-established biology of Basal-like breast cancer, but generated new insights and information regarding the role of stromal cell interactions in these pathways.
Basal-like cells respond to stromal interactions in our systems by increasing migration, while Luminal lines more frequently showed altered proliferation. One Basal-like cell line and a representative of the most aggressive, inflammatory breast cancers (40), SUM149, increased both proliferation and migration. Interestingly, the increased migratory capacity and upregulation of TWIST and TGF-beta, are characteristics consistent with increased epithelial-to-mesenchymal transition in Basal-like breast cancer cells interacting with fibroblasts. The induction of EMT/stem cell phenotypes by stromal interactions have been reported previously in other cancers (41, 42) and merits further investigation in breast cancer. These data also raise the hypothesis that invasive Basal-like breast cancers may progress by distinct microenvironment-mediated cellular mechanisms relative to Luminal cancers. For example, “self-seeding”, wherein circulating tumor cells colonize their tumors of origin and thereby increase their size (43), has been demonstrated compellingly with mouse xenograft models using the MDA-MB-231 (Basal-like or Basal B (25)) cell line model. This phenomenon was observed in the presence of stroma and many of the pathways that were altered specifically in Basal-like cocultures (e.g. IL-6, IL-8, oncostatin M signaling, and CXCL1) were important mediators of this effect (44). IL-8, in particular, has previously been associated with estrogen receptor negative cancers (45) and facilitates invasion among cell lines that possess cancer stem cell properties (46), however, its regulation has largely been studied in monoculture systems (47). A recent study has shown that regulation of IL-8 may occur through heterotypic signaling involving microRNA 17/20 (48). This suggests that coculture systems may represent important and convenient in vitro models for some of the cellular capabilities (e.g. migration) relevant to self-seeding phenomena. Because the genes involved in self-seeding appear to be upregulated by heterotypic interactions with Basal-like and not Luminal cell lines, this also raises the hypotheses that self-seeding may be characteristic of the more aggressive Basal-like breast cancer subtype. More broadly, these results suggest that targeting cancer cell capabilities other than mitosis, such as migration, may be an important strategy for halting progression of Basal-like breast cancer (43).
Future work should examine HER2-enriched (HER2E) subtype-specific interactions and also interactions between stroma and the emerging Claudin-low subtype of breast cancer (49). Our results suggest, based on both the SKBR3 coculture predictions and the predictions for HER2E tumors, that HER2E breast cancers may be associated with microenvironment conditions that are more similar to Luminal breast cancer microenvironments. However, a two-class predictor requires that a tumor be made to fit one of these categories, where certain signatures may be unique or unrecognized without explicitly studying this subtype. Likewise, the MDA-MB-231 and SUM159 may share some of the mesenchymal features of Claudin-low breast cancer (49) and so may be a strong model for this subtype. As models for this breast cancer subtype become more well-established, it will be possible to define unique microenvironment characteristics for Claudin-low tumors. It will also be important to study interindividual variation in fibroblasts (13) and how other mesenchymal cell types and endothelial cells interact with cancer cells.
Coculture systems serve as convenient in vitro models for studying molecular dynamics governing breast cancer subtype progression. Previously, we have used many of these cell lines to study chemosensitivity with proliferation as our primary endpoint (50). For drugs that target mitosis, gene expression profiles of drug-treated monocultures have been good predictors of mechanisms and sensitivity (51). However, as our appreciation for the importance of stem cells, microenvironment, and angiogenesis have grown and as new drugs targeting these pathways develop, novel approaches for understanding mechanism and predicting response will be needed. Coculture models have substantial advantages over monocultures for recapitulating many of the biological dynamics relevant to the mechanisms of action of these drugs. Pairing cocultures, computational deconvolution, and appropriate phenotypic endpoints (e.g. migration or invasion) allows important insights about mechanisms relevant to breast cancer biology and treatment.
Supplementary Material
Acknowledgments
We thank Dr. Ken. Hanson for image analysis of the migration assays. This research was supported by grants from National Cancer institute (R01 CA138255), National Institute of Environmental Health Sciences (R01 ES015739), the Center for Environmental Health and Susceptibility (P30 ES010126), and National Cancer Institute Breast SPORE Career Development Award (P50 CA58233-18), and the University Cancer Research Fund.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 3.Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 4.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 5.Hoadley KA, Weigman VJ, Fan C, Sawyer LR, He X, Troester MA, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyndi M, Sorensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol. 2008;26:1419–1426. doi: 10.1200/JCO.2007.14.5565. [DOI] [PubMed] [Google Scholar]
- 8.Heaphy CM, Griffith JK, Bisoffi M. Mammary field cancerization: molecular evidence and clinical importance. Breast Cancer Res Treat. 2009;118:229–239. doi: 10.1007/s10549-009-0504-0. [DOI] [PubMed] [Google Scholar]
- 9.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 11.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 12.Dong-Le Bourhis X, Berthois Y, Millot G, Degeorges A, Sylvi M, Martin PM, et al. Effect of stromal and epithelial cells derived from normal and tumorous breast tissue on the proliferation of human breast cancer cell lines in co-culture. Int J Cancer. 1997;71:42–48. doi: 10.1002/(sici)1097-0215(19970328)71:1<42::aid-ijc9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Wadlow RC, Wittner BS, Finley SA, Bergquist H, Upadhyay R, Finn S, et al. Systems-level modeling of cancer-fibroblast interaction. PLoS One. 2009;4:e6888. doi: 10.1371/journal.pone.0006888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [see comments] [DOI] [PubMed] [Google Scholar]
- 15.Buess M, Nuyten DS, Hastie T, Nielsen T, Pesich R, Brown PO. Characterization of heterotypic interaction effects in vitro to deconvolute global gene expression profiles in cancer. Genome Biol. 2007;8:R191. doi: 10.1186/gb-2007-8-9-r191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008 doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 17.Beck AH, Espinosa I, Gilks CB, van de Rijn M, West RB. The fibromatosis signature defines a robust stromal response in breast carcinoma. Lab Invest. 2008 doi: 10.1038/labinvest.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West RB, Nuyten DS, Subramanian S, Nielsen TO, Corless CL, Rubin BP, et al. Determination of stromal signatures in breast carcinoma. PLoS Biol. 2005;3:e187. doi: 10.1371/journal.pbio.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buess M, Rajski M, Vogel-Durrer BM, Herrmann R, Rochlitz C. Tumor-endothelial interaction links the CD44(+)/CD24(−) phenotype with poor prognosis in early-stage breast cancer. Neoplasia. 2009;11:987–1002. doi: 10.1593/neo.09670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troester MA, Lee MH, Carter M, Fan C, Cowan DW, Perez ER, et al. Activation of host wound responses in breast cancer microenvironment. Clin Cancer Res. 2009;15:7020–7028. doi: 10.1158/1078-0432.CCR-09-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson A, Brennan K, Cox A, Gee J, Harcourt D, Harris A, et al. Evaluation of the current knowledge limitations in breast cancer research: a gap analysis. Breast Cancer Res. 2008;10:R26. doi: 10.1186/bcr1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Troester MA, Hoadley KA, Sorlie T, Herbert BS, Borresen-Dale AL, Lonning PE, et al. Cell-type-specific responses to chemotherapeutics in breast cancer. Cancer Res. 2004;64:4218–4226. doi: 10.1158/0008-5472.CAN-04-0107. [DOI] [PubMed] [Google Scholar]
- 23.Proia DA, Kuperwasser C. Reconstruction of human mammary tissues in a mouse model. Nat Protoc. 2006;1:206–214. doi: 10.1038/nprot.2006.31. [DOI] [PubMed] [Google Scholar]
- 24.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Master SR, Chodosh LA. Computational expression deconvolution in a complex mammalian organ. BMC Bioinformatics. 2006;7:328. doi: 10.1186/1471-2105-7-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu P, Nakorchevskiy A, Marcotte EM. Expression deconvolution: a reinterpretation of DNA microarray data reveals dynamic changes in cell populations. Proc Natl Acad Sci U S A. 2003;100:10370–10375. doi: 10.1073/pnas.1832361100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher V, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, Samaha RR, et al. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 2006;7:127. doi: 10.1186/1471-2164-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troester MA, Millikan RC, Perou CM. Microarrays and epidemiology: ensuring the impact and accessibility of research findings. Cancer Epidemiol Biomarkers Prev. 2009;18:1–4. doi: 10.1158/1055-9965.EPI-08-0867. [DOI] [PubMed] [Google Scholar]
- 34.Stuelten CH, Busch JI, Tang B, Flanders KC, Oshima A, Sutton E, et al. Transient tumor-fibroblast interactions increase tumor cell malignancy by a TGF-Beta mediated mechanism in a mouse xenograft model of breast cancer. PLoS One. 5:e9832. doi: 10.1371/journal.pone.0009832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modur V, Feldhaus MJ, Weyrich AS, Jicha DL, Prescott SM, Zimmerman GA, et al. Oncostatin M is a proinflammatory mediator. In vivo effects correlate with endothelial cell expression of inflammatory cytokines and adhesion molecules. J Clin Invest. 1997;100:158–168. doi: 10.1172/JCI119508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douglas AM, Grant SL, Goss GA, Clouston DR, Sutherland RL, Begley CG. Oncostatin M induces the differentiation of breast cancer cells. Int J Cancer. 1998;75:64–73. doi: 10.1002/(sici)1097-0215(19980105)75:1<64::aid-ijc11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 38.Grant SL, Hammacher A, Douglas AM, Goss GA, Mansfield RK, Heath JK, et al. An unexpected biochemical and functional interaction between gp130 and the EGF receptor family in breast cancer cells. Oncogene. 2002;21:460–474. doi: 10.1038/sj.onc.1205100. [DOI] [PubMed] [Google Scholar]
- 39.Tiffen PG, Omidvar N, Marquez-Almuina N, Croston D, Watson CJ, Clarkson RW. A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo. Mol Endocrinol. 2008;22:2677–2688. doi: 10.1210/me.2008-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmeyer MR, Wall KM, Dharmawardhane SF. In vitro analysis of the invasive phenotype of SUM 149, an inflammatory breast cancer cell line. Cancer Cell Int. 2005;5:11. doi: 10.1186/1475-2867-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 42.Giannoni E, Bianchini F, Masieri L, Serni S, Torre E, Calorini L, et al. Reciprocal Activation of Prostate Cancer Cells and Cancer-Associated Fibroblasts Stimulates Epithelial-Mesenchymal Transition and Cancer Stemness. Cancer Res. doi: 10.1158/0008-5472.CAN-10-0785. [DOI] [PubMed] [Google Scholar]
- 43.Norton L, Massague J. Is cancer a disease of self-seeding? Nat Med. 2006;12:875–878. doi: 10.1038/nm0806-875. [DOI] [PubMed] [Google Scholar]
- 44.Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freund A, Chauveau C, Brouillet JP, Lucas A, Lacroix M, Licznar A, et al. IL-8 expression and its possible relationship with estrogen-receptor-negative status of breast cancer cells. Oncogene. 2003;22:256–265. doi: 10.1038/sj.onc.1206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freund A, Jolivel V, Durand S, Kersual N, Chalbos D, Chavey C, et al. Mechanisms underlying differential expression of interleukin-8 in breast cancer cells. Oncogene. 2004;23:6105–6114. doi: 10.1038/sj.onc.1207815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, et al. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Troester MA, Hoadley KA, Parker JS, Perou CM. Prediction of toxicant-specific gene expression signatures after chemotherapeutic treatment of breast cell lines. Environ Health Perspect. 2004;112:1607–1613. doi: 10.1289/txg.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [see comments] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




