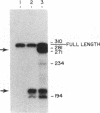
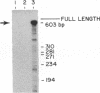
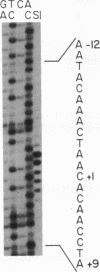
Abstract
Galactose utilization in Streptomyces lividans was shown to be controlled by an operon that is induced in the presence of galactose and repressed by glucose. Two promoters, galP1 and galP2, which direct transcription of two distinct polycistronic transcripts, have been identified. galP1 is located immediately upstream of the operon and is induced in the presence of galactose. This promoter directs transcription of the galT, galE, and galK genes. The second promoter, galP2, is located within the operon just upstream of the galE gene. This promoter is responsible for constitutive transcription of the galE and galK genes. Comparison of the S. lividans gal operon to the Escherichia coli gal operon indicates the presence of a constitutive promoter positioned upstream of galE in both operons. We suggest that coupling the operon's constitutive promoter to the galE gene fulfills a physiological requirement for constitutive UDPgalactose 4-epimerase expression in Streptomyces.
Full text
PDF

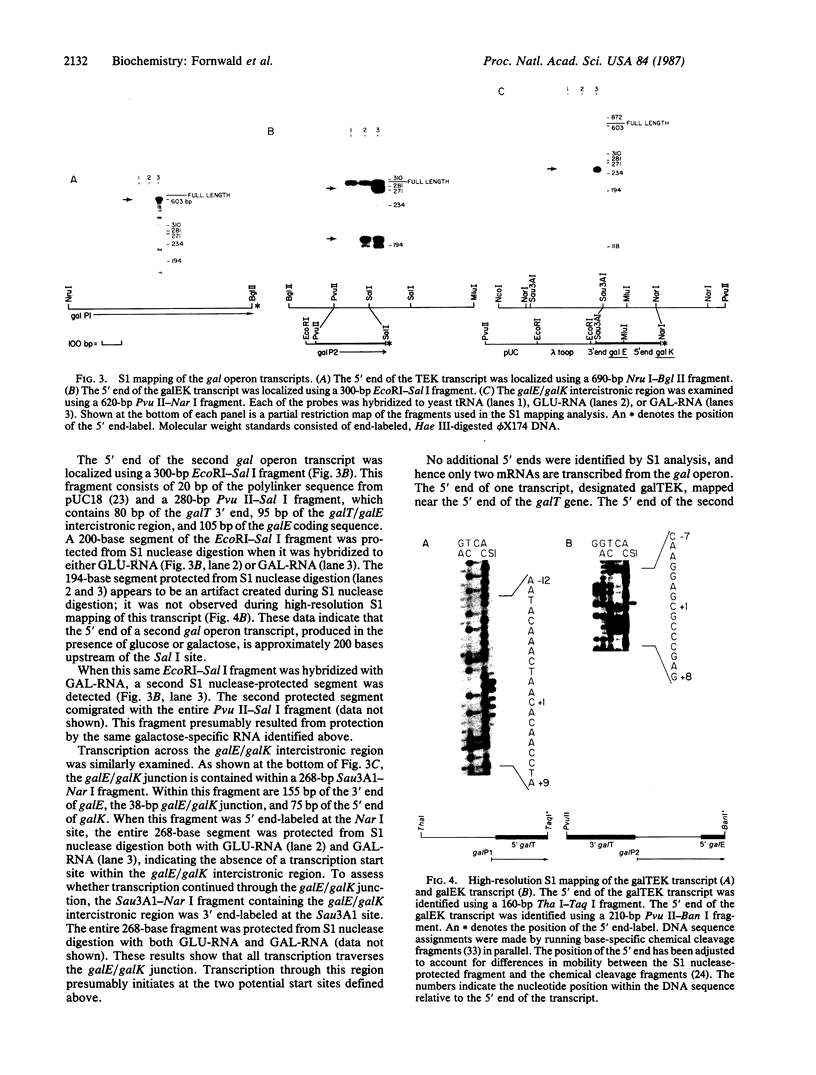


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barry G., Squires C. L., Squires C. Control features within the rplJL-rpoBC transcription unit of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4922–4926. doi: 10.1073/pnas.76.10.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawner M. E., Auerbach J. I., Fornwald J. A., Rosenberg M., Taylor D. P. Characterization of Streptomyces promoter sequences using the Escherichia coli galactokinase gene. Gene. 1985;40(2-3):191–201. doi: 10.1016/0378-1119(85)90042-3. [DOI] [PubMed] [Google Scholar]
- Cerretti D. P., Dean D., Davis G. R., Bedwell D. M., Nomura M. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res. 1983 May 11;11(9):2599–2616. doi: 10.1093/nar/11.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J. Resistance, regulatory and production genes for the antibiotic methylenomycin are clustered. EMBO J. 1985 Jul;4(7):1893–1897. doi: 10.1002/j.1460-2075.1985.tb03866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., Kopetzki E., Fröhlich K. U., Mecke D. Cloning of hexokinase isoenzyme PI from Saccharomyces cerevisiae: PI transformants confirm the unique role of hexokinase isoenzyme PII for glucose repression in yeasts. Mol Gen Genet. 1984;198(2):50–54. doi: 10.1007/BF00328699. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Wright H. M. Bacterial protoplast fusion: recombination in fused protoplasts of Streptomyces coelicolor. Mol Gen Genet. 1978 Jul 4;162(3):307–317. doi: 10.1007/BF00268856. [DOI] [PubMed] [Google Scholar]
- Jackson E. N., Yanofsky C. Internal promoter of the tryptophan operon of Escherichia coli is located in a structural gene. J Mol Biol. 1972 Aug 21;69(2):307–313. doi: 10.1016/0022-2836(72)90232-x. [DOI] [PubMed] [Google Scholar]
- Kolter R., Yanofsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- Lomovskaya N. D., Mkrtumian N. M., Gostimskaya N. L., Danilenko V. N. Characterization of temperate actinophage phi C31 isolated from Streptomyces coelicolor A3(2). J Virol. 1972 Feb;9(2):258–262. doi: 10.1128/jvi.9.2.258-262.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydiate D. J., Malpartida F., Hopwood D. A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35(3):223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- Malpartida F., Hopwood D. A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. 1984 May 31-Jun 6Nature. 309(5967):462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T., Oshima Y. Cyclic AMP may not be involved in catabolite repression in Saccharomyces cerevisiae: evidence from mutants unable to synthesize it. J Bacteriol. 1983 Nov;156(2):898–900. doi: 10.1128/jb.156.2.898-900.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Pastan I., Perlman R. Cyclic adenosine monophosphate in bacteria. Science. 1970 Jul 24;169(3943):339–344. doi: 10.1126/science.169.3943.339. [DOI] [PubMed] [Google Scholar]
- Seno E. T., Bruton C. J., Chater K. F. The glycerol utilization operon of Streptomyces coelicolor: genetic mapping of gyl mutations and the analysis of cloned gylDNA. Mol Gen Genet. 1984;193(1):119–128. doi: 10.1007/BF00327424. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Taylor W. E., Straus D. B., Grossman A. D., Burton Z. F., Gross C. A., Burgess R. R. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell. 1984 Sep;38(2):371–381. doi: 10.1016/0092-8674(84)90492-6. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. Cloning of antibiotic resistance and nutritional genes in streptomycetes. J Bacteriol. 1982 Aug;151(2):668–677. doi: 10.1128/jb.151.2.668-677.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]