Abstract
Osmotin is an abundant cationic multifunctional protein discovered in cells of tobacco (Nicotiana tabacum L. var Wisconsin 38) adapted to an environment of low osmotic potential. Beside its role as osmoregulator, it provides plants protection from pathogens, hence also placed in the PRP family of proteins. The osmotin induced proline accumulation has been reported to confer tolerance against both biotic and abiotic stresses in plants including transgenic tomato and strawberry overexpressing osmotin gene. The exact mechanism of induction of proline by osmotin is however, not known till date. These observations have led us to hypothesize that osmotin could be regulating these plant responses through its involvement either as transcription factor, cell signal pathway modulator or both in plants. We have therefore, undertaken the present investigation to analyze the osmotin protein as transcription factor using bioinformatics tools. The results of available online DNA binding motif search programs revealed that osmotin does not contain DNAbinding motifs. The alignment results of osmotin protein with the protein sequence from DATF showed the homology in the range of 0-20%, suggesting that it might not contain a DNA binding motif. Further to find unique DNA-binding domain, the superimposition of osmotin 3D structure on modeled Arabidopsis transcription factors using Chimera also suggested absence of the same. However, evidence implicating osmotin in cell signaling were found during the study. With these results, we therefore, concluded that osmotin is not a transcription factor, but regulating plant responses to biotic and abiotic stresses through cell signaling.
Keywords: Osmotin, Osmoregulator, DNA binding motifs, Alignment
Background
The growth and productivity of crop plants depend largely on their vulnerability to environmental stresses. High salinity, water deficit, and temperature stress are the major constraints that limit agricultural production [1]. Plants respond to these conditions with an array of biochemical and physiological adaptations, which involve the function of many stress-related genes. Hence, any attempt to improve the stress tolerance requires a better understanding of the underlying physiological, biochemical and molecular events [2].
A common adaptation in plants is the production of compatible solutes and they accumulate to high levels without disturbing intracellular biochemistry [3]. Compatible solutes have the capacity to persevere the activity of enzymes that are in saline solutions. These compounds have minimal effect on pH or charge balance of the cytosol or luminal compartments of organelles. Compatible solutes include low molecular weight sugars, organic acids, polyols and nitrogen containing compounds such as amino acids, amides, imino acids, proteins and quaternary ammonium compounds [4].
Osmotin, a stress responsive multifunctional protein, has been reported to be involved in osmotolerance of the plants [5, 6]. Singh et al. (1985) [7] isolated this protein and its gene from tobacco var. Wisconsin 38 that was cultured for several generations on the culture medium with high NaCl concentration. It is a basic protein with a molecular weight of 24 KDa that accumulated in the tobacco cells and imparted to them osmotic stress adaptation. This function of osmotin is linked to its action as compatible osmolyte that enhances osmotic potential of the cells. In addition to its role as osmoprotectant, osmotin is shown to exhibit antifungal activity against a broad range of fungal pathogens [8, 9]
Barthakur et al. (2001) [10] gave evidence that over expression of osmotin induces proline accumulation and confers tolerance to osmotic stress in transgenic tobacco. Biochemical analysis of tomato transformed with osmotin gene showed higher proline content in transformed plants under normal and stress conditions. 4 to 6 fold increase in proline content, over the wild type plants, was observed in leaves of strawberry plants transformed with osmotin gene even without subjecting them to the stress conditions [11]. It has been suggested that the synthesis of osmotin protein could induce synthesis and accumulation of certain solutes or could be involved in metabolic or structural changes of cellular proteins [12].
Based on the available literature and protein information on osmotin, we hypothesized that it may be involved in modulation of plant responses to biotic and abiotic stresses either directly by acting as transcriptional regulator for the genes encoding key enzymes of this pathway or as signaling molecule acting through intra cellular receptors. Being a 24 KDa protein, osmotin is within the range of low molecular weight transcription factors such as members of the GATA factor family of zinc finger transcription factors, WRKY transcription factors family etc. In the present study we therefore, evaluated osmotin protein for DNA binding domains and potential structural similarities with transcription factors available in databases.
Methodology
Analysis of osmotin protein for DNA-binding domain
Tobacco (Nicotiana tabacum) osmotin (1pcvA) (13) protein sequence and structural features were downloaded from RCSB-Protein Data Bank and UniProtKB/Swiss-Prot database for analysis. Amino acid sequence of osmotin protein in Fasta format was submitted to Compute pI/Mw at ExPASy Proteomics Server for calculating theoretical pI and molecular weight. The subcellular location of osmotin protein was predicted using TargetP 1.1 Server; http://www.cbs.dtu.dk/services/TargetP hosted at Technical University of Denmark, Denmark.
To identify the putative DNA binding sites on osmotin protein, amino acid sequence and structure were retrieved from NCBI databases and RCSBProtein Data Bank, respectively. GT1 protein of Trihelix family of transcription factors from Arabidopsis thialiana was taken as control for this study. The 3D structure of GT1 (2jmwA) was downloaded from RCSB-Protein Data Bank. Osmotin and GT1 amino acid sequences and structures (as required) were submitted to DNA-binding motif prediction softwares; 2-zip server, DBD transcription factor prediction database, GYM 2.0 and Predictdnahth [14, 15,16, 17].
Since these programs do not cover all types of DNA binding motifs especially those present in unique transcription factors, we also compared the osmotin protein sequence and structure with the transcription factors listed in Database of Arabidopsis transcription factors (DATF) [18] from Arabidopsis. To find unique DNA binding domain on osmotin protein, homology studies were carried out using sequence and structure of transcription factor families listed in DATF. For this, amino acid sequence of individual Arabidopsis transcription factor was taken from the DATF. At least, amino acid sequence of one and at maximum three transcription factors were retrieved from each of 64 families. Since the crystal structure of these transcription factors were not available, their nucleotide sequences were submitted to NCBI to blast against PDB to get a homolog on which modeling of these transcription factors can be performed. The best match was saved and used for homology modeling of the protein structure of transcription factors. Amino acid sequence of the representative transcription factor was submitted to SWISS-MODEL workspace, automated mode, for homology modeling of protein's structure [19]. The structures were visualized and analyzed using Chimera. SuperPose programs was used to study the superimposition of the structures of transcription factors with osmotin and GT.
Osmotin thaumatin like protein in Plants
The gene and protein sequences of osmotin and osmotin/thaumatin- like proteins were retrieved from published completed genomes of Arabidopsis lyrata, Arabidopsis thaliana, Carica papaya, Medicago truncatula, Glycine max, Cucumis sativus, Populus trichocarpa, Vitis vinifera, Oryza sativa, Zea mays and Sorghum bicolor. Superfamily server was used for structural domain analysis for Osmotin/thaumatin-like proteins family [20].
Results and Discussion
Evaluating osmotin protein using available bioinformatic tools, as direct/ indirect metabolic modulator (transcription factor/cell signal molecule), will help in understanding the molecular adaptations by plants against abiotic and biotic stresses. Motif-discovery is often one of the first steps performed during computational analysis of gene-regulation. For any protein to act as a gene regulator (transcription factor), it must contain a DNA binding motif. The results of transcription factor prediction softwares showed the presence of HTH motif on GT1 protein thus, suggesting that it binds to DNA molecule. However, no DNA binding domains were detected on 3D structure of osmotin (Table 1 see Table 1). Thus, overall results suggest that osmotin protein lacks leucine zippers and HTH motifs, the two predominant and important DNA binding motifs. The alignment results of osmotin with the protein sequence from DATF showed the homology in the range of 0-20%, suggesting that it does not contain a DNA binding motif. In order to find unique domain, the superimposition of osmotin 3D structure was done on modeled Arabidopsis transcription factors using Chimera. The transcription factor, GT1 sequence was used as the control protein to check the workability of the program. GT1 completely superimposed on transcription factors belonging to Tri-helix family, whereas no significant superimposition was found with osmotin to any protein in the database. These results thus, indicate that there are no DNA binding motifs present in osmotin, similar to those represented by our test set (DATF).
Osmotin/ thaumatin like proteins in Plants
Existing annotation and BLAST searches of well-characterized tobacco osmotin protein, used to identify osmotin/ thaumatin like protein homologs among the completed genome of Arabidopsis lyrata, Arabidopsis thaliana, Carica papaya, Medicago truncatula, Glycine max, Cucumis sativus, Populus trichocarpa, Vitis vinifera, Oryza sativa, Zea mays and Sorghum bicolor showed that it is present in all 11 plants, either alone or in combination of 5 different partner domains; Protein kinase-like (PK-like), Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin, Mitochondrial carrier, Class II aaRS ABD-related and ARM repeats (Table 2 see Table 2) . Each partner domain is shown to occur in diverse sets of protein families. Nature often brings several domains together to form multidomain and multifunctional proteins with a vast number of possibilities [22]. Proteins with multiple domains have advantages, providing increased stability and new cooperative functions [22]. In a multidomain protein, each domain may fulfill its own function independently, or in a concerted manner with its neighbours. Thus, the modular nature of osmotin has suggested the multiple roles for it.
Structural domain assignments from the superfamily database showed that A. thaliana genome contains 31 proteins belonging to osmotin/thaumatinlike protein superfamily. Osmotin/thaumatin domain together with 2 different partner domains; PK-like and Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin occur in seven different domain combinations, among the members of this family (Figure 1). Protein kinases catalyze the phosphotransfer reaction fundamental to most signaling and regulatory processes in the eukaryotic cell [23]. According to InterPro annotation, the three-dimensional fold of catalytic domain in
Figure 1.
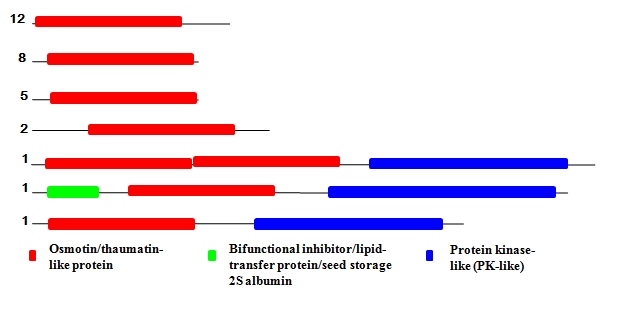
The domain architecture of Osmotin/thaumatin-like protein superfamily present in Arabidopsis thaliana
protein kinase is similar to domains found in proteins involved in cellular signalling including the catalytic domain of phosphoinositide-3-kinase (PI3K), the catalytic domain of the MHCK/EF2 kinase and choline kinase. They regulate a wide range of cellular process such as enzyme activation, assembly of macromolecules, protein localization and degradation. Three proteins in osmotin/thaumatin-like protein superfamily in A. thaliana contain this domain (AT4G18250.1, AT5G38280.1, AT1G70250) along with osmotin/thaumatin-like domain (Figure 2 and Figure 3). The third domain belongs to plant lipid-transfer and hydrophobic proteins family and is mainly involved in lipid metabolism.
Figure 2.
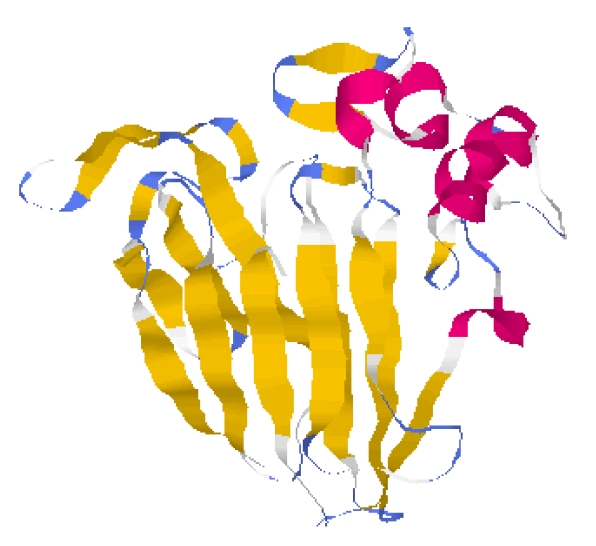
3D structure of 1pcvA downloaded from RCSB PDB and visualised by Rasmol
Figure 3.
The Protein kinases signature domains present in Osmotin/thaumatin-like protein superfamily
Responses to stress are generally complicated integrated circuits involving multiple pathways and cellular compartments [24]. The subcellular localization of a protein can provide important information about its function within the cell. The accurate function of proteins and their interaction networks relies greatly on the proper localization of each protein component. According to in silico estimates, N. tabacum osmotin is a cationic protein with theoretical pI of 8.13 and theoretical molecular weight of 26681.08 Da. The sequence alignment of N. tabacum osmotin (1pcvA) with Arabidopsis Osmotin/thaumatin-like protein superfamily shows that At4G 11650 (atosm34) is most similar to the N. tabacum osmotin, while AT4G 36000.1 is least. The score of location assignment program suggests N. tabacum osmotin to be a secretary protein i.e it is not bound to any membrane. According to the available evidences also, PR proteins are synthesized as precursors with an N-terminal signal peptide which mediates the transport. In general, the basic and neutral isoforms have been suggested to accumulate in vacuolar inclusion bodies [12]. Gau et al. (2004) [25] however, showed the accumulation of osmotin like protein in Malus domestica cv. Remo apoplast. Target P server results for 31 members of A. thaliana Osmotin/ thaumatin like protein superfamily suggests that all except seven (AT4G18250.1, AT1G20030.1, AT1G73620.1, AT1G77700.1, AT4G36000.1, AT4G38660.1 (M), AT4G38670.1 (C)) are secretory in nature. The secretory nature and multiple locations targeting of osmotin are in agreement to its multi functional role in plants exposed to biotic and abiotic stresses.
Apart from osmoregulation and functions involving partner domains, the osmotin could also act as modulator of transcription factors. This property of osmotin could be determined by the close protein–protein interactions with the transcription factors. Some In silico algorithms for studying protein‐protein interactions have been formulated over the years [26]. These however, also suffer from limitations of either over dependence on sequence-based information or knowledge of the tertiary structure of both partners prior to predicting the quaternary structure. The question of which residues in the protein‐protein interface actually interact often cannot be addressed and therefore, the exact function could be determined by the extensive studies involving labor-intensive experimental approaches. Consequently, experimental analyses are required to completely identify or assign a specific role for this protein.
The in silico analyses conducted in the present investigation indicate that osmotin/ thaumatin like protein superfamily is a large family of structurally similar proteins in the plants where osmotin/ thaumatin domain is present alone or with a partner domain. The specific function of this domain still needs to be addressed. The functions of partner domains however, are already determined. On this basis, we hypothesise that apart from osmoregulation, members of this super family are involved in diverse biological roles including cellular signal transduction.
Conclusion
With increase in the number of completed genomes being published, determining protein function is one of the most important problems of this post-genomic era. Osmotin is a cationic protein with theoretical pI of 8.13 and theoretical molecular weight of 26681.08 Da. No DNA-binding motif was detected by DNA-binding motif prediction softwares; 2-zip server, DBD transcription factor prediction database, GYM 2.0, HTH motif prediction, Predictdnahth. Alignment results of osmotin protein with the protein sequence from DATF showed the homology in the range of 0-20%, suggesting that it does not contain a DNA binding motif. Further to find unique DNA-binding domain, the superimposition of osmotin 3D structure, done on modeled Arabidopsis transcription factors using Chimera also suggested absence of the same. With these results, we concluded that osmotin does not contain a DNA-binding motif and may be regulating plant responses to biotic and abiotic stresses by acting as a modulator of transcription factors or metabolic signaling. Further work is progress in our laboratory to elucidate the exact function of osmotin in plants under stress conditions.
Supplementary material
Acknowledgments
This study was supported by a research grant to Prof. M. Z. Abdin from UGC, Govt. of India. The Project Fellowship awarded to Usha Kiran by UGC is gratefully acknowledged.
Footnotes
Citation:Abdin et al, Bioinformation 5(8): 336-340 (2011)
References
- 1.Cherian S, et al. Biologia Plantarum. 2006;50:481. [Google Scholar]
- 2.Cherian S, Reddy MP. Biol. Plant. 2003;46:193. [Google Scholar]
- 3.Bohnert HJ, et al. Plant Cell. 1995;7:1099. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashraf M, Harris PJC. Plant Sci. 2004;166:3. [Google Scholar]
- 5.Singh NK, et al. Plant Physiol. 1989;90:1096. doi: 10.1104/pp.90.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu B, et al. Plant Mol. Biol. 1995;28:17. doi: 10.1007/BF00042034. [DOI] [PubMed] [Google Scholar]
- 7.Singh NK, et al. Plant Physiol. 1985;79:126. doi: 10.1104/pp.79.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yun DJ, et al. Proc. Nat. Acad. Sci. 1997;94:7084. [Google Scholar]
- 9.Abad LR, et al. Plant Sci. 1996;118:11. [Google Scholar]
- 10.Barthakur S, et al. J. Plant Biochem. Biotechnol. 2001;10:31. [Google Scholar]
- 11.Husaini AM, Abdin MZ. Plant Sci. 2008;174:446. [Google Scholar]
- 12.Singh NK, et al. Plant Physiol. 1987;85:529. doi: 10.1104/pp.85.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min K, et al. Proteins. 2004;54:170. doi: 10.1002/prot.10571. [DOI] [PubMed] [Google Scholar]
- 14.Bornberg-Bauer E, et al. Nucleic Acids Res. 1998;26:2740. doi: 10.1093/nar/26.11.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson D, et al. Nucleic Acids Res. 2008;36:D88. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bu Narasimhan C, et al. J. of Computational Biol. 2002;9:707. doi: 10.1089/106652702761034145. [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin WA, Berman HM. J Mol Biol. 2003;27:43. doi: 10.1016/s0022-2836(03)00532-1. [DOI] [PubMed] [Google Scholar]
- 18.Guo A, et al. Bioinformatics. 2005;21:2568. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 19.Arnold K, et al. Bioinformatics. 2006;22:195. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Gough J, et al. J Mol Biol. 2001;313:903. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 21.Chothia C, et al. Nature. 1992;357:543. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 22.Ostermeier M, Benkovic SJ. Adv. Protein. Chem. 2000;55:29. doi: 10.1016/s0065-3233(01)55002-0. [DOI] [PubMed] [Google Scholar]
- 23.Engh RA, Bossemeyer D. Pharmacol Ther. 2002;93:99. doi: 10.1016/s0163-7258(02)00180-8. [DOI] [PubMed] [Google Scholar]
- 24.Dombrowski JE, et al. Plant Physiol. 2003;132:2098. doi: 10.1104/pp.102.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gau Achim E, et al. Eur J of Plant Pathology. 2004;110:703. [Google Scholar]
- 26.Ezkurdia I, et al. Brief Bioinform. 2009;10:233. doi: 10.1093/bib/bbp021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.