Abstract
Transmembrane signals initiated by a broad range of extracellular stimuli converge on nodes that regulate phospholipase C (PLC)–dependent inositol lipid hydrolysis for signal propagation. We describe how heterotrimeric guanine nucleotide–binding proteins (G proteins) activate PLC-βs and in turn are deactivated by these downstream effectors. The 2.7-angstrom structure of PLC-β3 bound to activated Gαq reveals a conserved module found within PLC-βs and other effectors optimized for rapid engagement of activated G proteins. The active site of PLC-β3 in the complex is occluded by an intramolecular plug that is likely removed upon G protein–dependent anchoring and orientation of the lipase at membrane surfaces. A second domain of PLC-β3 subsequently accelerates guanosine triphosphate hydrolysis by Gαq, causing the complex to dissociate and terminate signal propagation. Mutations within this domain dramatically delay signal termination in vitro and in vivo. Consequently, this work suggests a dynamic catch-and-release mechanism used to sharpen spatiotemporal signals mediated by diverse sensory inputs.
Phospholipase C (PLC) catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] to the second messengers inositol 1,4,5-trisphosphate [Ins(1,4,5)P3] and diacylglycerol in an essential step for the physiological action of many hormones, neurotransmitters, growth factors, and other extracellular stimuli (1–3). These cascades use signaling complexes consisting of Gα subunits of the Gq family (Gαq, 11, 14, and 16) of heterotrimeric guanine nucleotide–binding proteins (G proteins) and PLC-β isozymes (β1-4) (4–6). Agonist-stimulated receptors increase exchange of guanosine triphosphate (GTP) for guanosine diphosphate (GDP) on Gαq, GTP-bound Gαq engages and activates PLC-β, and PLC-β increases up to three orders of magnitude the rate of hydrolysis of GTP by its activating G protein (7–9). Coordination from upstream and downstream inputs sharpens time frame, amplitude, and on-off cycling of these signaling nodes. Although kinetic analyses revealed much about the dynamics of Gαq/PLC-β signaling complexes (10–12), how PLC-βs simultaneously act as effectors and GTPase activating proteins (GAPs) has remained unknown. Here, we describe the structure of PLC-β3 in an activated complex with Gαq, which together with supporting biochemical and physiological analyses reveals its mechanism of transmembrane signaling.
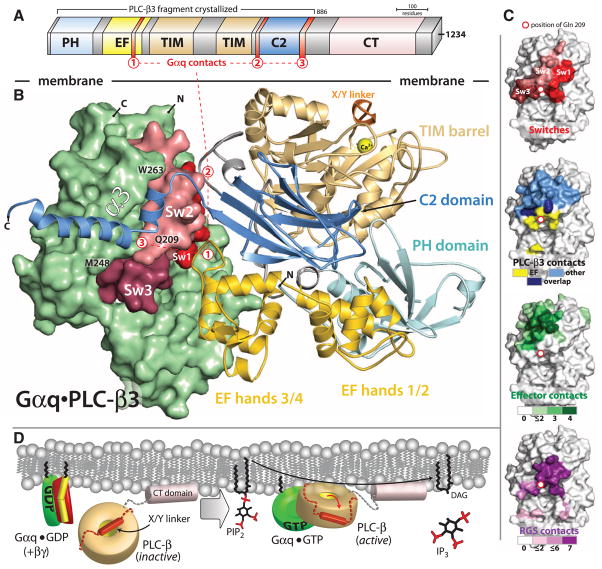
The three-dimensional structure of an AlF4−-dependent complex of Gαq bound to PLC-β3 was solved by molecular replacement using PLC-β2 [Protein Data Bank (PDB) code 2FJU] and Gαq (PDB 2BCJ) as search models and refined at 2.7-Å resolution (table S1). PLC-β3 engages Gαq through three distinct regions (Fig. 1, A and B). First, an extended loop between the third and fourth EF hands of PLC-β3 directly buttresses switch residues critical for GTP hydrolysis by Gαq. Second, the region of PLC-β3 that connects the catalytic TIM barrel and the C2 domain interacts with both switches 1 and 2 of Gαq. Third, a segment composed of a helix-turn-helix at the C terminus of the C2 domain resides primarily within a shallow declivity on the surface of Gαq formed by switch 2 and α3. Other effectors are known to engage this region within Gα subunits (Fig. 1C). GTP hydrolysis by Gα subunits is independently accelerated by a large family of regulator of G protein signaling (RGS) proteins (8, 13, 14), and PLC-β3 interacts with a surface on Gαq that overlaps almost completely with portions of Gα subunits needed for engagement of RGS proteins (Fig. 1C). Consistent with a biologically relevant interface (15), the complex of PLC-β3 and Gαq buries ~3200 Å2 of solvent-exposed surface area.
Fig. 1.
Structure of Gαq•PLC-β3. (A) Domain architecture of PLC-β3 drawn to scale and consisting of a N-terminal PH domain, a series of four EF hands, a catalytic TIM barrel, a C2 domain, and a carboxy-terminal (CT) domain. The CT domain is not necessary for Gαq binding (fig. S2), and, therefore, PLC-β3 truncated at residue 886 was used to facilitate crystallization. Three distinct regions of PLC-β3 that interact with Gq are indicated by red numerals. (B) Overall structure of the AlF4−-dependent complex of Gαq•PLC-β3 as viewed from the plane of the membrane. PLC-β3 is depicted as a ribbon cartoon with domains colored as in (A). Activated Gαq is depicted as a green surface with nucleotide-dependent switches (Sw1 to Sw3) in shades of red. Hα1/Hα2 (red 3) at the end of the C2 domain of PLC-β3 lies within the canonical effector-binding region of Gαq formed by α3 starting at M248 (21), the subsequent loop containing W263, and switch 2 containing Q209. The X/Y linker (orange) connects the two halves of the catalytic TIM barrel, and an ordered portion of the linker occludes the active site of the lipase highlighted by the Ca2+ (yellow ball) cofactor. (C) Surfaces of Gαq highlighting switches (top) in comparison to regions (lower images) of Gα subunits that interact with PLC-β3, other effectors, and RGS proteins. Interactions involving the EF3/4 loop are yellow except for overlap (dark blue) involving other regions of PLC-β3 (light blue). Gα subunits use a common interface (green) to engage four distinct effectors, and a different interface engages seven distinct RGS proteins (dark purple). Details of the analyses are supplied in (41). (D) Model for activation of PLC-β3 by GTP-activated Gαq. Gαq (green) bound to GDP is sequestered by Gβγ (red and yellow) and does not interact with PLC-β, depicted as a gold toroid except for its CT domain (light pink) and X/Y linker (orange cylinder and dotted lines). The CT domain basally associates with membranes, whereas the X/Y linker blocks the lipase active site. Upon activation of heterotrimeric Gq, Gαq-GTP dissociates from Gβγ and interacts with the main portion of PLC-β. Complex formation anchors and orients the lipase active site at membranes, leading to repulsion of the X/Y linker and freeing the active site for hydrolysis of PtdIns(4,5)P2 into diacylglycerol (DAG) and Ins(1,4,5)P3 (IP3).
Activated Gαq does not impinge on the active site of PLC-β3 and indeed is at least 40 Å from the calcium ion cofactor needed for PtdIns(4,5)P2 catalysis. Comparison of the structure of PLC-β3 in complex with Gαq to previous structures (16, 17) of PLC-β2 indicates that lipase activation does not involve Gαq-dependent propagation of a conformational change to the active site of the lipase (fig. S1A). Indeed, the active site of PLC-β3 is occluded by a portion of its X/Y linker (Fig. 1B): a poorly conserved loop that separates the two halves of the catalytic TIM barrel in all PLC isozymes. Our previous structural analyses illustrated that this region similarly occludes the active site of PLC-β2, and deletion of the negatively charged X/Y linker in PLC-β2, as well as in PLC-ε and -δ1, resulted in marked activation (17). Similarly, deletion of the highly negatively charged X/Y linker of PLC-β3 caused a large increase in lipase activity (fig. S1B), indicating that PLC-β3 also is robustly autoinhibited by its X/Y linker. Presumably, this region of PLC-β3 is forced out of the active site by steric and electrostatic repulsion mediated by the surface of the plasma membrane coupled to the engagement of Gαq (Fig. 1D). A similar mechanism was proposed previously for activation of PLC-β2 by Rac1, which binds entirely through the PH domain of PLC-β2 at substantial distance from the active site of the lipase (16). Consequently, Gαq, Rac1, and likely other activators such as Gβγ activate PLC isozymes by anchoring and orienting them at substrate membranes to release autoinhibition by the X/Y linker and promote access of PtdIns(4,5)P2 to the lipase active site.
PLC-β isozymes are effectors of Gαq as well as GAPs that enhance the intrinsic GTPase activity of the engaging Gα subunits. The structure of activated Gαq bound to PLC-β3 explains the integration of these reciprocal functions.
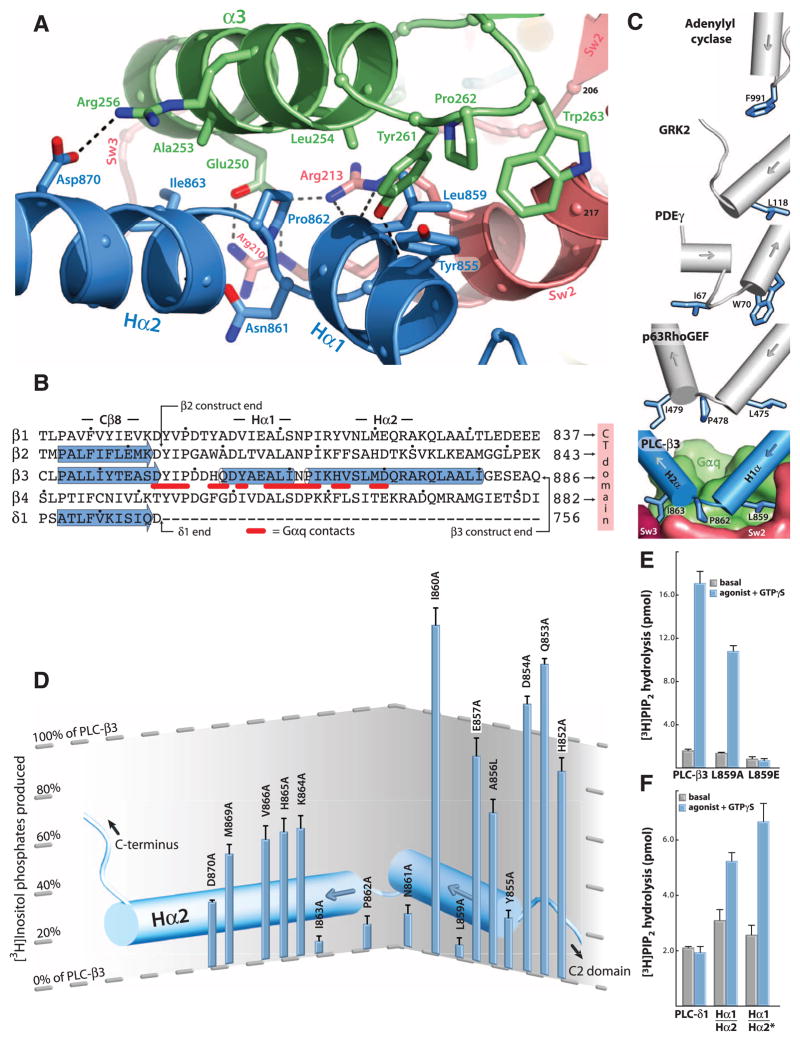
The catalytic core of the 13 mammalian PLC isozymes includes a pleckstrin homology (PH) domain, a set of four EF hands, a catalytic TIM barrel, and a C2 domain (18) (Fig. 1A). The canonical Gα effector-binding region of Gαq, located between α3 and switch 2, is occupied by a helix-turn-helix (Hα1/Hα2) that immediately follows the C2 domain of PLC-β3 (Fig. 2A). PLC-δ isozymes terminate immediately after their C2 domain, which is the last common domain found in all PLC isozymes (18). Thus, grafting of Hα1/Hα2 onto the C terminus of the C2 domain of PLC-β3 provides a large binding surface that makes numerous contacts with Gαq (~1750 Å2 of solvent accessible surface area buried). Only PLC-β isozymes are activated by Gαq, and the highly conserved Hα1/Hα2 motif is found in all PLC-βs (Fig. 2B) but not in other PLC isozymes. PLC-βs also contain a long C-terminal region that extends about 300 residues past the Hα1/Hα2 module. This long C-terminal extension previously was thought to be important for interaction with Gαq. However, absence of this region did not affect high affinity binding of PLC-β3 to Gαq [dissociation constant (Kd) ~200 nM; fig. S2], and it is not present in the PLC-β3 construct used for structure determination. The C-terminal domain is important for membrane association, but whether it has additional function(s) in the signaling complex remains unclear.
Fig. 2.
Structure of the effector binding interface of Gαq•PLC-β3. (A) Ribbon diagram of the interface between the Hα1/Hα2 region of PLC-β3 (blue) and the effector binding pocket of Gαq located between α3 (green) and Sw2 (pink). Interfacial residues (sticks) are labeled; hydrogen bonds are indicated by dashed lines. (B) Comparison of PLC sequences (21) at the end of the C2 domain (Cβ8) encompassing Hα1/Hα2. α helices (cylinders) and β sheets (arrows) were assigned by using crystal structures of PLC-β3 as reported here (PDB 3OHM), PLC-β2 (2ZKM), and PLC-δ1 (1DJX). C termini are indicated for full-length PLC-δ1 as well as the crystallized fragments of PLC-β2 and -β3. Residues in PLC-β3 that interact with Gαq are underlined in red. Dots indicate every 10th residue. (C) Comparison of effectors bound to Gα subunits. The major effector binding surface of Gαq (green with switches in red) engages Hα1/Hα2 (blue cylinders) of PLC-β3 through indicated residues (sticks) surrounding Pro862. Structurally analogous α helices (gray cylinders) and residues (blue sticks) in other effectors are highlighted after superimposition of bound Gα subunits (not shown). PDE-γ, cyclic GMP phosphodiesterase-γ. (D) Mutational analyses of Hα1/Hα2. PLC-β3 mutants harboring the indicated single substitutions were assessed for capacity to be activated upon cotransfection with Gαq in COS-7 cells as measured by [3H]inositol phosphate production. Further experimental details are described in figs. S3, A to C, and S4. (E) Requirement of Hα1/Hα2 for activation of PLC-β3 by Gαq assessed with purified proteins. [3H]PtdIns(4,5)P2-containing phospholipid vesicles reconstituted with purified P2Y1 receptor, Gαq, and Gβ1γ2 were used to assess the capacity of wild-type or PLC-β3 mutants (300 nM) to hydrolyze PtdIns(4,5)P2 in the absence (basal) or presence of a P2Y1 receptor agonist (2MeSADP, 1 μM) plus 100 nM GTPγS (agonist + GTPγS). Data are mean ± SEM from four independent experiments. (F) Grafting Hα1/Hα2 onto PLC-δ1 confers responsiveness to Gαq. Activities of purified proteins were compared as in (E). Residues 847 to 886 of PLC-β3 were added to the end of PLC-δ1 to create PLC-δ1(Hα1/Hα2); starred variant consists of PLC-δ1(Hα1/Hα2) with additional substitutions (D610R and N612D) of PLC-δ1 to analogous PLC-β3 residues (see domain architectures in fig. S7A).
Pro862 of PLC-β3 lies within the turn between Hα1 and Hα2, makes extensive contacts with multiple residues of Gαq, and forms the center of a Gαq-binding interface (Fig. 2A). The side chain of the preceding Asn861 supports this turn by forming a hydrogen bond with the backbone amide of Lys864. This Asn-Pro couplet is preserved in three of the four PLC-β isozymes (it is Asp-Pro in PLC-β4) and presumably defines the turn because of helix capping and breaking propensities of Asn/Asp and Pro, respectively. The turn is bracketed by Leu859, which inserts into a hydrophobic pocket formed by residues in α3 and switch 2, and by Ile863, which interacts with tandem glutamates in α3. Tyr855 in Hα1 and Asp870 in Hα2 also support the binding interface at the periphery. The binding of Gαq to Hα1/Hα2 of PLC-β3 recapitulates almost entirely the interaction of Gαq with several guanine nucleotide exchange factors (GEFs) for Rho, including p63RhoGEF, Trio, and Kalirin, which use a helix-turn-helix grafted onto the end of a DH/PH cassette to bind the α3/Sw2 declivity of Gαq (19, 20) (Fig. 2C). PLC-β3 and p63RhoGEF use identical residues in their primary interfaces with Gαq, and other effectors also engage this region of Gα subunits in similar fashion (Fig. 2C).
The role of Hα1/Hα2 residues in Gαq-mediated activation was examined by mutational analyses. Whereas expression of Gαq or PLC-β3 alone in COS-7 cells had no effect, their coexpression resulted in a large increase in inositol lipid hydrolysis (fig. S3A). In contrast, coexpression of PLC-β3 with mutation L859→E859 (21) [PLC-β3(L859E)] with Gαq had no effect over a broad range of conditions (fig. S3B). Gβγ independently activates PLC-β3, and coexpression of either PLC-β3 or PLC-β3(L859E) with Gβ1γ2 resulted in similar levels of activation (fig. S3C). Mutation of the analogous residue (Leu810) in PLC-β1 also completely abrogated Gαq-dependent stimulation (fig. S3D).
The contribution of residues across Hα1/Hα2 of PLC-β3 was examined (Fig. 2D and fig. S4). In each case, the relative sensitivity of the PLC-β3 mutant to activation by Gαq versus Gβ1γ2 was compared under conditions where maximal response to each activator was observed. Whereas single substitutions throughout Hα1/Hα2 did not affect Gβγ-stimulated activity (fig. S4), certain of these mutations (Y855A, L859A, N861A, P862A, and I863A) resulted in substantial or complete loss of the capacity of Gαq to promote PLC-β3–dependent increases in inositol phosphate accumulation (Fig. 2D).
The binding and lipase activities of PLC-β3 mutants also were tested by using purified proteins (fig. S5). PLC-β3, PLC-β3(L859E), and PLC-β3(L859A) exhibited similar basal lipase activities (fig. S5A) and were similarly activated by Gβ1γ2 (fig. S5B). However, the binding affinity of PLC-β3(L859A) for Gαq in the presence of AlF4− was sevenfold lower than PLC-β3, and no AlF4−-dependent binding of PLC-β3(L859E) was observed (fig. S5C). Activities of PLC-β3 isozymes mutated in Hα1/Hα2 also differed markedly in a signaling complex reconstituted with purified P2Y1 receptor, heterotrimeric Gq, and PLC-β3. The P2Y1 receptor agonist, 2MeSADP, promoted robust activation of PLC-β3, but PLC-β3(L859E) was completely refractory to activation and intermediate activation was observed with PLC-β3(L859A) (Fig. 2E).
Alanine-scanning mutagenesis previously identified two small segments (residues 243 to 245 and 256 to 257) of Gαq necessary for elevated production of inositol phosphates (22). These regions contribute to interactions with Hα1/Hα2 (fig. S6A). Additional alanine substitutions were made in Gαq, and, of those Gαq mutants that expressed as stable trypsin-resistant proteins, most exhibited a predictable loss in capacity to activate PLC-β3 (fig. S6B).
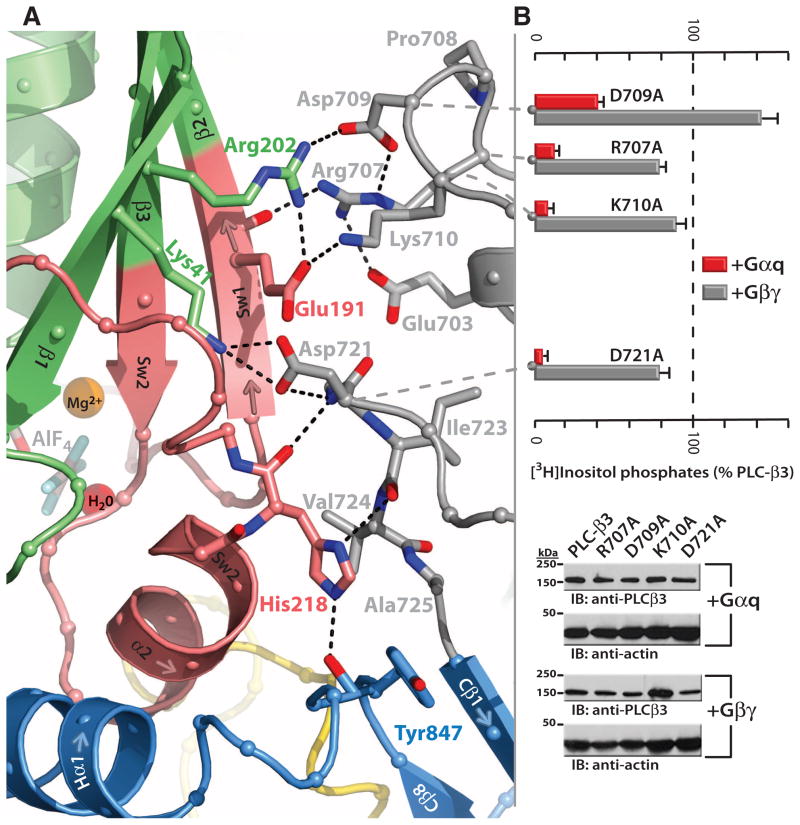
The loop between the end of the TIM barrel and the beginning of the C2 domain comprises a second distinct segment of PLC-β3 that makes extensive contacts with active Gαq, including switches 1 and 2 (Fig. 3A). This interface includes a series of interdigitated pairs of charged residues, specifically (in PLC-β3/Gαq) Asp709/Arg202, Lys710/Glu191, and Asp721/Lys41; these in turn are supported by additional charged residues (Glu703 and Arg707) of PLC-β3. Alanine substitution of several of these residues in PLC-β3 compromised the capacity of Gαq, but not Gβ1γ2, to activate PLC in COS-7 cells (Fig. 3B).
Fig. 3.
Secondary Gαq•PLC-β3 interface. (A) Ribbon diagram highlighting residues (gray) preceding the C2 domain (light blue) of PLC-β3 that interact with Sw1 and 2 (pink) of activated Gαq (green). AlF4− (gray cross-stick), Mg2+ (orange ball), and the catalytic water (red ball) within the nucleotide-binding pocket also are shown. (B) Mutational analysis of the Gαq•PLC-β3 binding interface. (Top) Activation of the indicated mutants of PLC-β3 in the presence of cotransfected Gαq (red) or Gβ1γ2 (gray) was determined by quantification of [3H]inositol phosphate accumulation in COS-7 cells. Data are mean ± SEM from four independent experiments. (Bottom) Relative expression of PLC-β3 and mutant forms was quantified under each transfection condition by using a PLC-β3–specific antibody. Actin immunoblots (IB) included as loading controls.
Residues adjacent to both borders of the C2 domain (Val724 and Tyr847) converge to envelop His218 of Gαq, which is wedged between the afore-mentioned interface and the start of Hα1/Hα2 to anchor two of the three major interfaces within the Gαq•PLC-β3 complex (Fig. 3A). Mutation of His218 results in loss of capacity of Gαq to activate PLC-β3 (fig. S6B).
PLC-δ isozymes are not regulated by Gαq, presumably because of lack of both Hα1/Hα2 and the Gαq-interacting residues found in PLC-β isozymes between the TIM barrel and the C2 domain. Thus, we hypothesized that G protein–dependent regulation could be engineered into PLC-δ1 (fig. S7A). Surface plasmon resonance (SPR) analyses revealed that, whereas PLC-δ1 did not exhibit AlF4−-dependent binding to Gαq, introduction of the Hα1/Hα2 segment of PLC-β3 into PLC-δ1 conferred binding (fig. S7B). Moreover, receptor- and guanine nucleotide–stimulated lipase activity was observed with the chimeric isozymes but not PLC-δ1 (Fig. 2F), and the median effective concentration (EC50) of GTPγS for activation of PLC-δ1(Hα1/Hα2) by GTPγS was 50 nM (fig. S7C). Thus, Hα1/Hα2 is a small, linear module used for functional engagement of Gαq.
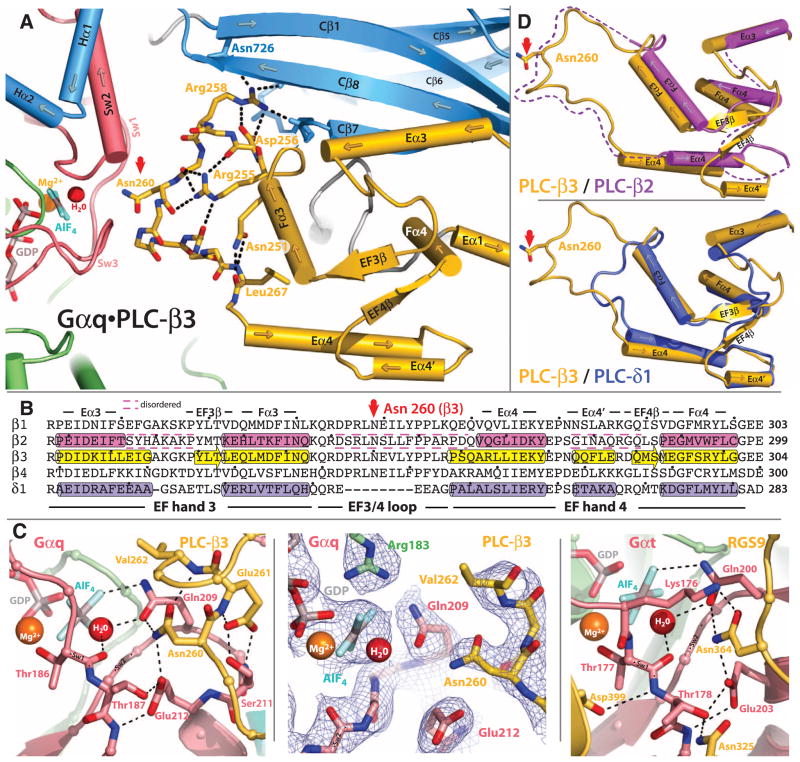
An extended loop between EF hands 3 and 4 of PLC-β3 interacts with the GTP-binding region of Gαq (Fig. 4A). This loop is highly conserved in all PLC-β isozymes, is not found in PLC-δ1 (Fig. 4B) or other PLC isozymes, and interacts with the active site of Gαq. Asn260 of the EF3/4 loop promotes GTP hydrolysis by interaction with the side chain of Gln209 of Gαq (Fig. 4C), which rearranges during GTP hydrolysis to stabilize the transition state mimicked by GDP•AlF4−•H20. Asn260 also interacts with Glu212 to stabilize switch 1 for GTP hydrolysis. The interactions of Asn260 of PLC-β3 with Gαq are recapitulated by a functionally equivalent asparagine in RGS9 (23) (Fig. 4C) and other RGS proteins (24, 25).
Fig. 4.
Interaction of the EF3/4 loop of PLC-β3 with switch residues critical for GTP hydrolysis by Gαq. (A) Ribbon and cylinder diagram highlighting conserved interactions within EF hands 3 and 4 (yellow) of PLC-β3 needed for the optimal positioning of Asn260 (red arrow) within the guanine nucleotide binding pocket of Gαq. Sw1 to Sw3 are pink; other portions of Gαq are green. The C2 domain and adjacent Hα1/Hα2 of PLC-β3 are light blue; key PLC-β3 residues (sticks) and hydrogen bonds (dotted lines) that support Asn260 (red arrow) are highlighted. The guanine nucleotide binding pocket contains GDP and AlF4− (sticks) as well as the Mg2+ cofactor (orange ball) and catalytic water (red ball). (B) Sequence alignment comparing EF hands 3 and 4 of PLC-βs with equivalent region of PLC-δ1. α helices (cylinders) and β sheets (arrows) assigned from crystal structures (PLC-β3, 3OHM; PLC-β2, 2ZKM; δ1, 1DJX); dashed lines bracket disordered regions. The asparagine (Asn260 in PLC-β3) that is positioned for promotion of GTP hydrolysis by Gαq is indicated by a red arrow. The colors correspond to those of the structures depicted in (D) below. Dots indicate every 10th residue. (C) Comparison of the GTP-binding sites of Gαq•PLC-β3 and Gαt•RGS9. Left image depicts portions of the EF3/4 loop (yellow) of PLC-β3 contacting Sw1 and 2 (light red) of Gαq. Other portions of Gαq are shown as in (A). Middle image highlights electron density (composite simulated annealing omit map contoured at 1.2 σ) centered on Asn260 of Gαq•PLC-β3. Right image depicts analogous portions of RGS9 (yellow) bound to Gαt as revealed in the crystal structure determined by Slep et al. (23). (D) Ribbon and cylinder diagrams comparing EF hands 3 and 4 of PLC-β3 (yellow) with PLC-β2 (top, magenta) and PLC-δ1 (bottom, purple). Asn260 highlighted with red arrow, and dotted lines indicate disordered portions of PLC-β2.
Asn260 is positioned at the active site of Gαq as part of a tight turn (residue 260 to 264) of PLC-β3 that is stabilized by Glu261 and underpinned by an extensive series of hydrogen bonds principally mediated by Asp256 and Arg255 and Arg258 (Fig. 4A). These residues are highly conserved in all PLC-βs, as are Asn251 and Leu267, which appear crucial in stabilizing the ends of the loop (Fig. 4B). The EF3/4 loop as well as other portions of EF hands 3 and 4 are disordered in the crystal structure of PLC-β2 (Fig. 4D). A likely scenario is that Gαq initially engages the EF3/4 loop of PLC-β3 to nucleate the underlying hydrogen bonding network and promote cooperative ordering of EF hands 3 and 4.
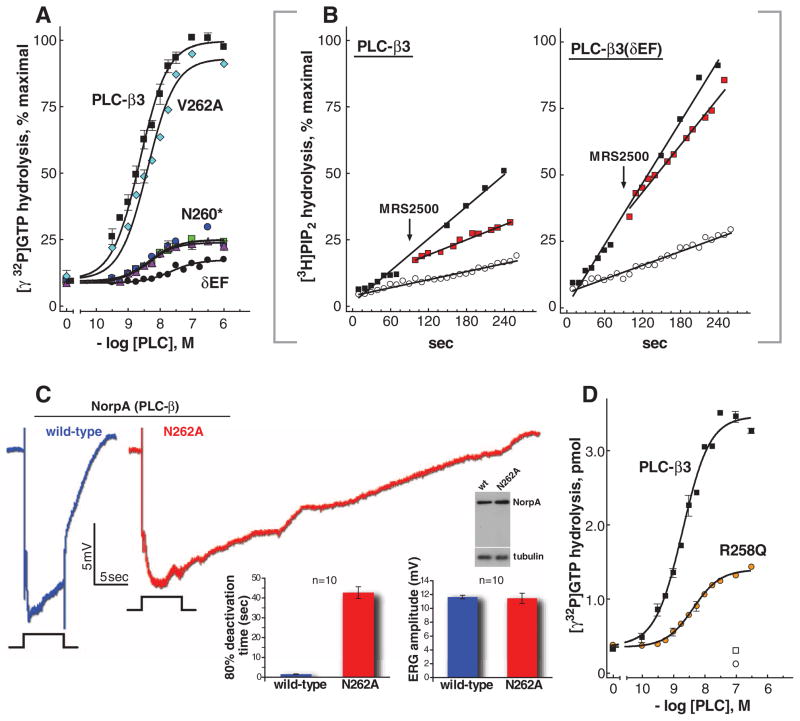
To directly examine the role of the EF3/4 region of PLC-β3 in mediating inactivation of its activating G protein, we quantified GTP hydrolysis by Gαq in the presence of purified PLC-β3 mutants (fig. S8A) by using phospholipid vesicles reconstituted with the P2Y1 receptor and heterotrimeric Gq. In the presence of receptor agonist, PLC-β3 promoted up to 100-fold stimulation of GTP hydrolysis (Fig. 5A and fig. S8B), and activation occurred with an EC50 ~ 3 nM (table S2). A chimeric PLC-β3 replacing the EF3/4 loop with the analogous region of PLC-δ1 was severely crippled in its capacity to accelerate GTP hydrolysis by Gαq (Fig. 5A). Similarly, substitution of Asn260 dramatically reduced the capacity of PLC-β3 to promote GTP hydrolysis, whereas substitution of Val262 had negligible effect. Importantly, basal and Gβγ-stimulated lipase activities of these purified mutants were unaffected (fig. S8, A and C). The EC50 values of mutant and wild-type PLC-β3 for stimulation of GTP hydrolysis also were similar (table S2). In contrast, substitution of Leu859 to Ala859 within Hα1/Hα2, which reduced the binding affinity of the complex by ~sevenfold (fig S5C), also increased the EC50 for stimulation of GTP hydrolysis by ~10-fold (table S2). Thus, the EF3/4 loop of PLC-β3 is crucial for stimulation of GTP hydrolysis by Gαq but contributes minimally to forming the signaling complex.
Fig. 5.
Contribution of the EF hand region to GAP activity of PLC-β3. (A) The GAP activity of purified wild-type PLC-β3 is compared with that of mutant PLC-β3 isozymes. Steady-state GTP hydrolysis was quantified with phospholipid vesicles reconstituted with purified P2Y1 receptor, Gαq, and Gβ1γ2. Assays were in the presence of the P2Y1 receptor agonist 2MeSADP (3 μM) and the indicated concentrations of purified PLC-β3; PLC-β3(δEF); PLC-β3(V262A); or PLC-β3(N260A), PLC-β3(N260G), or PLC-β3(N260S) [all designated as PLC-β3(N260*)] as described in (41). Data are plotted as percent of maximal response obtained with PLC-β3. Data are mean ± SEM of three experiments. (B) Deficiency in termination of Gαq-stimulated PLC activity of a GAP-deficient mutant of PLC-β3. PLC activity was quantified with [3H]PtdIns(4,5)P2-containing phospholipid vesicles reconstituted with purified P2Y1 receptor, Gαq, and Gβ1γ2. Vesicles were incubated with 300 nM PLC-β3 or PLC-β3(δEF) in the absence (open circles) or presence of the P2Y1 receptor agonist 2MeSADP (300 nM; black squares) and either 30 μM GTP or 100 nM GTPγS for 90 s before addition of P2Y1 receptor antagonist MRS2500 (50 μM; red squares) or vehicle. Incubations were continued for an additional 165 s. Data are plotted as percent of the maximal response observed with either PLC-β3 or PLC-β3(δEF) in the presence of agonist plus GTPγS. (C) Delayed termination of the photoresponse in Drosophila expressing a GAP-deficient mutant of PLC-β. Electro-retinograms from flies harboring wild-type PLC-β (NORPA, blue) or a mutant form (NORPAN262A, red) deficient in capacity to accelerate the GTPase activity of Gαq. Flies ~1 day posteclosion were dark-adapted for 2 min before exposure to 5-s pulses of orange light indicated by the event marker below each electroretinogram. At right are plotted deactivation rates and maximal amplitudes for the average of ten individual electroretinograms. Error bars indicate SEM. Expression of the norpA transgenes was confirmed by immunoblot (gel) of head extracts prepared from flies ~1 day posteclosion. (D) GAP activity of a mutant of PLC-β found in pancreatic cancer. PLC-β3 was mutated at a position (R258) (21) equivalent to a homozygous substitution identified in PLC-β4 during genome-wide profiling of pancreatic cancers (30). GAP activity of PLC-β3(R258Q) was compared with that of PLC-β3 as described in (A) above. Data are mean ± SEM of three experiments.
Loss of capacity of PLC-β3 to promote GTP hydrolysis by Gαq should decrease its capacity to turn off subsequent to Gαq-mediated activation. This idea was first tested in vitro with use of purified proteins. Addition of a P2Y1 receptor antagonist (fig. S8, B and D) to an agonist pre-activated signaling complex of the P2Y1 receptor, heterotrimeric Gαq, and wild-type PLC-β3 resulted in a rapid decline of lipase activity to levels similar to those observed in the absence of agonist (Fig. 5B). In contrast, little reversal of lipase activity occurred upon addition of P2Y1 receptor antagonist to a similarly preactivated complex containing PLC-β3(δEF) (Fig. 5B).
Rhodopsin-initiated phototransduction in Drosophila melanogaster is mediated by Gαq-dependent activation of PLC-β (26). To examine the role of the EF3/4 loop of PLC-β in a physiological system, we replaced wild-type PLC-β (NORPA) in Drosophila with a version mutated to alanine in the conserved Asn (N262) demonstrated above to be required for PLC-β-promoted GTP hydrolysis by Gαq. Flies expressing wild-type NORPA or NORPAN262A exhibited similar amplitudes of the light-induced photoresponse (Fig. 5C). In contrast, whereas termination of light resulted in rapid termination of photoresponse in wild-type flies, we observed a marked defect in recovery with norpAN262A.
PLC-β3 is a tumor suppressor, and its disruption in humans contributes to lymphomas and other myeloid malignancies (27, 28). Similarly, Gαq is an oncogene, and its constitutive activation drives ~50% of all uveal melanomas (29). Signaling through the Gαq/PLC-β axis is important for regulation of cell proliferation, and other disruptions in this node can be expected to contribute to cancer. In this regard, homozygous substitution of Arg254 within the EF3/4 loops of PLC-β4 was found in a pancreatic tumor during genome-wide profiling (30). The equivalent substitution in PLC-β3 resulted in a decrease in capacity to accelerate GTP hydrolysis by Gαq (Fig. 5D and fig. S8E).
The high-resolution structure of Gαq•PLC-β highlights a dynamic interplay between regions of the complex needed to coordinate rapid activation and inactivation of this signaling node required for highly responsive, low-noise signal transduction. We propose that a conformationally flexible Hα1/Hα2 samples a relatively large volume to maximize probability of encountering Gαq, and transient interactions with Gαq guide the final folding of Hα1/Hα2. The process of coupling folding with binding to increase the rate of formation of the final encounter complex has been described for other protein complexes (31, 32) and is referred to as fly-casting. A subset of Dbl-family RhoGEFs typified by p63RhoGEF also apparently use fly-casting to engage Gαq (19, 20). In particular, p63RhoGEF uses a helix-turn-helix immediately adjacent to a conserved PH domain to engage Gq in a fashion that is recapitulated almost identically in Gαq•PLC-β. Thus, an independent module is grafted onto PLC-βs and RhoGEFs to confer binding to Gαq. The Hα1/Hα2 module in PLC-βs is encoded by a single exon, which suggests that these signaling proteins acquired capacity to bind Gαq through intergenic exon shuffling.
Engagement of PLC-β by Gαq is intimately coupled to inactivation of the complex. A primordial PLC-δ acquired an extended loop between EF hands 3 and 4 (Fig. 4D) that directly engages the switch regions of Gαq to stabilize the transition state for GTP hydrolysis. This EF3/4 loop and Hα1/Hα2 are linked evolutionarily, because both motifs are found in the two PLC-β isozymes of Caenorhabditis elegans. Indeed, they are not found separately in any PLC-β and therefore are the defining motifs of members of the PLC-β family. Taken together, we propose that Gαq is “caught” by a flycast from Hα1/Hα2 and is “released” by EF3/4 loop-mediated stimulation of GTP hydrolysis, which results in a conformational change in Sw2 and abrogation of the binding sites for both the EF3/4 loop and the Hα1/Hα2 segment. We also note that p115RhoGEF binds to Gα13 and promotes GTP hydrolysis through two different domains (33).
Rapid cycling of effector engagement and GTP hydrolysis favors the maintenance of heterotrimeric Gq/effector complexes necessary for signal acuity in a process generally referred to as kinetic scaffolding (9, 10, 34, 35). Phototransduction requires high signal amplification in rapid cycles of activation/deactivation in a signaling system organized for suppression of noise and therefore provides an excellent model for comparison of signaling mediated by PLC-βs and other effectors. Although Gαq-promoted activation of PLC-β mediates phototransduction in some metazoans such as fruit flies, mammalian rod and cone phototransduction involves Gαt-mediated activation of cyclic guanosine monophosphate (GMP) phosphodiesterase (PDE) (36). This PDE is not a GAP, and acceleration of GTP hydrolysis evolved in a separate protein, RGS9, which nonetheless stabilizes the switch regions of Gαt in much the way the EF3/4 loop of PLC-β stabilizes Gαq (23) (Fig. 4C). The binding of PDE to the effector pocket of Gαt allosterically increases binding of RGS9 (23, 37). G protein–coupled receptor kinase 2 (GRK2) and, to a lesser extent, p63RhoGEF enhance the GAP activity of RGS4 in a complex with Gαq (38). This allostery is inherent in the catch-and-release mechanism used by PLC-βs. Ablation of GAP function of PLC-β markedly prolongs deactivation of phototransduction in Drosophila (Fig. 5C), and disruption of RGS9 in mice (39) and mutation of RGS9 in human disease (40) produce analogous phenotypes.
Supplementary Material
Acknowledgments
This research was supported by NIH grants GM38213 and GM57391 (J.S. and T.K.H.), GM61454 and GM074001 (T.K.), and EY010852 (C.M.). T.K.R. was supported by a Ruth L. Kirschstein National Service Award F31 Predoctoral Fellowship and a United Negro College Fund Merck Graduate Science Research Dissertation Fellowship. K.T. was supported by a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science. We acknowledge the outstanding help with structural analyses by B. Temple, L. Betts, J. Vanhooke, T. Charpentier, V. Arshavsky, and M. Kosloff; with analysis of linker-deleted PLC-β3 by N. Vincent Jordan; with SPR analyses by A. Kimple; with [γ32P]GTP purification by E. Lazarowski and the insightful comments on the manuscript by H. Dohlman, R. Nicholas, E. Ross, and K. Slep. Coordinates and structure factors for Gαq•PLC-β3 have been deposited under the PDB accession code 3OHM.
Footnotes
References and Notes
- 1.Berridge MJ, Irvine RF. Nature. 1984;312:315. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 2.Michell RH. Biochim Biophys Acta. 1975;415:81. doi: 10.1016/0304-4157(75)90017-9. [DOI] [PubMed] [Google Scholar]
- 3.Nishizuka Y. Science. 1992;258:607. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 4.Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Science. 1991;251:804. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 5.Taylor SJ, Chae HZ, Rhee SG, Exton JH. Nature. 1991;350:516. doi: 10.1038/350516a0. [DOI] [PubMed] [Google Scholar]
- 6.Waldo GL, Boyer JL, Morris AJ, Harden TK. J Biol Chem. 1991;266:14217. [PubMed] [Google Scholar]
- 7.Singer WD, Brown HA, Sternweis PC. Annu Rev Biochem. 1997;66:475. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- 8.Ross EM, Wilkie TM. Annu Rev Biochem. 2000;69:795. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 9.Ross EM. Curr Biol. 2008;18:R777. doi: 10.1016/j.cub.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biddlecome GH, Berstein G, Ross EM. J Biol Chem. 1996;271:7999. doi: 10.1074/jbc.271.14.7999. [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay S, Ross EM. Proc Natl Acad Sci USA. 1999;96:9539. doi: 10.1073/pnas.96.17.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turcotte M, Tang W, Ross EM. PLoS Comput Biol. 2008;4:e1000148. doi: 10.1371/journal.pcbi.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neubig RR, Siderovski DP. Nat Rev Drug Discov. 2002;1:187. [Google Scholar]
- 14.Dohlman HG, Thorner J. J Biol Chem. 1997;272:3871. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 15.Chakrabarti P, Janin J. Proteins. 2002;47:334. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 16.Jezyk MR, et al. Nat Struct Mol Biol. 2006;13:1135. doi: 10.1038/nsmb1175. [DOI] [PubMed] [Google Scholar]
- 17.Hicks SN, et al. Mol Cell. 2008;31:383. doi: 10.1016/j.molcel.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harden TK, Sondek J. Annu Rev Pharmacol Toxicol. 2006;46:355. doi: 10.1146/annurev.pharmtox.46.120604.141223. [DOI] [PubMed] [Google Scholar]
- 19.Rojas RJ, et al. J Biol Chem. 2007;282:29201. doi: 10.1074/jbc.M703458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutz S, et al. Science. 2007;318:1923. doi: 10.1126/science.1147554. [DOI] [PubMed] [Google Scholar]
- 21.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 22.Venkatakrishnan G, Exton JH. J Biol Chem. 1996;271:5066. doi: 10.1074/jbc.271.9.5066. [DOI] [PubMed] [Google Scholar]
- 23.Slep KC, et al. Nature. 2001;409:1071. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 24.Slep KC, et al. Proc Natl Acad Sci USA. 2008;105:6243. doi: 10.1073/pnas.0801569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tesmer JJG, Berman DM, Gilman AG, Sprang SR. Cell. 1997;89:251. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 26.Wang T, Montell C. Pfugers Arch. 2007;454:821. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- 27.Mullighan CG, et al. Nature. 2007;446:758. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 28.Xiao W, et al. Cancer Cell. 2009;16:161. doi: 10.1016/j.ccr.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Raamsdonk CD, et al. Nature. 2009;457:599. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones S, et al. Science. 2008;321:1801. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci USA. 2000;97:8868. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugase K, Dyson HJ, Wright PE. Nature. 2007;447:1021. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Singer WD, Sternweis PC, Sprang SR. Nat Struct Mol Biol. 2005;12:191. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- 34.Doupnik CA, Davidson N, Lester HA, Kofuji P. Proc Natl Acad Sci USA. 1997;94:10461. doi: 10.1073/pnas.94.19.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong H, et al. J Biol Chem. 2003;278:7278. doi: 10.1074/jbc.M208819200. [DOI] [PubMed] [Google Scholar]
- 36.Arshavsky VY, Lamb TD, Pugh EN., Jr Annu Rev Physiol. 2002;64:153. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 37.Skiba NP, Hopp JA, Arshavsky VY. J Biol Chem. 2000;275:32716. doi: 10.1074/jbc.C000413200. [DOI] [PubMed] [Google Scholar]
- 38.Shankaranarayanan A, et al. J Biol Chem. 2008;283:34923. doi: 10.1074/jbc.M805860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CK, et al. Nature. 2000;403:557. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- 40.Nishiguchi KM, et al. Nature. 2004;427:75. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- 41.Materials and methods are available as supporting material on Science Online.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.