SUMOylation is an essential modification that regulates predominantly nuclear proteins. Here we describe two pathways for the generation of nuclear SUMO E1 enzyme, import of individual subunits, and transport of the holo-enzyme. The NLS in Uba2 is required for transport of the complex; the c-Myc–like NLS in Aos1 functions only in the free subunit.
Abstract
SUMOylation, reversible attachment of small ubiquitin-related modifier (SUMO), serves to regulate hundreds of proteins. Consistent with predominantly nuclear targets, enzymes required for attachment and removal of SUMO are highly enriched in this compartment. This is true also for the first enzyme of the SUMOylation cascade, the SUMO E1 enzyme heterodimer, Aos1/Uba2 (SAE1/SAE2). This essential enzyme serves to activate SUMO and to transfer it to the E2-conjugating enzyme Ubc9. Although the last 40 amino acids in yeast Uba2 have been implicated in its nuclear localization, little was known about the import pathways of Aos1, Uba2, and/or of the assembled E1 heterodimer. Here we show that the mammalian E1 subunits can be imported separately, identify nuclear localization signals (NLSs) in Aos1 and in Uba2, and demonstrate that their import is mediated by importin α/β in vitro and in intact cells. Once assembled into a stable heterodimer, the E1 enzyme can still be efficiently imported by importin α/β, due to the Uba2 NLS that is still accessible. These pathways may serve distinct purposes: import of nascent subunits prior to assembly and reimport of stable E1 enzyme complex after mitosis.
INTRODUCTION
Posttranslational modification with ubiquitin-related proteins of the SUMO (small ubiquitin-related modifier) family is an essential mechanism in most eukaryotes. It regulates fate and function of many target proteins by changing their interactions with other macromolecules (reviewed in Hay, 2005; Geiss-Friedlander and Melchior, 2007; Wilkinson and Henley, 2010). Covalent attachment of SUMO requires an enzymatic cascade consisting of a single E1-activating enzyme (see below), a single E2-conjugating enzyme (Ubc9), and one of several E3 ligases that facilitate transfer of SUMO from Ubc9 to the substrate. Specific SUMO isopeptidases make this modification reversible and highly dynamic (reviewed in Kim and Baek, 2009). The majority of known SUMO targets are nuclear proteins, and, consistent with this, most of the known SUMO enzymes are enriched in this compartment.
The SUMO E1-activating enzyme is a heterodimer consisting of the two subunits: Aos1 (SAE1) and Uba2 (SAE2) (Dohmen et al., 1995; Johnson et al., 1997; Desterro et al., 1999; Gong et al., 1999; Okuma et al., 1999). Aos1 resembles the N-terminal, and Uba2 the C-terminal, part of the ubiquitin E1 enzyme (Lois and Lima, 2005; Schulman and Harper, 2009). In addition, Uba2 contains a C-terminal extension of ∼80 amino acids. This region contains a cluster of basic amino acids reminiscent of a classical nuclear localization signal (NLS), and its deletion in Saccharomyces cerevisiae leads to cytoplasmic mislocalization (Dohmen et al., 1995). Although S. cerevisiae Uba2 (S.c. Uba2) is essential, partial or complete deletion of its C-terminal extension has only minor or no growth defects (Dohmen et al., 1995; del Olmo et al., 1997; Lois and Lima, 2005). Consistent with this, SUMOylation patterns are largely unchanged (Lois and Lima, 2005). One possible explanation for this finding is that sufficient levels of Uba2 still reach the nuclear compartment, for example, in association with Aos1. This is consistent with the finding that some Uba2 was still present in the yeast nuclei (Dohmen et al., 1995). An NLS in Aos1, however, has not been described, and it is unknown whether the SUMO E1 enzyme heterodimer assembles before or after nuclear import. To gain insights into these questions, we decided to characterize the nuclear import of mammalian SUMO E1 subunits and of the holo-enzyme.
Translocation of proteins through nuclear pore complexes (NPCs) is mediated by import and export receptors and requires the GTPase Ran (reviewed in Mattaj and Englmeier, 1998; Fried and Kutay, 2003; Weis, 2003; Pemberton and Paschal, 2005; Conti et al., 2006; Stewart, 2007). Whereas nuclear import via classical NLSs (one or two short clusters of basic amino acids; first identified in SV40 large T antigen [Kalderon et al., 1984] and in nucleoplasmin [Robbins et al., 1991], respectively) is mediated by importin β together with the adaptor protein importin α (Adam and Gerace, 1991; Gorlich et al., 1995; Moroianu et al., 1995; Weis et al., 1995), cargos bearing a nonclassical NLS (e.g., an M9 [Siomi and Dreyfuss, 1995], BIB [Jäkel and Gorlich, 1998], or RS domain [Kataoka et al., 1999; Lai et al., 2001]) can directly interact with distinct members of the importin β family.
Here we show that Aos1 and Uba2 have distinct functional NLSs and demonstrate that the Uba2 NLS is required for import of the holo-enzyme. Importin α/β functions as the main transport receptor for both individual subunits and the assembled E1 enzyme.
RESULTS
Aos1 and Uba2 both contain distinct functional NLSs
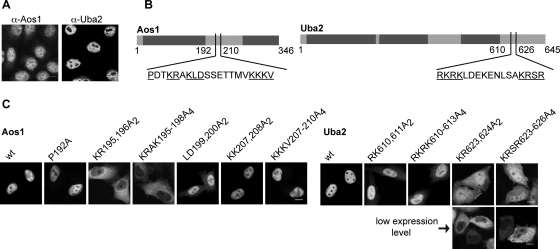
Endogenous Aos1 and Uba2 are highly concentrated in the nuclei of mammalian cells (Azuma et al., 2001; Pichler et al., 2002). Although some Aos1 can be detected in the cytoplasm of adherent HeLa cells, virtually no Uba2 is detected in this compartment (Figure 1A; confocal microscopy). Based on gel filtration experiments, both subunits are expressed at comparable levels and form a stable complex, at least in cell extracts (Azuma et al., 2001). Consequently, intranuclear localization of both subunits may be due to import of the complex.
FIGURE 1:
Aos1 and Uba2 both contain functional NLSs. (A) Localization of endogenous Aos1 and Uba2 in HeLa cells, detected by indirect immunofluorescence with affinity-purified polyclonal goat-α-Aos1 and goat-α-Uba2 antibodies. Bar, 20 μm. (B) Schematic illustration of potential NLSs in Aos1 and Uba2. Underlined are amino acids that were changed to alanines in Aos1 and Uba2 mutants. (C) HeLa cells were transiently transfected with pET28a-CFP-Aos1 (left panel) or pET28b-Uba2-YFP (right panel) variants. At 24 h posttransfection, intracellular localization was detected by fluorescence microscopy. For two Uba2 variants, different expression levels are shown, as these influenced localization. Bar, 10 μm.
However, overexpression of either subunit (CFP-tagged Aos1 or Uba2-YFP) in HeLa cells resulted in clear intranuclear localization (Figure 1C, wt). No cytoplasmic pool of CFP-Aos1 could be detected. Endogenous Aos1 has a size (38 kDa) compatible with slow diffusion into and out of the nucleus, but the CFP-tagged version should only be transported by active processes. Taken together, these findings suggest that Aos1 and Uba2 may have distinct NLSs.
A cluster of basic amino acids reminiscent of an NLS has already been identified in S.c. Uba2 and was implicated in nuclear transport due to partial mislocalization of a GFP-tagged C-terminal deletion fragment (Δ551–626) in the cytoplasm (Dohmen et al., 1995). This cluster is conserved among Uba2 proteins from different species (Supplemental Figure 1) and consists of two stretches of basic amino acids, reminiscent of a classical bipartite NLS. Mutational analysis of Uba2-YFP (Figure 1B, underlined residues) revealed distinct roles for both clusters of basic amino acids, RKRK610–613 and KRSR623–626, in mediating nuclear import (Figure 1C). Whereas replacement of the first cluster by alanines resulted only in partial mislocalization of Uba2, Uba2-KR623, 624A2, and Uba2-KRSR623–626A4 showed strong cytoplasmic accumulation. This was most apparent in cells expressing low levels of the protein.
An NLS in Aos1 has not yet been described. Sequence alignment of Aos1 from different species (Supplemental Figure 2) revealed two conserved clusters of basic amino acids, again matching the characteristics of classical NLSs. To test whether these conserved residues are indeed required for nuclear import of CFP-Aos1, we substituted amino acids of the potential NLSs by alanines (Figure 1B, underlined residues). Analysis upon transfection revealed that CFP-Aos1-KR195196A2 and CFP-Aos1-KRAK195–198A4 strongly mislocalized to the cytoplasm (Figure 1C). In clear contrast, CFP-Aos1-KKKV207–210A4 showed no localization defect. These findings implicate the first basic cluster in nuclear import of Aos1 and point toward a monopartite NLS. Comparison with known import sequences revealed a striking similarity of the identified region in Aos1 with conserved residues of the NLS in c-Myc, 320PxxKRxKLD328 (Stone et al., 1987; Dang and Lee, 1988; Makkerh et al., 1996). This NLS deviates from typical monopartite (SV40-type) and bipartite (nucleoplasmin-type) types of NLSs, as it contains only three basic residues and requires proline, leucine, and aspartate residues for efficient import. To explore the possibility that the Aos1 NLS contains characteristics of a c-Myc-type NLS, we also analyzed Aos1-LD199200A2, which indeed showed partial mislocalization. Aos1-P192A, on the other hand, had no apparent localization defect. Taken together, these findings point to an NLS in Aos1 (amino acids 195–200) that largely matches the NLS of c-Myc.
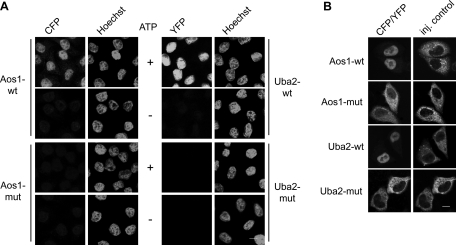
Intracellular distribution of proteins 24 h after transfection reflects a steady-state situation that does not allow conclusions about efficiency of nuclear import. Moreover, multiple point mutations may cause folding problems that may influence import indirectly or artificially. We therefore turned next to in vitro nuclear import assays (Adam et al., 1990). For this, we generated recombinant CFP-Aos1 and Uba2-YFP wild-type (wt) and mutant variants from bacteria. Wild-type and mutant proteins behaved identically during expression and purification, which included gel filtration, excluding significant folding defects. On purification, they were analyzed for their accumulation in nuclei of semipermeable HeLa cells in the presence of cytosol and ATP. As shown in Figure 2A, wt proteins were efficiently imported in dependence of ATP, but the double mutants CFP-Aos1-KR195, 196A2 and Uba2-KR623, 624A2-YFP both failed to be imported.
FIGURE 2:
Aos1 and Uba2 are independently imported into HeLa cell nuclei. (A) In vitro import of CFP-Aos1 or Uba2-YFP using semipermeable HeLa cells and cytosolic extract. Nuclear accumulation of wt proteins (CFP-Aos1-wt, Uba2-wt-YFP) and of NLS mutants (CFP-Aos1-KR195, 196A2, Uba2-KR623, 624A2-YFP) was analyzed by fluorescence microscopy. +ATP, in the presence of ATP-regenerating system; –ATP, in the presence of ATP-depleting system. Bar,10 μm. (B) Microinjection of CFP-Aos1 or Uba2-YFP into the cytoplasm of HeLa cells. Wild-type proteins, Aos1-KR195, 196A2, and Uba2-KR623, 624A2 were injected together with TRITC-dextran, incubated for 30 min, fixed, and analyzed by fluorescence microscopy. Bar, 10 μm.
To confirm these findings also in the context of intact cells, we then used microinjection experiments (Figure 2B). Recombinant wt proteins were fully competent for rapid nuclear import after injection into the cytoplasm of adherent Hela cells, whereas the CFP-Aos1 and the Uba2-YFP mutants remained in the cytoplasm throughout the course of the experiment. In conclusion, Aos1 and Uba2 both have functional nuclear import sequences.
The importin α/β heterodimer recognizes both Aos1 and Uba2
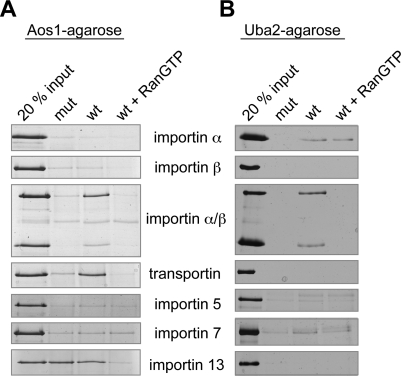
To identify transport receptors capable of recognizing the SUMO E1 subunits, we performed pull-down assays with His-CFP-Aos1 and Uba2-YFP-His immobilized on CNBr activated sepharose, and a variety of soluble import receptors (Figure 3, A and B). To control for specificity of the detected interactions, binding assays were carried out in the absence or presence of RanGTP. Inhibition of binding in the presence of RanGTP is characteristic for bona fide import receptor/cargo complexes (Rexach and Blobel, 1995; Izaurralde et al., 1997). To test whether the interactions depend on the putative NLSs identified in Aos1 and Uba2, binding was tested for both wt E1 subunits and for the import-deficient double mutants (Aos1-KR195, 196A2 and Uba2-KR623, 624A2). Figure 3A shows the analysis of Aos1 interactions: Heterodimeric importin α/β (Gorlich et al., 1995), transportin (Pollard et al., 1996), and importin 13 (Mingot et al., 2001) interacted with Aos1 in a RanGTP-sensitive manner. Importin β alone did not bind. Although importin 13 bound to wt and to mutant Aos1, transportin and importin α/β did not interact with the import-incompetent variant. Taken together, these findings pointed to both transportin and importin α/β as potentially relevant for import of Aos1 in HeLa cells; moreover, importin 13 could contribute to Aos1 import, even if it would not play a major role in HeLa cells. However, neither transportin nor importin 13 was capable of mediating import of CFP-Aos1 in vitro (Supplemental Figures 3 and 4A). Finally, we tested whether importin 13 could synergize with importin α/β to make transport more efficiently. Precedence for cooperation of import receptors comes, for example, from the finding that histone H1 can be imported by an importin β/importin 7 heterodimer (Jäkel et al., 1999). However, importin 13 impaired, rather than stimulated, CFP-Aos1 import under rate-limiting concentrations of importin α/β (Supplemental Figure 4B).
FIGURE 3:
Importin β binds to Aos1 and to Uba2 via the adaptor importin α. Immobilized Aos1 wt and the NLS mutant Aos1-KR195, 196A2 (A) or Uba2 and the Uba2 mutant Uba2-KR623, 624A2 (B) were incubated with recombinant import factors importin α, β, α/β, transportin, importin 5, 7, or 13. Bound proteins were eluted by SDS sample buffer and compared with 20% of the input by SDS–PAGE and silverstaining. To control for specificity of binding, experiments were also performed in the presence of RanGTP, which interferes with formation of receptor–cargo complexes.
As shown in Figure 3B, Uba2 wt was specifically recognized by the importin α/β heterodimer. This interaction was inhibited in the presence of RanGTP and required the intact NLS in Uba2. Although importin β alone did not bind, a weak but specific interaction was also observed for importin α alone, pointing to a rather strong NLS in Uba2. As expected, the interaction of the adaptor protein importin α with Uba2 was not inhibited by RanGTP.
Importin α/β mediates nuclear import of Aos1 and Uba2 in vitro and in vivo
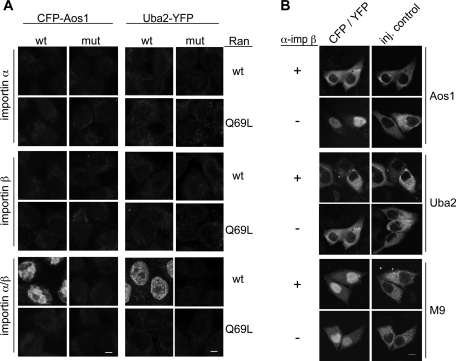
To test whether importin α/β can indeed mediate import of Aos1 and Uba2, we again carried out in vitro import assays but replaced cytosol with the GTPase Ran and recombinant transport receptors (Figure 4A). As a control for active transport, the hydrolysis-deficient Ran mutant RanQ69L was included in some experiments. Whereas importin α and importin β alone were not sufficient for nuclear accumulation of CFP-Aos1 and Uba2-YFP, the heterodimeric importin α/β complex mediated efficient import of both SUMO E1 subunits in the presence of wt Ran. In contrast to wt proteins, neither Aos1-KR195, 196A2 nor Uba2-KR623, 624A2 enriched in the nucleus. Taken together, these experiments reveal that the putative NLSs of Aos1 and Uba2 are indeed functional signals for import by the heterodimeric importin α/β transport receptor.
FIGURE 4:
Importin α/β mediates nuclear import of Aos1 and Uba2. (A) In vitro import of CFP-Aos1 or Uba2-YFP using semipermeable HeLa cells and recombinant import factors (importin α, importin β, or importin α/β). Nuclear accumulation of wt proteins and NLS mutants (Aos1-KR195, 196A2, Uba2-KR623, 624A2) was analyzed by fluorescence microscopy. Experiments were performed in the presence of wt Ran or the hydrolysis-defective mutant RanQ69L. Bar, 10 μm. (B) Microinjection: Inhibitory α-importin β antibodies interfere with nuclear import of CFP-Aos1 or Uba2-YFP in HeLa cells. Import cargo, TRITC-dextran and either monoclonal inhibitory anti–importin β antibody 3E9 (α−imp β+) or transport buffer (α-imp β−) were injected into the cytoplasm of HeLa cells. After 30 min, cells were fixed and analyzed by fluorescence microscopy. The transportin-dependent cargo YFP-M9 (Siomi and Dreyfuss, 1995; Nakielny et al., 1996) was included as a control for importin β–independent transport cargo. Bar, 10 μm.
We next wanted to find out whether transport by importin α/β is the main import pathway for SUMO E1 subunits in intact HeLa cells. To address this question, we made use of a monoclonal anti–importin β antibody that had previously been described to inhibit importin β–dependent nuclear import (Chi et al., 1995). CFP-Aos1 and Uba2-YFP were microinjected with or without this inhibitory antibody into the cytoplasm of HeLa cells and analyzed for intranuclear accumulation. As shown in Figure 4B, import of both E1 subunits was completely abolished in the presence of the anti–importin β antibody. Translocation of YFP-M9, a cargo for the import receptor transportin (Siomi and Dreyfuss, 1995; Nakielny et al., 1996), was not affected by the antibodies (Figure 4B, bottom panel). Together with the in vitro finding that importin β binds and imports Aos1 or Uba2 only in the presence of importin α (Figures 3 and 4), these data demonstrate that efficient nuclear import of the SUMO E1 subunits in cells is mediated via the importin α/β import pathway.
The SUMO E1 holo-enzyme can be imported via the NLS of Uba2
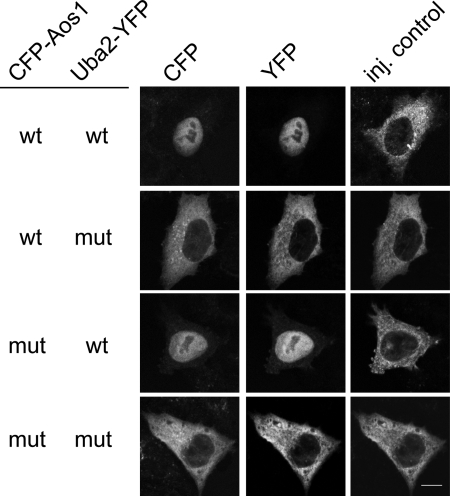
Although import of single subunits may be sufficient to explain intracellular localization of the SUMO E1 enzyme, this would require dissociation of the E1 complex for reimport after mitosis. Therefore, we wanted to test whether the assembled Aos1/Uba2 heterodimer is also competent for nuclear import. For this, we reconstituted the heterodimer from recombinant CFP-Aos1 and Uba2-YFP and purified the stable complex by gel filtration. It was then used as cargo for in vitro import assays in the presence of HeLa cytosol (Supplemental Figure 5) and for microinjection studies (Figure 5B). These experiments showed rapid accumulation of both subunits in the nucleus, suggesting that the E1 enzyme can be imported as a holo-enzyme. However, these experiments did not fully rule out the possibility that the E1 enzyme dissociated prior to import of individual subunits. We addressed this issue by using different combinations of wt and import-incompetent subunits of the E1 enzyme. When the complex was assembled from mutant Aos1 and wt Uba2, both subunits entered the nucleus; in contrast, when wt Aos1 was combined with mutant Uba2, both subunits remained in the cytoplasm (Figure 5 and Supplemental Figure 5). These findings demonstrate that the holo-enzyme remains stable throughout the experiments and reveal that the NLS of Uba2, but not of Aos1, is required for nuclear import of the E1 complex.
FIGURE 5:
The Uba2 NLS is required and sufficient for import of the SUMO E1 holo-enzyme. Microinjection: E1 complexes reconstituted from wt proteins (CFP-Aos1-wt, Uba2-wt-YFP) and/or NLS mutants (CFP-Aos1-KR195, 196A2, Uba2-KR623, 624A2-YFP) were injected into the cytoplasm of HeLa cells with TRITC-dextran. After incubation for 30 min, cells were fixed and analyzed by fluorescence microscopy. Bar, 10 μm.
Importin α/β mediates nuclear import of the SUMO E1 holo-enzyme
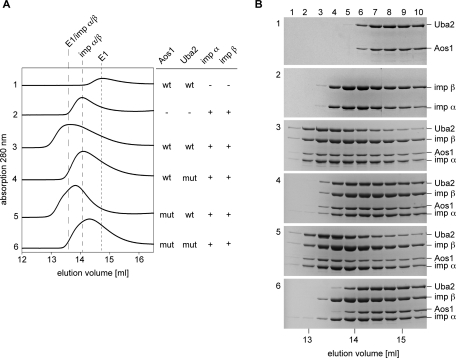
The finding that the Uba2 NLS is required for import of the Aos1/Uba2 complex suggested that the Uba2 NLS, but not the Aos1 NLS, is accessible for import receptor binding in the heterodimer. To test this directly, we incubated different variants of purified E1 complexes with a twofold molar excess of recombinant importin α/β and tested for formation of complexes by analytical gel filtration. Fractions of 300 μl were collected and subsequently analyzed by SDS–PAGE (Figure 6). Whereas an E1 complex in which both NLSs had been mutated failed to interact significantly with importin α/β (Figure 6B, compare panels 1 and 2 with bottom panel), complexes that contained wt Uba2-YFP and either wt or mutant Aos1 clearly formed a complex with importin α/β, as indicated by the appearance of all four proteins in lanes 2–4 (Figure 6B, panels 3 and 5). Consistent with this, importin α/β mediates efficient import of SUMO E1 complexes that contain wt Uba2 (in vitro import assays; Supplemental Figure 6). Gel-filtration experiments involving a complex formed from wt Aos1 and mutant Uba2 (Figure 6B, panel 4) suggest that the Aos1-NLS might still be partially accessible in the complex; the E1 complex shifts by two fractions in the presence of importin α/β, whereas transport receptor elution appears largely unaffected. This weak interaction at high protein concentrations, however, does not suffice for nuclear protein import by importin α/β (Supplemental Figure 6). Taken together, these data reveal that the Uba2 NLS is necessary and sufficient for recognition and import of the E1 complex by importin α/β in vitro.
FIGURE 6:
Importin α/β binds to the Uba2 NLS of the SUMO E1 holo-enzyme. Wild-type and mutant CFP-Aos1/Uba2-YFP complexes (5 μM) were incubated with a twofold molar excess of importins α and β (10 μM each), and subjected to gel filtration. Aos1 mut, Aos1-KR195, 196A2; Uba2 mut, Uba2-KR623, 624A2. (A) Elution profiles from the superose6-HR10/30 column were recorded by the Äkta purifier system (GE Healthcare) and processed with sigma plot 8.02 (Systat Software). (B) Fractions were analyzed by SDS–PAGE and Coomassie staining.
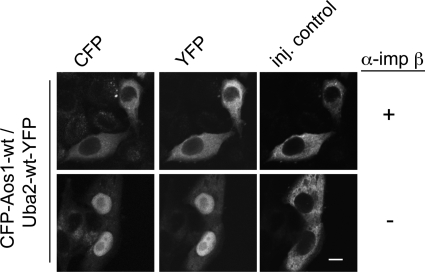
Finally, we wanted to test whether the importin α/β pathway is also the main import pathway for assembled E1 in intact cells. For this, we again turned to microinjection in the presence of inhibitory antibodies. Indeed, import of the assembled E1 complex (Figure 7) was efficiently blocked when anti–importin β antibodies were coinjected. This suggests that the importin α/β pathway is the major import pathway not only for single E1 subunits but also for the holo-enzyme.
FIGURE 7:
Inhibition of importin β prevents nuclear import of SUMO E1 holo-enzyme. Microinjection: Mixtures of CFP-Aos1/Uba2-YFP complex, TRITC-dextran, and either inhibitory anti–importin β antibody (+) or transport buffer (−) were injected into the cytoplasm of HeLa cells. After incubation for 30 min, cells were fixed and analyzed by fluorescence microscopy. Bar, 10 μm.
DISCUSSION
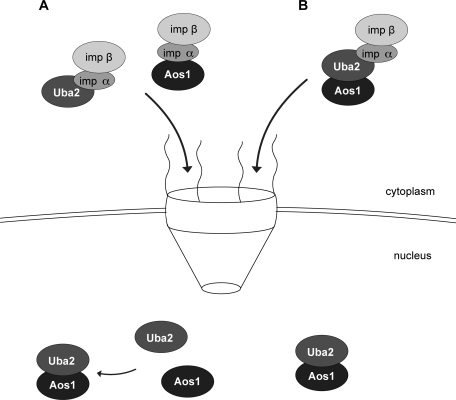
The findings described here reveal two pathways for the generation of intranuclear E1 enzyme, both of which depend on importin α/β (Figure 8). First, both subunits contain an NLS and can be transported separately, prior to heterodimerization in the nucleus (left). Second, E1 complexes may already assemble in the cytoplasm, prior to being imported. Although the NLS in Aos1 is not functional in the heterodimer, the NLS in Uba2 is available for recognition and import by importin α/β (right). Because E1 complexes don’t seem to dissociate in cells, the latter pathway is required at least for reimport of the E1 complex after mitosis.
FIGURE 8:
Two mechanisms for generation of active nuclear E1 complex. (A) The single subunits Aos1 and Uba2 are independently imported by importin α/β via their distinct NLSs. After translocation into the nucleus, active SUMO E1 complex is formed by heterodimerization. (B) Alternatively, E1 enzyme assembles in the cytoplasm, or reaches the cytoplasm upon nuclear envelope breakdown, and is imported as a complex by importin α/β. This mechanism requires the NLS of Uba2, whereas the NLS of Aos1 is masked.
A strong NLS in Uba2
Although deletion analysis of yeast Uba2 had already implicated a basic cluster within the last 82 amino acids in import (Dohmen et al., 1995), detailed analysis of its NLS had not yet been carried out. Sequence alignment of Uba2 from yeast to human (Supplemental Figure 1) revealed two short stretches of basic amino acid residues separated by a short linker of 9–12 amino acids, suggesting a nucleoplasmin-type bipartite NLS. However, Uba2 from Drosophila melanogaster contains two clusters separated by 26 amino acids, perhaps more consistent with one or two independent monopartite NLSs. Our analysis revealed unequal contributions of both clusters to intranuclear localization of human Uba2, with mutations in the first cluster causing only a minor, and mutations in the second cluster causing a major, defect. Binding assays, in vitro import analysis, and import after microinjection all demonstrated that mutation of two amino acids in the second cluster (K623 and R624) abolish interaction with, and import by, importin α/β. The NLS in Uba2 seems remarkably strong, based on our observation that importin α stably interacts with Uba2 even in the absence of importin β (Figure 3B). Interaction of importin α with targets usually requires the presence of importin β, which prevents autoinhibition of importin α (Kobe, 1999). Structural analysis of the Uba2/importin α interaction could provide interesting insights into the exact nature (monopartite vs. bipartite) of this efficient NLS.
The NLS in human Aos1 resembles the NLS of c-Myc
With a molecular weight of 38 kDa, Aos1 could slowly diffuse into the nucleus or may be imported in association with Uba2. Human Aos1 can indeed be efficiently imported via association with Uba2 (Figure 5 and Supplemental Figures 5 and 6). However, we also identified an NLS in Aos1 that facilitates efficient import of the isolated subunit by importin α/β. This NLS is conserved in orthologues from mouse, zebrafish, Xenopus, and Drosophila, but is not detectable in Aos1 from S. cerevisiae. Whether S.c. Aos1 can also be imported as a single subunit remains to be tested. The NLS in human Aos1 (PDTKRAKLD) fullfills the minimal sequence requirements for cNLSs, K-K/R-x-K/R (reviewed in Lange et al., 2007). Interestingly, it shares additional features with the NLS of the oncoprotein c-Myc (320PAAKRVKLD328) (Stone et al., 1987; Dang and Lee, 1988), a hydrophobic amino acid that interrupts the basic cluster, and a C-terminal leucine and aspartate motif. Whereas the latter are required for efficient import of both, c-Myc and CFP-Aos1, the N-terminal proline is required only for nuclear accumulation of c-Myc (Makkerh et al., 1996; Figure 1C). This difference may be due to structural features: Pro-320 in c-Myc may serve to ensure structural flexibility required for interaction with importin α, whereas in Aos1 this function may be carried out by other flanking residues. In line with this, crystallographic analysis revealed that the main interactions between the c-Myc NLS and importin α occur via the cluster 323KRVKL327, whereas the side chain of Pro-320 does not contribute directly to binding (Conti and Kuriyan, 2000). The NLS in Aos1, however, is located in a disordered region that starts several residues N-terminal of Pro-192 (amino acids 178–203), based on the crystal structure of the Aos1/Uba2 complex (Lois and Lima, 2005). Identification of a c-Myc-related NLS in human Aos1 raises the interesting possibility that this NLS variant might be more frequently used than previously anticipated.
Efficient import of the stable holo-enzyme via the NLS of Uba2
The SUMO E1 enzyme is a stable complex in vitro and in cells (see above). It can be efficiently imported by importin α/β, which specifically binds to the Uba2 NLS. The C-terminus of Uba2 (amino acids 551–640 in human) is specific for the SUMO E1 enzyme and is not part of the core catalytic domain that Aos1/Uba2 shares with other E1 enzymes. It was not resolved in the crystal structure of the SUMO E1 complex (Lois and Lima, 2005), indicating structural flexibility. In contrast to the Uba2 NLS, which is always accessible for importin α binding, the NLS in Aos1 is not productively recognized in the context of the holo-enzyme. The most likely interpretation for this is that it is largely inaccessible in the heterodimeric complex. Indeed, while residues spanning the Aos1 NLS were disordered in the crystal, its predicted position is in a cavity-like topology formed by both subunits (Lois and Lima, 2005).
Alternative pathways for Uba2 import?
Although in vitro import assays and microinjection studies clearly showed that the Uba2 NLS is necessary for efficient import of the E1 complex into the nucleus, some intranuclear localization of overexpressed Uba2 NLS mutants was detectable 24 h after transfection (Figure 1C). Moreover, a C-terminal deletion fragment of yeast Uba2 that lacks both clusters of basic amino acids also remains partially nuclear (Dohmen et al., 1995). This cannot be explained by import in association with Aos1, because its c-Myc-like NLS, which is masked in the E1 complex, seems to be the only relevant NLS, at least in HeLa cells. Alternative explanations may be additional, less efficient import pathways for Uba2 or “piggy-back” transport in association with an unknown binding partner. Whether this is physiologically relevant, however, is unclear.
Is the intranuclear localization of the E1 enzyme essential?
SUMOylation is an essential process in most eukaryotes (including bakers yeast and humans) and affects predominantly nuclear proteins. Consistent with this, the E1 enzyme is highly enriched in the nuclear compartment. The finding that yeast Uba2 can be largely mislocalized to the cytoplasm without severe phenotypes (Dohmen et al., 1995; del Olmo et al., 1997; Lois and Lima, 2005) is therefore rather surprising. One explanation that could be addressed experimentally (e.g., by replacing endogenous S.c. Uba2 by a variant that is stably anchored in the cytoplasm) is that residual nuclear Uba2 is sufficient for essential functions. An intriguing alternative is that the SUMO E1 enzyme may carry out its essential function of SUMO activation and E2 (Ubc9) loading also in the cytoplasm. If Ubc9 itself is able to shuttle efficiently between both compartments even when charged with SUMO, the localization of the E1 enzyme may not be rate limiting for efficient SUMOylation in either compartment. Precedence for this idea, which we will follow up in the future, comes from the finding that several class III ubiquitin E2 enzymes require ubiquitin charging for nuclear import by importin 11 (Plafker et al., 2004).
MATERIALS AND METHODS
Plasmid constructs
For generation of pET28a-His6-CFP-Aos1, CFP was PCR-amplified from pECFP-C1 (Clontech, Mountain View, CA) and cloned into NheI and BamHI sites of pET28a (Novagen, Madison, WI). Aos1 was PCR-amplified from pET28a-Aos1 (Pichler et al., 2002) and cloned into EcoRI and HindIII sites of pET28a-His6-CFP. pcDNA3.1-CFP-Aos1 was generated by cloning the NheI–BamHI fragment from pET28a-His6-CFP-Aos1 into pcDNA3.1(−) (Invitrogen, Carlsbad, CA). For generation of pET28b-Uba2-YFP-His6, YFP was PCR-amplified from pEYFP-C1 (Clontech) and cloned into NheI and BamHI sites of pET28b (Novagen). Uba2 was PCR-amplified from pET11d-Uba2 (Pichler et al., 2002) and cloned into NcoI and NheI sites of pET28b-YFP-His6. pcDNA3.1-Uba2-YFP was generated by PCR amplification of Uba2-YFP from pET28b-Uba2-YFP-His6 and cloning into XhoI and EcoRI sites of pcDNA3.1(−). Single-point mutants were generated by site-directed mutagenesis. For generation of pGEX-6P-1-Ubc9, mouse Ubc9 coding sequence was PCR-amplified from pET23a-mUbc9 (Pichler et al., 2002) and cloned into BamHI and XhoI sites of pGEX-6P-1 (GE Healthcare, Uppsala, Sweden). The importin β open reading frame was cloned into pET23a. Bacterial expression plasmids pQE32-His transportin, pRSETb-His importin α, pQE80-His importin13, and pET28a-His-YFP-M9 were kindly provided by Ralph Kehlenbach (University of Göttingen).
Protein expression and purification
YFP- and CFP-tagged SUMO E1 subunits were expressed and purified as described for His-Aos1 and Uba2-His (Werner et al., 2009). Briefly, His-CFP-Aos1 expression in Escherichia coli BL21(DE3) was induced with 1 mM IPTG for 6 h at 25°C. Bacteria were lysed in buffer A (50 mM Na-phosphate, pH 8.0, 300 mM NaCl, 10 mM imidazole, 1 mM β-mercaptoethanol, 1 mg/ml each of aprotinin, leupeptin, and pepstatin, and 0.1 mM phenylmethylsulfonyl fluoride) using an EmulsiFlex (Avestin, Ottawa, Ontario, Canada). After centrifugation at 100,000 × g at 4°C for 1 h, the supernatant was subjected to ProBond Nickel-chelatin resin (Invitrogen). Bead-bound proteins were eluted with 250 mM imidazole. Final purification was by gel filtration (HiLoad 26/60 Superdex 200 pg column; GE Healthcare) in buffer B (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM dithiothreitol [DTT], 1 mg/ml each of aprotinin, leupeptin, and pepstatin). Expression and purification of Uba2-YFP-His followed the same procedure but included an additional purification step: After gel filtration, Uba2-containing fractions were subjected to a MonoQ anion-exchange column (GE Healthcare) in buffer B and eluted with 50–500 mM NaCl. His-CFP-Aos1/Uba2-YFP-His complexes were obtained either by mixing equimolar amounts of subunits and subsequent gel filtration or by incubating Uba2-YFP-His with 1.5-fold excess of His-CFP-Aos1 on ice for 2 h, followed by MonoQ ion exchange chromatography and gel filtration (Figure 6). Purified subunits and complexes were dialyzed against transport buffer (20 mM HEPES, pH 7.3, 110 mM KOAc, 2 mM Mg(OAc)2, 1 mM EDTA), flash-frozen in liquid nitrogen, and stored at −80°C. Purification of His-tagged transport receptors, His-YFP-M9, and GST-Ubc9 followed standard procedures and included a final gel filtration step (S200 analytical column) in transport buffer. GST-Ubc9 was labeled with Alexa Fluor 488 following manufacturer’s instructions (Molecular Probes, Eugene, OR).
Binding assays
Pull-down assays. His-CFP-Aos1 or Uba2-YFP-His was immobilized on CNBr-activated sepharose (Sigma Aldrich, St. Louis, MO) according to manufacturer’s instructions at concentrations of 1 μg/μl beads (Aos1) or 1.3 μg/μl beads (Uba2). Then 15 μl beads was incubated for 1.5 h at 4°C in 500 μl pull-down buffer (50 mM Tris, pH 7.4, 200 mM NaCl, 1 mM MgCl2, 5% glycerol) with 2 mg/ml bovine serum albumin (BSA) or ovalbumin and 1 μM (final concentration) recombinant import receptor. Where indicated, reactions contained 1 μM RanQ69L loaded with GTP (Kehlenbach et al., 1999). After washing with pull-down buffer, bound proteins were eluted with SDS-sample buffer and subjected to SDS–PAGE followed by silver staining.
Solution assays. In these assays, 5 μM purified His-CFP-Aos1/Uba2-YFP-His complex was incubated with 10 μM of recombinant importin α and importin β in a total volume of 250 μl transport buffer with 1 mM DTT overnight at 4°C, prior to analytical gel-filtration on a Superose6-HR10/30 column (GE Healthcare). Then, 300 μl fractions were collected, separated on 5–20% SDS–PAGE gradient gels, and stained with Coomassie.
Mammalian cell culture
HeLa cells were cultured in DMEM (Life Technologies, Carlsbad, CA) containing 10% fetal bovine serum (FBS) and maintained in a humidified incubator with 5% CO2 at 37°C.
Immunofluorescence
Transfection of fluorescent proteins. HeLa cells were grown on 12-mm glass coverslips 24 h prior to transfection with 0.75 μg DNA using Polyfect transfection reagent (QIAGEN, Valencia, CA) according to manufacturer’s instructions. After 24 h, cells were fixed for 15 min in 3.7% formaldehyde, and proteins were visualized by confocal microscopy with a Zeiss confocal microscope (LSM 510meta) using a Plan-Neofluar 63×/1.3 Imm Korr DIC objective.
Indirect immunofluorescence. HeLa cells grown on coverslips were fixed with 3.7% formaldehyde for 15 min, washed, and permeabilized with 0.2% Triton X-100. After blocking with 2% BSA in phosphate-buffered saline (PBS) for 30 min, cells were incubated with 10 μg/ml affinity-purified anti-Aos1 or anti-Uba2 antibody (Bossis and Melchior, 2006) in PBS/BSA for 1 h at room temperature, washed, and incubated for 1 h with 2 μg/ml Alexa488-coupled donkey anti–goat antibody (Molecular Probes). Fluorescence was visualized by confocal microscopy (Zeiss LSM 510meta) using a 63× objective (Plan-Neofluar 63×/1.3 Imm Korr DIC).
Microinjection
HeLa cells were seeded on 12-mm glass coverslips 24 h prior to microinjection, and medium was changed to CO2-independent medium (Life Technologies) 1 h before the experiment. Microinjection with an Eppendorf Femtojet was carried out with 1.4–2.4 μM His-CFP-Aos1/Uba2-YFP-His complexes and 1.5 μM TRITC-Dextran, or with 4.5 μM TRITC-Dextran and 17 μM His-CFP-Aos1 or 8 μM Uba2-YFP-His. Inhibition experiments with monoclonal anti–importin β antibody clone 3E9 (Abcam, Boston, MA) were carried out with 2:1:1 (vol:vol:vol) mixtures of antibody, TRITC-dextran (final 1.6 μM) and His-CFP-Aos1 (final 5.6 μM), Uba2-YFP-His (final 3.3 μM), His-YFP-M9 (final 8.3 μM), or His-CFP-Aos1/Uba2-YFP-His complex (final 3 μM). Cells were incubated for 30 min at 37°C and 5% CO2 in DMEM (Life Technologies) containing 10% FBS, prior to fixation and analysis with a Zeiss confocal microscope (LSM510meta, Plan-Neofluar 63×/1.3 Imm Korr DIC objective).
In vitro nuclear import assays
In vitro import reactions were performed using semipermeabilized HeLa cells as described (Adam et al., 1990). In brief, HeLa cells grown on 12-mm glass coverslips to 40–70% confluency were washed with transport buffer supplemented with 1 mg/ml AP, LP, and 2 μg/ml DTT, and permeabilized with digitonin. Permeabilized cells were incubated for up to 45 min at 30°C or 37°C with 30 μl of a mix containing fluorescently labeled cargo protein (5 μM of single E1 subunits or 1 μM of E1 complexes), 15 μl cytosolic HeLa extract, and either 1.5 μl of an energy-regenerating system (final concentrations 1 mM ATP, 5 mM creatinephosphate and 100 U/ml creatine phosphokinase) or 2 μl of an energy-depleting system (final concentration 16 U hexokinase and 5 mM glucose in transport buffer). Import assays with recombinant import receptors were performed with 1 μM cargo protein, 1.5 μM importin α, and/or 1 μM importin β and either 12 μM Ran or 11.6 μM RanQ69L, loaded with GTP as previously described (Kehlenbach et al., 1999). After import, nuclear accumulation was analyzed by fluorescence microscopy with a Zeiss confocal microscope (LSM 510meta) using a Plan-Neofluar 63×/1.3 Imm Korr DIC objective, or a Leica SP2 confocal microscope using an HCX PL APO 63×/1.4 OIL BD UV objective.
Supplementary Material
Acknowledgments
We thank Ralph Kehlenbach (University Göttingen) for the generous gift of recombinant import receptors and expression plasmids and acknowledge Annette Flotho and Achim Werner for critical reading of the manuscript. Special thanks go to Andreas Werner, Annette Flotho, and Achim Dickmanns for invaluable discussions and to Anja Schreieck and Heidi Ehret for expert help with protein purifications. This work was supported by funding of the DFG within Graduate Program 521.
Abbreviations used:
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- SUMO
small ubiquitin-related modifier
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-05-0461) on January 5, 2011.
REFERENCES
- Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Tan SH, Cavenagh MM, Ainsztein AM, Saitoh H, Dasso M. Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 2001;15:1825–1827. doi: 10.1096/fj.00-0818fje. [DOI] [PubMed] [Google Scholar]
- Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8:4048–4054. doi: 10.1128/mcb.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Olmo M, Mizrahi N, Gross S, Moore CL. The Uba2 and Ufd1 proteins of Saccharomyces cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of cell extracts. Mol Gen Genet. 1997;255:209–218. doi: 10.1007/s004380050491. [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gong L, Li B, Millas S, Yeh ET. Molular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448:185–189. doi: 10.1016/s0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S, Albig W, Kutay U, Bischoff FR, Schwamborn K, Doenecke D, Görlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–23. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Baek SH. Emerging roles of desumoylating enzymes. Biochim Biophys Acta. 2009;1792:155–162. doi: 10.1016/j.bbadis.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci USA. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkerh JP, Dingwall C, Laskey RA. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Mingot JM, Kostka S, Kraft R, Hartmann E, Gorlich D. Importin 13: a novel mediator of nuclear import and export. EMBO J. 2001;20:3685–3694. doi: 10.1093/emboj/20.14.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Siomi MC, Siomi H, Michael WM, Pollard V, Dreyfuss G. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp Cell Res. 1996;229:261–266. doi: 10.1006/excr.1996.0369. [DOI] [PubMed] [Google Scholar]
- Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Plafker SM, Plafker KS, Weissman AM, Macara IG. Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J Cell Biol. 2004;167:649–659. doi: 10.1083/jcb.200406001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Stone J, de Lange T, Ramsay G, Jakobovits E, Bishop JM, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Weis K, Mattaj IW, Lamond AI. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- Werner A, Moutty MC, Moller U, Melchior F. Performing in vitro sumoylation reactions using recombinant enzymes. Methods Mol Biol. 2009;497:187–199. doi: 10.1007/978-1-59745-566-4_12. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.