Abstract
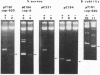
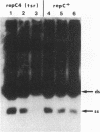
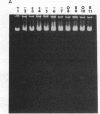
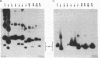
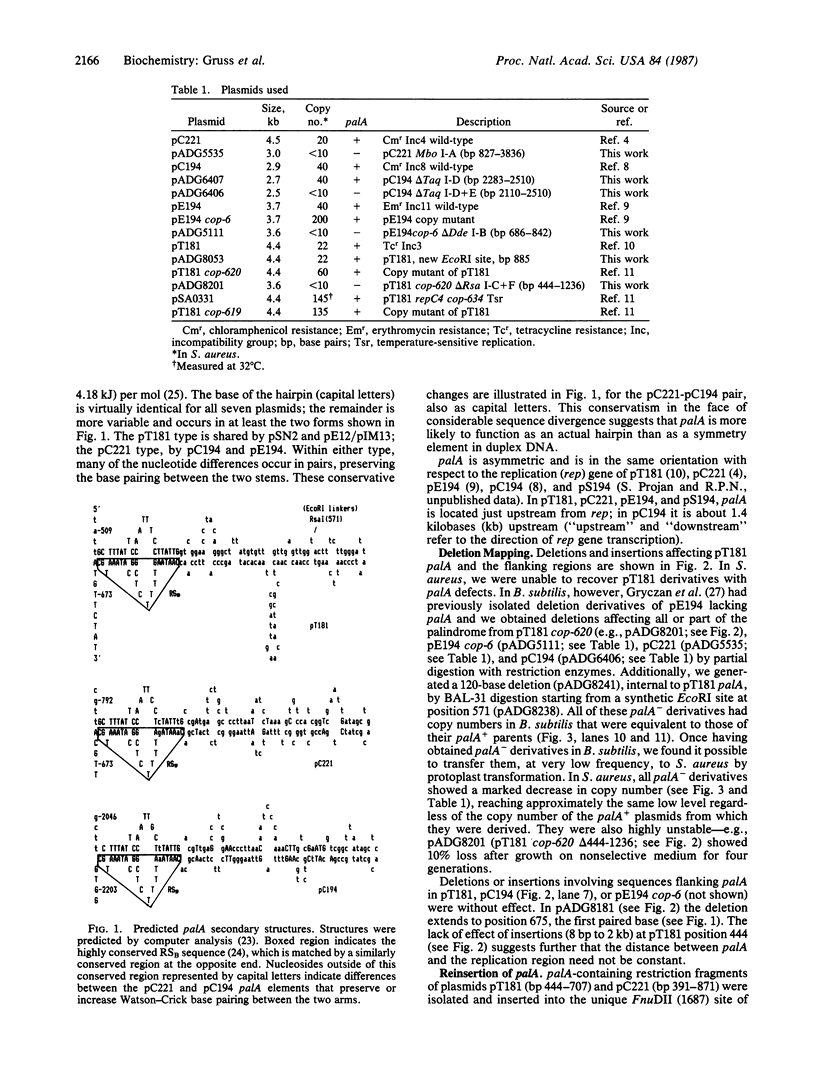
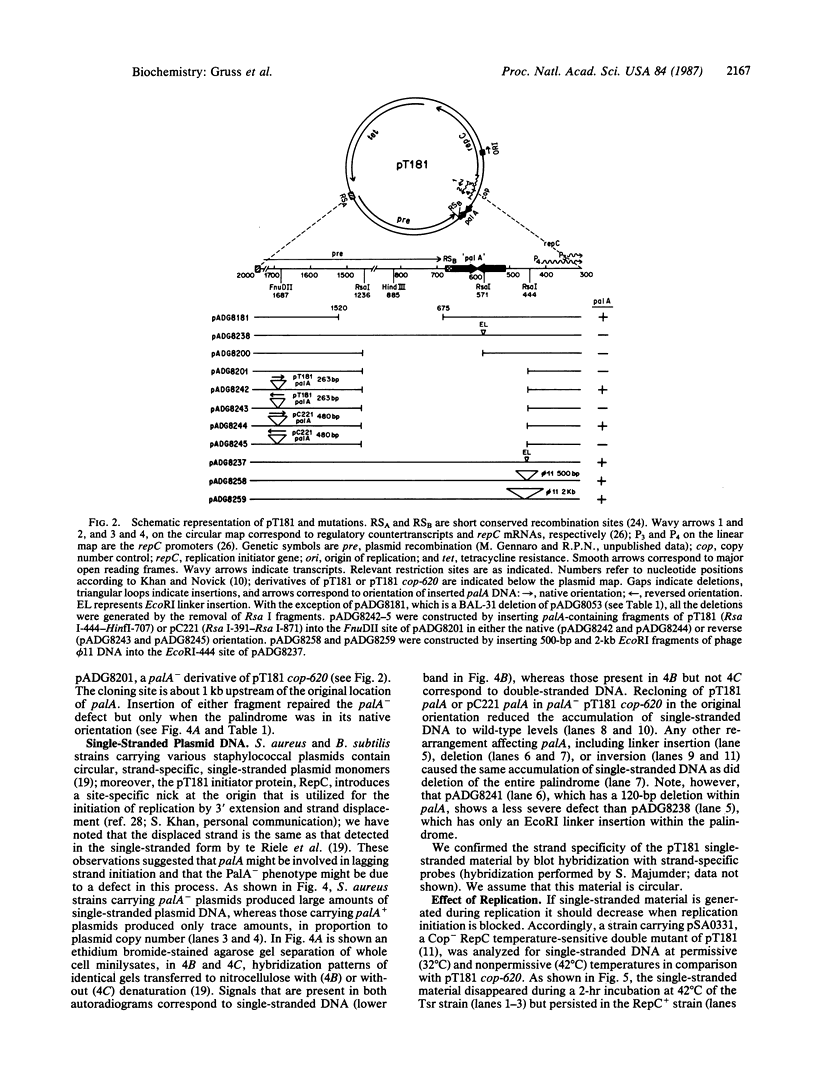
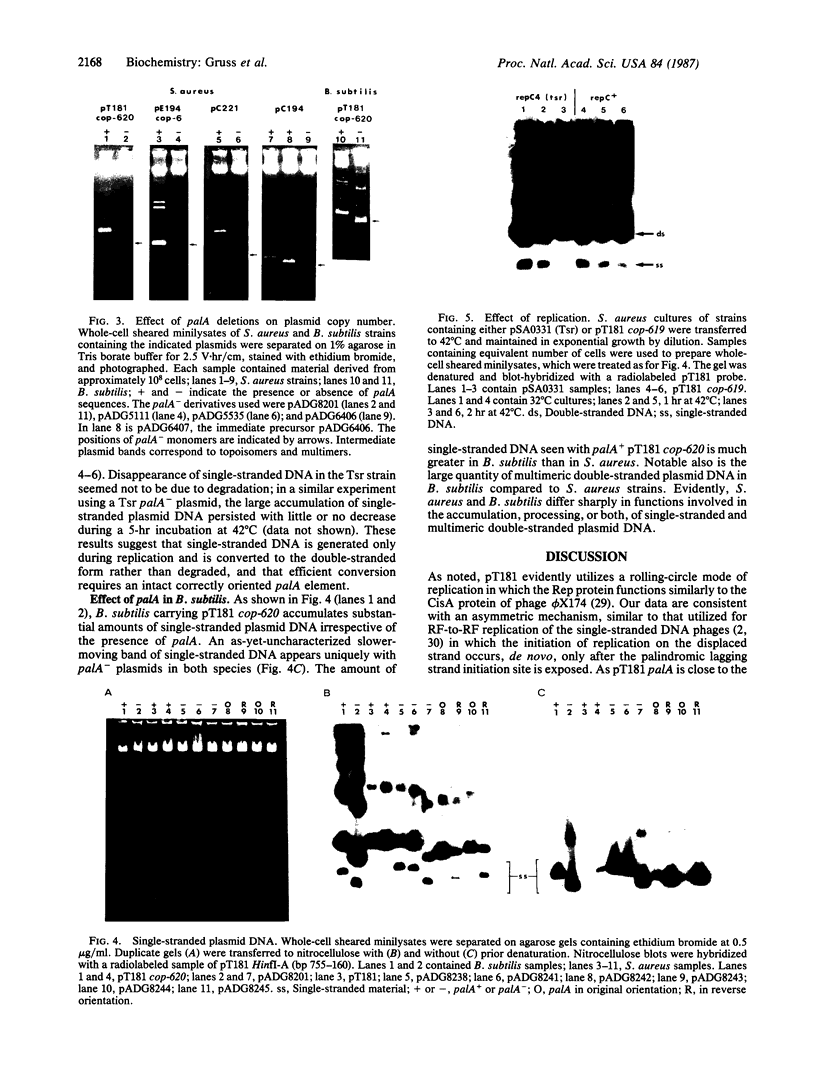
Most small multicopy antibiotic-resistance plasmids of Staphylococcus aureus contain a major axis of hyphenated dyad symmetry (palA) that is required for normal replication and stability, although located outside of the minimal replicon. Rearrangements affecting palA cause plasmid instability, a marked reduction in copy number, and the accumulation of large quantities of strand-specific circular single-stranded plasmid DNA. In view of the recent observation that pT181 initiates replication by a nick and 3'-extension mechanism (S. Khan, personal communication), it is suggested that these plasmids replicate by an asymmetric rolling-circle mechanism in which the displaced plus strand remains single stranded until palA is exposed, forming a hairpin that serves as the lagging strand origin.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burger K. J. Removal of symmetrical sequences from the origin region of plasmid R1 reduces its replication efficiency. EMBO J. 1983;2(5):657–662. doi: 10.1002/j.1460-2075.1983.tb01480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton S., Projan S. J., Highlander S. K., Moghazeh S. M., Novick R. P. Control of pT181 replication II. Mutational analysis. EMBO J. 1984 Oct;3(10):2407–2414. doi: 10.1002/j.1460-2075.1984.tb02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Fiddes J. C., Barrell B. G., Godson G. N. Nucleotide sequences of the separate origins of synthesis of bacteriophage G4 viral and complementary DNA strands. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1081–1085. doi: 10.1073/pnas.75.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall C. M., Cross T. A., DiVerdi J. A., Opella S. J. Protein dynamics by solid-state NMR: aromatic rings of the coat protein in fd bacteriophage. Proc Natl Acad Sci U S A. 1982 Jan;79(1):101–105. doi: 10.1073/pnas.79.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Hobom G. DNA sequences and structural homologies of the replication origins of lambdoid bacteriophages. Nature. 1979 Feb 22;277(5698):621–627. doi: 10.1038/277621a0. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Hahn J., Contente S., Dubnau D. Replication and incompatibility properties of plasmid pE194 in Bacillus subtilis. J Bacteriol. 1982 Nov;152(2):722–735. doi: 10.1128/jb.152.2.722-735.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Structural analysis of plasmid pSN2 in Staphylococcus aureus: no involvement in enterotoxin B production. J Bacteriol. 1982 Feb;149(2):642–649. doi: 10.1128/jb.149.2.642-649.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsel R. R., Murray R. W., Rosenblum W. D., Khan S. A. The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6845–6849. doi: 10.1073/pnas.82.20.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C. C., Novick R. P. Plasmid pT181 replication is regulated by two countertranscripts. Proc Natl Acad Sci U S A. 1985 Feb;82(3):638–642. doi: 10.1073/pnas.82.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986 Mar;15(2):93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- Monod M., Denoya C., Dubnau D. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J Bacteriol. 1986 Jul;167(1):138–147. doi: 10.1128/jb.167.1.138-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Majumder S., Khan S. A., Carleton S., Rosenblum W. D., Iordanescu S. Coding sequence for the pT181 repC product: a plasmid-coded protein uniquely required for replication. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4108–4112. doi: 10.1073/pnas.79.13.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Murphy E., Gryczan T. J., Baron E., Edelman I. Penicillinase plasmids of Staphylococcus aureus: restriction-deletion maps. Plasmid. 1979 Jan;2(1):109–129. doi: 10.1016/0147-619x(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Rosenblum W., Edelman I. Staphylococcal plasmid cointegrates are formed by host- and phage-mediated general rec systems that act on short regions of homology. Mol Gen Genet. 1984;195(1-2):374–377. doi: 10.1007/BF00332777. [DOI] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967 Sep;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Carleton S., Novick R. P. Determination of plasmid copy number by fluorescence densitometry. Plasmid. 1983 Mar;9(2):182–190. doi: 10.1016/0147-619x(83)90019-7. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Kornblum J., Moghazeh S. L., Edelman I., Gennaro M. L., Novick R. P. Comparative sequence and functional analysis of pT181 and pC221, cognate plasmid replicons from Staphylococcus aureus. Mol Gen Genet. 1985;199(3):452–464. doi: 10.1007/BF00330758. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Zipursky S. L., Weisbeek P., Brown D., Hurwitz J. Studies on the phi X174 gene A protein-mediated termination of leading strand DNA synthesis. J Biol Chem. 1983 Jan 10;258(1):529–537. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer-Abramowitz J., Gryczan T. J., Dubnau D. Origin and mode of replication of plasmids pE194 and pUB110. Plasmid. 1981 Jul;6(1):67–77. doi: 10.1016/0147-619x(81)90054-8. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J., Dressler D. Site-specific initiation of a DNA fragment: nucleotide sequence of the bacteriophage G4 negative-strand initiation site. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3094–3098. doi: 10.1073/pnas.75.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Itoh T. The importance of RNA secondary structure in CoIE1 primer formation. Cell. 1982 Dec;31(3 Pt 2):575–583. doi: 10.1016/0092-8674(82)90313-0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]