Abstract
Staphylococcal chromosomal cassette mec (SCCmec) is a mobile genetic element that carries resistance genes for beta-lactam antibiotics. Coagulase-negative staphylococci, such as S. epidermidis, are thought to be a reservoir of diverse SCCmec elements that can spread to the most virulent staphylococcal species, S. aureus, but very little is known about the extent of cross-species spread of these elements in natural populations or their dynamics in different species. We addressed these questions by using a sample of 86 S. aureus and S. epidermidis isolates with SCCmec type IV that were collected from a single hospital over a period of six months. To subtype SCCmec IV, we used multiplex PCR to detect structural variations and we used sequences from a fragment of the ccrB gene and from the dru repeats to detect additional variations. Multiplex PCR had significantly lower typeability than ccrB:dru sequencing, due to more nontypeable isolates among S. epidermidis. No statistically significant differences in diversity were detected by subtyping method or species. Interestingly, while only 4 of 24 subtypes (17%) were shared between species, these so-called shared subtypes represented 58 of 86 isolates (67%). The shared subtypes differed significantly between species in their frequencies. The shared subtypes were also significantly more concordant with genetic backgrounds in S. aureus than in S. epidermidis. Moreover, the shared subtypes had significantly higher minimum inhibitory concentrations to oxacillin in S. aureus than in S. epidermidis. This study has identified particular SCCmec IV subtypes with an important role in spreading beta-lactam resistance between species, and has further revealed some species differences in their abundance, linkage to genetic background, and antibiotic resistance level.
Keywords: Staphylococcus aureus, Staphylococcus epidermidis, SCCmec, strain typing, horizontal genetic transfer
1. Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) infections are a global health problem (Grundmann et al., 2006). Resistance to beta-lactam antibiotics is mediated by the mecA gene, which is carried on a mobile genetic element known as the staphylococcal chromosomal cassette mec (SCCmec) (Katayama et al., 2000). In addition, resistance genes for aminoglycosides, macrolides, tetracyclines, and heavy metals such as cadmium and mercury, can accrue on SCCmec elements. Eight different SCCmec types have been defined, all of which are chromosomally integrated close to the origin of replication at an open reading frame of unknown function called orfX (IWG-SCC, 2009). SCCmec type IV is among the shortest types at 20.9 to 24.2 kb and is thought to be relatively more mobile and to have a relatively lower cost on strain fitness in comparison to some other types (Lee et al., 2007; Okuma et al., 2002; Robinson and Enright, 2003). SCCmec IV occurs frequently in both hospital-acquired and community-acquired MRSA. For example, it currently resides in the most prevalent MRSA clone in the United States, ST8-MRSA-IV (USA300) (Tenover et al., 2006). Eight subtypes of SCCmec IV have been defined (Milheiriço et al., 2007). These subtypes differ from each other structurally in the length of the 3′ end of the element known as the J1 region. Additionally, SCCmec IVa carries pUB110 that confers bleomycin resistance and SCCmec IVc carries Tn4001 that confers broad aminoglycoside resistance.
S. epidermidis is a coagulase-negative staphylococci that has been identified previously as a potential source of conjugative plasmids and other mobile genetic elements that can spread to S. aureus (Archer et al., 1994; McDonnell et al., 1983, Mongkolrattanothai et al., 2004). Several observations are consistent with the hypothesis that S. epidermidis is a reservoir of diverse SCCmec elements: i) it is ubiquitous on human skin, ii) >50% of its isolates are consistently resistant to antistaphylococcal penicillins regardless of geographic locale, and iii) >10% of its isolates yield nontypeable SCCmec elements (Diekema et al., 2001; Garza-González et al., 2010; Hanssen et al., 2007; Ibrahem et al., 2009; Miragaia et al., 2005, 2007; Ruppé et al., 2009). In particular, S. epidermidis may be a reservoir of SCCmec IV. Although the origin of SCCmec IV has not been identified, it has been found among S. epidermidis isolates that were collected in the 1970s, whereas some of the earliest reported S. aureus isolates with SCCmec IV are from the 1980s (Wisplinghoff et al., 2003). Among more recent isolates, SCCmec IVa is common in both S. aureus and S. epidermidis in Japan and France (Barbier et al., 2010; Jamaluddin et al., 2008). Wisplinghoff et al. (2003) indicated that these two species could carry SCCmec IV elements with 98–99% nucleotide identity to each other. Furthermore, Bloemendaal et al. (2010) confirmed an in vivo transfer event of SCCmec IVa from S. epidermidis to S. aureus in which the elements differed by only a single bp (Bloemendaal et al., 2010; Wielders et al., 2001).
The occurrence of very similar SCCmec elements in different staphylococcal species is more parsimoniously explained by their relatively recent horizontal genetic transfer between species rather than their convergent or parallel evolution within species. While these observations provide evidence that cross-species spread of SCCmec can occur in nature, they do not provide an indication of its contribution to the pool of SCCmec elements found in circulating MRSA. In some geographic locales, such as in Norway, the prevalence of MRSA infections is low (<1%) and cross-species spread of SCCmec may be an important mechanism for generating sporadic MRSA (Hanssen et al., 2004). However, in other locales, such as in the United States, the prevalence of MRSA infections is high (>50%) and both coagulase-negative staphylococci as well as other MRSA might be donors of SCCmec. In this study, we characterized a local population of staphylococci from the United States to determine how often isolates carry SCCmec elements that occur in both S. aureus and coagulase-negative staphylococci. We found that a small proportion of the total number of SCCmec IV subtypes were common to both S. aureus and S. epidermidis, but these so-called shared subtypes represented a majority of isolates in both species. We also found that these shared subtypes differed in several characteristics between the two species.
2. Materials and methods
2.1. Bacterial isolates
The Microbiology Laboratory of Westchester Medical Center in Valhalla, NY provided weekly samples of both S. aureus and coagulase-negative staphylococci from clinical specimens from January through June 2007. The isolates were sampled from multiple patients and without regard to their clinical relevance. Upon receipt, the isolates were purified and examined for beta hemolysis and mannitol fermentation phenotypes on appropriate agar plates. Routine bacterial growth was done overnight on tryptic soy agar plates at 37°C. Long-term storage was at −80°C in a solution of tryptic soy broth and 15% glycerol. Bacterial genomic DNA was isolated with a DNeasy kit (Qiagen), according to the manufacturer’s instructions. Species identification was done by PCR amplification and sequencing of both strands of a fragment of the tuf gene, as described previously (Heikens et al., 2005).
tuf gene sequencing identified 12 different species in the isolate collection (181 S. aureus, 3 S. auricularis, 7 S. capitis, 1 S. cohnii, 160 S. epidermidis, 6 S. haemolyticus, 40 S. hominis, 2 S. lugdunensis, 1 S. saprophyticus, 1 S. sciuri, 3 S. simulans, 5 S. warneri). Subsequent SCCmec typing revealed that only SCCmec IV was present in sufficient numbers in S. aureus and coagulase-negative staphylococci, S. epidermidis in this sample, to allow comparisons. For example, there were 50 S. aureus isolates with SCCmec II but only single isolates of S. capitis, S. cohnii, and S. hominis with this type.
Multiple isolates were available from some patients because either more than one colony was picked from an agar plate or specimens were available on more than one date. To avoid bias in frequency-sensitive calculations, we included a single isolate where multiple, genetically indistinguishable isolates were recovered from the same patient. In total, the study sample included 26 S. aureus and 60 S. epidermidis isolates with SCCmec IV. For these isolates, oxacillin minimum inhibitory concentrations (MICs) were determined by Etest (AB Biodisk), according to the manufacturer’s instructions. Characteristics of these isolates are listed in Table S1.
2.2. Characterization of genetic backgrounds and SCCmec elements
Multilocus sequence typing (MLST) was used to identify the genetic backgrounds of the isolates. MLST was done according to published methods for S. aureus (Enright et al., 2000) and S. epidermidis (Thomas et al., 2007). Briefly, fragments of seven housekeeping genes were amplified by PCR and sequenced on both strands. Alleles and sequence types (STs) were determined using the S. aureus and S. epidermidis MLST databases (http://www.mlst.net).
SCCmec IV was identified using PCR methods that score the mec class and ccr allelic group, as done previously (Robinson and Enright, 2003). These results were confirmed using multiplex PCR reactions 1 and 2 of Kondo et al. (2007). SCCmec IV was subtyped using the multiplex PCR method of Milheirço et al. (2007), which was supplemented with a separate PCR reaction to detect the dcs region (Oliveira and de Lencastre, 2002), as recommended. Control strains used to represent the multiplex PCR subtypes included JCSC4744 (SCCmec IVa), CDC OR-9 (SCCmec IVb), JCSC4788 (SCCmec IVc), and JCSC4469 (SCCmec IVd). SCCmec IV was also subtyped by PCR amplification and sequencing on both strands of a fragment of the ccrB gene and the direct repeat units (dru repeats), as done previously (Smyth et al., 2010). The combined subtyping data of multiplex PCR and ccrB:dru sequencing identified indistinguishable SCCmec IV elements that occurred in both species; throughout this study, these elements were referred to as shared subtypes.
2.3. Statistical analyses
Typeability was defined as the proportion of typeable isolates, p. Assuming a binomial sampling distribution, as is commonly done with allele frequency data, the variance of p was calculated as p(1−p)/n, where n is the number of isolates tested. 95% confidence intervals (CI) were constructed as p +/− 2√Var(p).
Diversity was measured with Simpson’s index (Hunter and Gaston, 1988) as well as the ke3 estimator of the effective number of types (Nielsen et al., 2003). 95% CI for these two measures of diversity were calculated as described previously (Grundmann et al., 2001; Smyth et al., 2010). Simpson’s index provides an estimate of the probability that two isolates picked at random from the population belong to different types, and the effective number of types provides an estimate of the number of equally frequent types that will produce the observed diversity.
Differentiation in type frequencies between species was measured with Jost’s D (Jost, 2008). We used the SPADE computer program of Chao and Shen (2003) to calculate the D estimator in equation 13 of Jost (2008) and to calculate 95% CI based on bootstrap resampling with 1000 replicates. Jost’s D will be 0 when subpopulations are completely undifferentiated (i.e. identical) in type frequencies and 1 when subpopulations are completely differentiated. When used simply as a measure of differentiation, D does not require knowledge of the processes that underlie variation in the markers and therefore may be applicable to a variety of typing datasets.
The distributions of MICs were represented with box-and-whisker plots. Differences in median oxacillin MICs were tested with a two-tailed Mann-Whitney U test, using InStat v3.1 (GraphPad Software).
2.4. Nucleotide sequence accession numbers
The sequences of each different ccrB allele has been deposited in the GenBank database with accession numbers HQ236719-21. Unique dru repeats and dru types were deposited in a publicly available dru typing database (http://www.dru-typing.org).
3. Calculation
To measure the concordance between genetic backgrounds and SCCmec IV subtypes, we cross-classified all pairs of isolates according to whether they matched or mismatched for MLST-defined STs and SCCmec IV subtypes. For n isolates, there are N = (n2−n)/2 such pairwise comparisons. When comparing two markers, X and Y, these pairwise comparisons can be summarized in a 2×2 table accordingly: a, number of isolate pairs that match for both markers; b, number of isolate pairs that match for marker X but mismatch for marker Y; c, number of isolate pairs that mismatch for marker X but match for marker Y; d, number of isolate pairs that mismatch for both markers. The Rand index (Rand, 1971) represents the proportion of concordant pairs and is calculated as
Carriço et al. (2006) noted that this measure does not take into account the concordance due to chance and may therefore overestimate the concordance between markers. The adjusted Rand index (Hubert and Arabie, 1985) was recommended for typing datasets to correct for chance concordance and can be calculated with a general formula (Albatineh et al., 2006) as
The maximum RI is always 1. The expected RI due to chance concordance can be calculated from the marginal frequencies of the 2×2 table as
Substituting these into the numerator of the observed RI, we have
With the observed, maximum, and expected RI, we can then calculate the ARI as indicated above. We point out that the Wallace index, which is another measure of concordance recommended for typing datasets (Carriço et al., 2006), is also influenced by chance concordance and should be correctable with an analogous procedure as outlined above. In fact, we find that the expected Wallace index due to chance concordance is very similar, if not identical, to the Wi of Pinto et al. (2008). Since the Wallace index has been used to assess the value of including additional typing methods in studies of both S. aureus and S. epidermidis (Faria et al., 2008; Miragaia et al., 2008; Shore et al., 2010), an adjusted version of this index should be examined with some priority; however, such a development is beyond the scope of this work.
In order to make statistical comparisons of the ARI for different markers, we developed and partially validated a jackknife resampling procedure. We initially investigated several bootstrap resampling procedures, but found that they performed poorly with our datasets. We note that the ARI, like Simpson’s index (Grundmann et al., 2001), uses a sample to make an inference about a population parameter. In this case, the population parameter is the unknown concordance between markers in the larger population. Thus, the ARI is subject to sampling variation and our jackknife procedure attempts to account for that source of variation. It remains to be determined whether the ARI is the preferred population parameter to be estimated and whether these sorts of cross-classification indices are most appropriate for comparing all classes of markers; for example, linkage disequilibrium indices may be more appropriate when the markers consist of discrete genetic loci. Nonetheless, our jackknife procedure is based on a general approach (Crowley, 1992). To begin, each of the n isolates are removed in turn from the dataset and the ARI for each of these n datasets are calculated. Next, n pseudovalues are calculated as
where ARIo is the observed ARI with all isolates included, and ARI−i is the ARI with isolate i removed (i=1, 2, 3, …n). The jackknife estimator of the ARI is the average of the pseudovalues
Its variance and 95% CI are calculated as
To examine the validity of this approach, we first simulated two artificial populations of 1000 isolates each, where each isolate had three markers and each marker had ten types in the population. Marker A was made with 820 isolates of one type followed by 20 isolates each of the remaining nine types. Marker B was made with 460 isolates of one type followed by 60 isolates each of the remaining nine types. Marker C was made with 100 isolates for each of the ten types. Simpson’s index for these three markers was 0.324, 0.757, and 0.901, respectively. In the second population, the types for each marker were randomized among the isolates 100 times to simulate recombination and lower the concordance. Separate random samples of 25, 50, and 100 isolates were taken from each population, and this sampling was repeated 100 times. We then counted the number of samples where the jackknife confidence intervals included the population ARI. With one exception, which achieved 93.3% accuracy, the confidence intervals included the population ARI in 95% or more of the samples.
4. Results
4.1. Typeability and diversity of SCCmec IV
Comparisons of typeability revealed an overall statistically significant difference between the multiplex PCR and ccrB:dru sequencing methods (Table 1); that is, the 95% CI for the proportion of typeable isolates for these two methods did not overlap. The typeability of the multiplex PCR method was relatively low because of the many nontypeable S. epidermidis isolates. For the isolates of both species that were nontypeable by multiplex PCR, only the mecA positive control gene was amplified on repeated attempts. This finding indicated that more uncharacterized structural variations existed in the SCCmec IV elements of S. epidermidis than S. aureus. On the other hand, the ccrB:dru sequencing method yielded a single isolate of S. epidermidis that was nontypeable for its ccrB gene, and the dru repeats were typeable for all 86 staphylococcal isolates (Table 1).
Table 1.
Typeability of SCCmec IV subtyping methods.
| Species | No. of isolates tested | Proportion of typeable isolates by method (95% CI) |
|
|---|---|---|---|
| multiplex PCR | ccrB:dru | ||
| S. aureus | 26 | 0.885 (0.759, 1.000) | 1.000 (1.000, 1.000) |
| S. epidermidis | 60 | 0.683 (0.563, 0.803) | 0.983 (0.950, 1.000) |
| Total | 86 | 0.744 (0.650, 0.838) | 0.988 (0.965, 1.000) |
The diversity of genetic backgrounds, as reflected by MLST, did not differ significantly between the two species (Table 2) even though this sample of SCCmec IV-carrying isolates included more than twice as many S. epidermidis as S. aureus. Moreover, there were no significant differences in the diversity of SCCmec IV based on subtyping method or species. These observations were confirmed using both measures of diversity (Table 2).
Table 2.
Diversity of genetic backgrounds and SCCmec IV subtypes.
| Species | Method | No. of typesa | Simpson’s index of diversity (95% CI) | Effective no. of types (95% CI) |
|---|---|---|---|---|
| S. aureus | MLST | 6 | 0.511 (0.294, 0.728) | 2.04 (0.60, 3.48) |
| multiplex PCR | 4 | 0.403 (0.178, 0.628) | 1.67 (0.92, 2.42) | |
| ccrB:dru | 7 | 0.563 (0.351, 0.775) | 2.28 (0.45, 4.11) | |
| S. epidermidis | MLST | 13 | 0.794 (0.707, 0.881) | 4.84 (2.73, 6.95) |
| multiplex PCR | 6 | 0.636 (0.547, 0.725) | 2.75 (2.11, 3.38) | |
| ccrB:dru | 11 | 0.561 (0.418, 0.704) | 2.28 (0.64, 3.91) |
S. aureus STs (no. isolates): ST1 (1), ST5 (4), ST8 (18), ST59 (1), ST72 (1), ST1183 (1)
S. epidermidis STs (no. isolates): ST2 (6), ST5 (25), ST6 (1), ST16 (7), ST23 (1), ST59 (1), ST69 (5), ST83 (6), ST89 (3), ST175 (1), ST177 (1), ST179 (2), ST180 (1)
multiplex PCR and ccrB:dru subtypes are given in Table 3
4.2. Species distributions of SCCmec IV subtypes
Multiplex PCR subtype IVa was the most prevalent subtype in both species (Table 3). This observation extends the reports that IVa is highly prevalent in both species (Barbier et al., 2010; Jamaluddin et al., 2008). Jost’s D revealed that, with these sample sizes, S. aureus and S. epidermidis were not differentiated from each other based on the frequencies of their multiplex PCR subtypes (Table 3). Although there was a large difference between the two species in the number of nontypeable isolates, this difference was not large enough to genetically differentiate between the two species.
Table 3.
Differentiation between S. aureus and S. epidermidis based on SCCmec IV subtypes.
| multiplex PCR subtypes | No. of isolates |
ccrB:dru subtypes | No. of isolates |
shared subtypes | No. of isolates |
||||
|---|---|---|---|---|---|---|---|---|---|
| S.a. | S.e. | S.a. | S.e. | S.a. | S.e. | ||||
| IVa | 20 | 31 | 3:10a | 4 | 39 | IVa:3:10a | 1 | 18 | |
| IVb | 0 | 2 | 3:10m | 0 | 1 | IVa:3:9g | 17 | 3 | |
| IVc | 0 | 3 | 3:11b | 0 | 7 | IVg:3:10a | 1 | 3 | |
| IVd | 2 | 0 | 3:11q | 0 | 1 | IVnt:3:10a | 1 | 14 | |
| IVg | 1 | 3 | 3:12f | 0 | 1 | Total | 20 | 38 | |
| IVh | 0 | 2 | 3:7f | 0 | 1 | ||||
| IVnt | 3 | 19 | 3:7n | 0 | 1 | ||||
| Total | 26 | 60 | 3:9g | 17 | 6 | ||||
| 3:9q | 0 | 1 | |||||||
| 7:10a | 0 | 1 | |||||||
| nt:8a | 0 | 1 | |||||||
| 3:10e | 1 | 0 | |||||||
| 3:10y | 1 | 0 | |||||||
| 3:8q | 1 | 0 | |||||||
| 3:9d | 1 | 0 | |||||||
| 6:7j | 1 | 0 | |||||||
| Total | 26 | 60 | |||||||
| Jost’s D (95% CI): | 0.093 (0.000, 0.244) | 0.622 (0.367, 0.878) | 0.789 (0.591, 0.987) | ||||||
Note: S.a. is S. aureus, S.e. is S. epidermidis, nt is nontypeable
The ccrB:dru sequencing revealed two prevalent subtypes, 3:10a and 3:9g (Table 3). In contrast to the multiplex PCR method, the ccrB:dru sequencing method was able to differentiate between S. aureus and S. epidermidis, as revealed by a significant non-zero Jost’s D. This result was partly due to the two prevalent ccrB:dru subtypes having different frequencies in the two species. Note that both species also had unique ccrB:dru subtypes, but these were represented by single isolates in all except one subtype (3:11b) (Table 3).
When both the multiplex PCR and ccrB:dru sequencing data was combined, a total of 24 subtypes were identified. While only four of these subtypes occurred in both species, these so-called shared subtypes represented two-thirds of all isolates (Table 3). Furthermore, these shared subtypes represented more than half of each species’ isolates: 20/26 (77%) in S. aureus and 38/60 (63%) in S. epidermidis. By Jost’s D, the shared subtypes alone were able to differentiate between the two species (Table 3). These findings indicated that the shared subtypes were among the most successful elements overall and differed in frequency between species.
4.3. Linkage of SCCmec IV subtypes to genetic backgrounds
The concordance between genetic backgrounds and SCCmec IV subtypes was compared using a new jackknife estimator of the ARI and its confidence interval (see Calculation section). Within species, there were no significant differences in concordance between the different markers (Table 4). The concordance between genetic backgrounds and multiplex PCR subtypes was similar for both species. However, genetic backgrounds were significantly more concordant with ccrB:dru subtypes and with the shared subtypes in S. aureus than in S. epidermidis (Table 4). The multiplex PCR subtypes were also significantly more concordant with ccrB:dru subtypes in S. aureus than in S. epidermidis. These findings revealed that marker concordance was much higher in S. aureus than in S. epidermidis, on average 7.5x higher.
Table 4.
Concordance between genetic backgrounds and SCCmec IV subtypes.
| Species | Comparisona | Observed Rand index | Expected Rand index | Adjusted Rand index | Jackknife estimator (95% CI) |
|---|---|---|---|---|---|
| S. aureus | MLST, multiplex PCR | 0.677 | 0.498 | 0.357 | 0.370 (−0.009, 0.749) |
| MLST, ccrB:dru | 0.818 | 0.501 | 0.636 | 0.652 (0.323, 0.982) | |
| multiplex PCR, ccrB:dru | 0.803 | 0.488 | 0.616 | 0.628 (0.281, 0.975) | |
| MLST, shared subtypes | 0.911 | 0.559 | 0.797 | 0.821 (0.416, 1.000) | |
| S. epidermidis | MLST, multiplex PCR | 0.630 | 0.580 | 0.119 | 0.119 (0.021, 0.217) |
| MLST, ccrB:dru | 0.569 | 0.536 | 0.072 | 0.072 (−0.065, 0.208) | |
| multiplex PCR, ccrB:dru | 0.501 | 0.517 | −0.032 | −0.034 (−0.139, 0.071) | |
| MLST, shared subtypes | 0.642 | 0.569 | 0.169 | 0.171 (−0.003, 0.346) |
Marker A, Marker B
We note that the crude ARI and the jackknife estimator produced results that were very similar to each other (Table 4). Although sufficient to distinguish between species, the confidence intervals for the S. aureus data were very wide, which might be due to the relatively small sample sizes available for that species (n=20 for the shared subtypes, n=26 for other data). This observation would argue against the use of thresholds to interpret the ARI as “moderate”, “good”, and so on, because the uncertainty in the estimate may span several thresholds.
4.4. Oxacillin susceptibilities of SCCmec IV subtypes
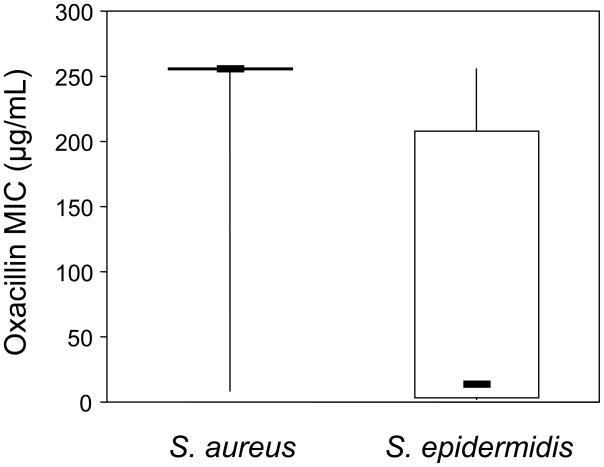
MICs to oxacillin were determined for all isolates with the goal of assessing whether different SCCmec IV subtypes had different levels of resistance. The distributions of MICs for these two species showed mostly high MICs for S. aureus and a wide range of MICs for S. epidermidis (Figure 1). The median MIC for S. aureus isolates was >256 μg/mL compared to 14 μg/mL for S. epidermidis isolates, which is a statistically significant difference (Mann-Whitney U=269.5, P<0.0001). These same patterns extended to the shared subtypes, where S. aureus isolates had a median MIC of >256 μg/mL compared to 12 μg/mL for S. epidermidis isolates (Mann-Whitney U=93.5, P<0.0001).
Figure 1.
Distribution of oxacillin MICs for S. aureus and S. epidermidis isolates with SCCmec IV. Top and bottom lines indicate maximum and minimum values, top and bottom of the boxes indicate the 75th and 25th percentiles, and the dark horizontal lines represents the medians.
The most prevalent shared subtype in S. aureus, identified as IVa:3:9g (Table 3), had a median MIC of >256 μg/mL. Its genetic background included ST8 and a single locus variant of ST8 that was newly identified from this population, ST1183 (Wong et al., 2010). These isolates are closely related to the USA300 MRSA clone that is currently prevalent in the United States (Tenover et al., 2006). In S. epidermidis, this same shared subtype occurred in two STs that differed from each other at three MLST loci and its median MIC was 8 μg/mL. Likewise, the most prevalent shared subtype in S. epidermidis, identified as IVa:3:10a (Table 3), was spread among six STs that differed from each other at an average of 2.6 MLST loci and had a median MIC of 12 μg/mL. In S. aureus, this same shared subtype occurred in a single isolate of ST59 and had an MIC of >256 μg/mL. These results demonstrated that the dynamics of SCCmec IV subtypes that have spread between species can differ in different species; in S. aureus, the shared subtypes have spread among a few genetic backgrounds with high oxacillin MICs whereas, in S. epidermidis, they have spread to more genetic backgrounds with lower MICs.
5. Discussion
The estimated number of acquisitions of SCCmec elements by S. aureus has been upwardly revised with advancements in typing technology (Branger and Goullet, 1987; Enright et al., 2002, Musser and Kapur, 1992; Nübel et al., 2008; Robinson and Enright, 2003). These estimates may differ substantially between clonal groups, from zero acquisitions for much of the current genetic diversity found in S. aureus, to a single acquisition in the ST239 clonal group (Smyth et al., 2010), to a couple of dozen acquisitions in the ST5 clonal group (Nübel et al., 2008). However, with some notable exceptions (Blomendaal et al., 2010; Hanssen et al., 2004), we have very little insight into where the acquired elements come from in individual cases.
Here, we provide new information about the extent of cross-species spread of SCCmec type IV and some dynamics of this element in S. aureus and S. epidermidis. In summary, only a small proportion of the total number of SCCmec IV subtypes circulating in the population was found in both species but, importantly, these shared subtypes were the most prevalent subtypes in the population. Since these shared subtypes are indistinguishable in the two species based on several genetic markers, we infer that they have spread between species. We conclude that these particular SCCmec IV subtypes have an important role in spreading beta-lactam resistance between species and in the population at large. We further determined that these shared subtypes differed significantly between the two species in several characteristics, including abundance, linkage to genetic background, and antibiotic resistance level. While the species differences in abundance might be used to infer the direction of spread (e.g. high abundance indicating the donor and low abundance indicating the recipient), we avoid such inferences in this study because there is the potential of selection for the different MICs to impact abundances. The species differences in the linkage of shared subtypes to strain genetic backgrounds might be explained by differences in the relative clonality of these species, but this result was not entirely expected since the dynamics of mobile genetic elements can be independent of their hosts.
Almost one-third of the S. epidermidis isolates were nontypeable by the multiplex PCR method. These data indicated that uncharacterized variations existed in the S. epidermidis elements, which is consistent with the hypothesis that this species is a reservoir of diverse SCCmec IV elements. However, there was no excess of nontypeable elements by the ccrB:dru sequencing method and the overall diversities of the elements in the two species did not differ significantly. Moreover, 14/19 (74%) of the S. epidermidis isolates with nontypeable elements by multiplex PCR were of the same ccrB:dru subtype (IVnt:3:10a; Table 3), indicating that they probably do not represent an extremely diverse collection of SCCmec IV elements but rather a few uncharacterized structural variants. This observation also justifies our use of the nontypeable isolates as a discrete type during the analyses.
Variation at the ccrB locus is part of the definition of SCCmec types, and its sequence variation appears to agree qualitatively with PCR-detected variation (Lina et al., 2006; Oliveira et al., 2006). The dru sequencing method has been used previously to improve discrimination among MRSA isolates with SCCmec IV from Scottish and Irish hospitals (Goering et al., 2008; Larsen et al., 2009; Shore et al., 2010). We previously compared a multiplex PCR method with the ccrB:dru sequencing method among global strains of the ST239-MRSA-III clonal group, which all carry SCCmec type III (Smyth et al., 2010). Within that clonal group, variations in the ccrB:dru sequences reflected the phylogenetic history of the strains. In contrast, the multiplex PCR method reflected homoplasious (i.e. non-treelike) variations representing independent losses of accessory regions of the element in different strains. In the current study, we observed a difference in the ability of the multiplex PCR and ccrB:dru sequencing methods to differentiate between staphylococcal species. This ability might reflect different mutation rates for these markers. Length variation in the J1 region of SCCmec IV elements is the primary basis for the different multiplex PCR subtypes. Variation at the dru locus, which arises by slipped-strand mispairing and nucleotide substitutions, might accumulate more rapidly than does variation among the multiplex PCR targets or at the ccrB locus. Our results demonstrate that sequence-based subtyping of SCCmec IV has advantages in typeability and differentiation ability over multiplex PCR subtyping in a population-based sample of staphylococci.
We developed a new estimator of the ARI and its confidence interval using a jackknife procedure. This innovation allowed us to make statistical comparisons of the concordance between genetic backgrounds and SCCmec IV subtypes. Based on previous MLST analyses (Feil et al., 2003, Kozitskaya 2005; Miragaia et al., 2007; Ruimy et al., 2008), S. aureus appears to undergo effectively less strain-to-strain recombinations in core genes than does S. epidermidis, resulting in S. aureus having a relatively more clonal population structure than S. epidermidis. Although evidence for multiple acquisitions of SCCmec within diverse samples of both species has been presented (Miragaia et al., 2007; Robinson and Enright, 2003), it has been unknown whether SCCmec was more tightly linked to genetic background in S. aureus or in S. epidermidis. Since mobile genetic elements are often acquired through nonhomologous recombinations mediated by recA-independent transposases or recombinases, it is not obvious whether a correlation should be expected between homologous background recombinations and nonhomologous acquisitions of mobile elements. Our results suggest such a correlation may exist in staphylococci because the concordance between genetic backgrounds and SCCmec IV subtypes was much higher in S. aureus than in S. epidermidis. We find this result surprising given that there were no significant differences in the diversity of these species in terms of their genetic backgrounds or SCCmec IV subtypes.
While the occurrence of indistinguishable subtypes in different species is consistent with a history of cross-species spread of SCCmec IV, the shared subtypes defined here on the basis of all available subtyping data probably represent the most recent subtypes to be spread between these species. Consider that subtypes that both differ slightly from each other and that occur in different species might have accumulated their differences after having been spread between species. Phylogenetic analyses of complete SCCmec IV sequences might be able to identify related elements that have crossed species boundaries, but the markers used here are not ideal for conducting such analyses. On the other hand, previous studies have indicated that variations in the excision of SCCmec IV from the chromosome may influence the propensity of different elements to spread between strains (Higgins et al., 2009; Jansen et al., 2006; Noto and Archer, 2006). Accordingly, the shared subtypes could represent the more transferrable subtypes. Further work is necessary to address these issues.
Questions also remain about where and how frequently SCCmec IV subtypes are spread between species; our data address the impact of cross-species spread on standing diversity of SCCmec IV but not these other questions. The actual horizontal genetic transfer events that resulted in the shared subtypes need not have taken place in the population that we sampled. In fact, the most prevalent shared subtypes from both species occur in MRSA isolated outside our local population (unpublished data). In addition, even though the shared subtypes are the most prevalent in our local population, the frequency of transfer in nature could be very low due to, for example, barriers to horizontal genetic transfer (Corvaglia et al., 2010; Marraffini and Sontheimer, 2008; Waldron and Lindsay, 2006).
Finally, it is clear that in the presence of beta-lactam antibiotics, SCCmec IV could provide a selective advantage to staphylococci. Some data suggest that the cost of carrying type IV is lower than the cost of carrying some other SCCmec types (Lee et al., 2007). Other data indicate that the oxacillin resistance levels conferred by the element have the major effect on fitness, with an inverse relationship between MIC and growth rate (Ender et al., 2004). We observed that the shared subtypes differed significantly in oxacillin MICs between the species. For example, subtype IVa:3:10a was rare in S. aureus where it had a high MIC, whereas it was common in S. epidermidis where it had a low MIC. The same type of pattern, with S. aureus having the higher MIC, was observed for subtype IVa:3:9g even though its frequencies in the two species were reversed. It has been reported that S. epidermidis tends to display a broader range of oxacillin MICs than does S. aureus (McDonald et al., 1995); however, to our knowledge, the results presented here are the first to show that this pattern extends to isolates that share indistinguishable SCCmec subtypes. It is possible that this pattern reflects species differences in the numerous non-SCCmec loci that are known to impact oxacillin MICs (Berger-Bächi et al., 2009). However, it is also possible that this pattern reflects species differences in the intensity of selection, which could be driven by antibiotic treatment of the more aggressive infections that S. aureus causes. These explanations are not necessarily mutually exclusive, since a favorable MIC could be reached by selection on multiple loci.
Supplementary Material
Acknowledgments
We thank Maria Aguero-Rosenfeld, Jon Wegienek, and the technical staff of the Microbiology Laboratory of Westchester Medical Center for providing isolates. This work was supported by a grant from the American Heart Association and by NIH grant GM080602 (to D.A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albatineh AN, Niewiadomska-Bugaj M, Mihalko D. On similarity indices and correction for chance agreement. J Classification. 2006;23:301–313. [Google Scholar]
- Archer GL, Niemeyer DM, Thanassi JA, Pucci MJ. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier F, Ruppé E, Hernandez D, Lebeaux D, Francois P, Felix B, Desprez A, Maiga A, Woerther PL, Gaillard K, Jeanrot C, Wolff M, Schrenzel J, Andremont A, Ruimy R. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202:270–281. doi: 10.1086/653483. [DOI] [PubMed] [Google Scholar]
- Berger-Bächi B, Senn MM, Ender M, Seidl K, Hübscher J, Schulthess B, Heusser R, Meier PS, McCallum N. Resistance to beta-lactam antibiotics. In: Crossley KB, Archer G, Jefferson K, Fowler V, editors. Staphylococci in Human Disease. 2. Wiley-Blackwell; New Jersey: 2009. pp. 170–192. [Google Scholar]
- Bloemendaal AL, Brouwer EC, Fluit AC. Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS One. 2010;5:e11841. doi: 10.1371/journal.pone.0011841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branger C, Goullet P. Esterase electrophoretic polymorphism of methicillin-sensitive and methicillin-resistant strains of Staphylococcus aureus. J Med Microbiol. 1987;24:275–281. doi: 10.1099/00222615-24-3-275. [DOI] [PubMed] [Google Scholar]
- Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol. 2006;44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Shen TJ. Program SPADE (Species Prediction And Diversity Estimation) 2003. [Google Scholar]
- Corvaglia AR, François P, Hernandez D, Perron K, Linder P, Schrenzel J. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc Natl Acad Sci USA. 2010;107:11954–11958. doi: 10.1073/pnas.1000489107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PH. Resampling methods for computation-intensive data analysis in ecology and evolution. Annu Rev Ecol Syst. 1992;23:405–447. [Google Scholar]
- Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M SENTRY Partcipants Group. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32:S114–132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- Ender M, McCallum N, Adhikari R, Berger-Bächi B. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:2295–2297. doi: 10.1128/AAC.48.6.2295-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci USA. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NA, Carriço JA, Oliveira DC, Ramirez M, de Lencastre H. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol. 2008;46:136–144. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, Peacock SJ, Smith JM, Murphy M, Spratt BG, Moore CE, Day NP. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-González E, López D, Pezina C, Muruet W, Bocanegra-García V, Muñoz I, Ramírez C, Llaca-Díaz JM. Diversity of staphylococcal cassette chromosome mec structures in coagulase-negative staphylococci and relationship to drug resistance. J Med Microbiol. 2010;59:323–329. doi: 10.1099/jmm.0.015800-0. [DOI] [PubMed] [Google Scholar]
- Goering RV, Morrison D, Al-Doori Z, Edwards GF, Gemmell CG. Usefulness of mec-associated direct repeat unit (dru) typing in the epidemiological analysis of highly clonal methicillin-resistant Staphylococcus aureus in Scotland. Clin Microbiol Infect. 2008;14:964–969. doi: 10.1111/j.1469-0691.2008.02073.x. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001;39:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H, Aires de Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- Hanssen AM, Kjeldsen G, Sollid JU. Local variants of Staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother. 2004;48:285–296. doi: 10.1128/AAC.48.1.285-296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen AM, Sollid JU. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob Agents Chemother. 2007;51:1671–1677. doi: 10.1128/AAC.00978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikens E, Fleer A, Paauw A, Florijn A, Fluit AC. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol. 2005;43:2286–2290. doi: 10.1128/JCM.43.5.2286-2290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins PG, Rosato AE, Seifert H, Archer GL, Wisplinghoff H. Differential expression of ccrA in methicillin-resistant Staphylococcus aureus strains carrying staphylococcal cassette chromosome mec type II and IVa elements. Antimicrob Agents Chemother. 2009;53:4556–4558. doi: 10.1128/AAC.00395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert L, Arabie P. Comparing partitions. J Classification. 1985;2:193–218. [Google Scholar]
- Hughes AL, Friedman R. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J Bacteriol. 2005;187:2698–2704. doi: 10.1128/JB.187.8.2698-2704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahem S, Salmenlinna S, Virolainen A, Kerttula AM, Lyytikäinen O, Jägerroos H, Broas M, Vuopio-Varkila J. Carriage of methicillin-resistant staphylococci and their SCCmec types in a long-term-care facility. J Clin Microbiol. 2009;47:32–37. doi: 10.1128/JCM.01085-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) Ito T, Hiramatsu K, Oliveira DC, de Lencastre H, Zhang K, Westh H, O’Brien F, Giffard PM, Coleman D, Tenover FC, Boyle-Vavra S, Skov RL, Enright MC, Kreiswirth B, Ko KS, Grundmann H, Laurent F, Sollid JE, Kearns AM, Goering R, John JF, Daum R, Soderquist B. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaluddin TZ, Kuwahara-Arai K, Hisata K, Terasawa M, Cui L, Baba T, Sotozono C, Kinoshita S, Ito T, Hiramatsu K. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. J Clin Microbiol. 2008;46:3778–3783. doi: 10.1128/JCM.02262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen WT, Beitsma MM, Koeman CJ, van Wamel WJ, Verhoef J, Fluit AC. Novel mobile variants of staphylococcal cassette chromosome mec in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:2072–2078. doi: 10.1128/AAC.01539-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43:4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen AR, Goering R, Stegger M, Lindsay JA, Gould KA, Hinds J, Sørum M, Westh H, Boye K, Skov R. Two distinct clones of methicillin-resistant Staphylococcus aureus (MRSA) with the same USA300 pulsed-field gel electrophoresis profile: a potential pitfall for identification of USA300 community-associated MRSA. J Clin Microbiol. 2009;47:3765–3768. doi: 10.1128/JCM.00934-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bächi B, Cook GM. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother. 2007;51:1497–1499. doi: 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Durand G, Berchich C, Short B, Meugnier H, Vandenesch F, Etienne J, Enright MC. Staphylococcal chromosome cassette evolution in Staphylococcus aureus inferred from ccr gene complex sequence typing analysis. Clin Microbiol Infect. 2006;12:1175–1184. doi: 10.1111/j.1469-0691.2006.01548.x. [DOI] [PubMed] [Google Scholar]
- Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, Daum RS, Hiramatsu K. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CL, Maher WE, Fass RJ. Revised interpretation of oxacillin MICs for Staphylococcus epidermidis based on mecA detection. Antimicrob Agents Chemother. 1995;39:982–984. doi: 10.1128/aac.39.4.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell RW, Sweeney HM, Cohen S. Conjugational transfer of gentamicin resistance plasmids intra- and interspecifically in Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1983;23:151–160. doi: 10.1128/aac.23.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheiriço C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother. 2007;60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- Miragaia M, Carriço JA, Thomas JC, Couto I, Enright MC, de Lencastre H. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: proposal for clone definition. J Clin Microbiol. 2008;46:118–129. doi: 10.1128/JCM.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragaia M, Couto I, de Lencastre H. Genetic diversity among methicillin-resistant Staphylococcus epidermidis (MRSE) Microb Drug Resist. 2005;11:83–93. doi: 10.1089/mdr.2005.11.83. [DOI] [PubMed] [Google Scholar]
- Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189:2540–2552. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:1823–1836. doi: 10.1128/AAC.48.5.1823-1836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Kapur V. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol. 1992;30:2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Tarpy DR, Reeve HK. Estimating effective paternity number in social insects and the effective number of alleles in a population. Mol Ecol. 2003;12:3157–3164. doi: 10.1046/j.1365-294x.2003.01994.x. [DOI] [PubMed] [Google Scholar]
- Noto MJ, Archer GL. A subset of Staphylococcus aureus strains harboring staphylococcal cassette chromosome mec (SCCmec) type IV is deficient in CcrAB-mediated SCCmec excision. Antimicrob Agents Chemother. 2006;50:2782–2788. doi: 10.1128/AAC.00032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2008;105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O’Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, Tiensasitorn C, Ito T, Hiramatsu K. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40:4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, Milheiriço C, Vinga S, de Lencastre H. Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J Antimicrob Chemother. 2006;58:23–30. doi: 10.1093/jac/dkl208. [DOI] [PubMed] [Google Scholar]
- Pinto FR, Melo-Cristino J, Ramirez M. A confidence interval for the wallace coefficient of concordance and its application to microbial typing methods. PLoS One. 2008;3:e3696. doi: 10.1371/journal.pone.0003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand WM. Objective criteria for the evaluation of clustering methods. J Amer Stat Assoc. 1971;66:846–850. [Google Scholar]
- Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:3926–3934. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruimy R, Maiga A, Armand-Lefevre L, Maiga I, Diallo A, Koumaré AK, Ouattara K, Soumaré S, Gaillard K, Lucet JC, Andremont A, Feil EJ. The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J Bacteriol. 2008;190:3962–3968. doi: 10.1128/JB.01947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppé E, Barbier F, Mesli Y, Maiga A, Cojocaru R, Benkhalfat M, Benchouk S, Hassaine H, Maiga I, Diallo A, Koumaré AK, Ouattara K, Soumaré S, Dufourcq JB, Nareth C, Sarthou JL, Andremont A, Ruimy R. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob Agents Chemother. 2009;53:442–449. doi: 10.1128/AAC.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore AC, Rossney AS, Kinnevey PM, Brennan OM, Creamer E, Sherlock O, Dolan A, Cunney R, Sullivan DJ, Goering RV, Humphreys H, Coleman DC. Enhanced discrimination of highly clonal ST22-methicillin-resistant Staphylococcus aureus IV isolates achieved by combining spa, dru, and pulsed-field gel electrophoresis typing data. J Clin Microbiol. 2010;48:1839–1852. doi: 10.1128/JCM.02155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DS, McDougal LK, Gran FW, Manoharan A, Enright MC, Song JH, de Lencastre H, Robinson DA. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS One. 2010;5:e8582. doi: 10.1371/journal.pone.0008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006;44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol. 2007;45:616–619. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron DE, Lindsay JA. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol. 2006;188:5578–5585. doi: 10.1128/JB.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielders CL, Vriens MR, Brisse S, de Graaf-Miltenburg LA, Troelstra A, Fleer A, Schmitz FJ, Verhoef J, Fluit AC. In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet. 2001;357:1674–1675. doi: 10.1016/s0140-6736(00)04832-7. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Rosato AE, Enright MC, Noto M, Craig W, Archer GL. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob Agents Chemother. 2003;47:3574–3579. doi: 10.1128/AAC.47.11.3574-3579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob Agents Chemother. 2010;54:742–748. doi: 10.1128/AAC.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.