Abstract
The tobacco (Nicotiana tabacum) agglutinin or Nictaba is a member of a novel class of plant lectins residing in the nucleus and the cytoplasm of tobacco cells. Since tobacco lectin expression is only observed after the plant has been subjected to stress situations such as jasmonate treatment or insect attack, Nictaba is believed to act as a signaling protein involved in the stress physiology of the plant. In this paper, a nuclear proteomics approach was followed to identify the binding partners for Nictaba in the nucleus and the cytoplasm of tobacco cv Xanthi cells. Using lectin affinity chromatography and pull-down assays, it was shown that Nictaba interacts primarily with histone proteins. Binding of Nictaba with histone H2B was confirmed in vitro using affinity chromatography of purified calf thymus histone proteins on a Nictaba column. Elution of Nictaba-interacting histone proteins was achieved with 1 m N-acetylglucosamine (GlcNAc). Moreover, mass spectrometry analyses indicated that the Nictaba-interacting histone proteins are modified by O-GlcNAc. Since the lectin-histone interaction was shown to be carbohydrate dependent, it is proposed that Nictaba might fulfill a signaling role in response to stress by interacting with O-GlcNAcylated proteins in the plant cell nucleus.
For a long time, it was believed that plant lectins were abundant proteins confined to the vacuolar and extracellular compartments of plant cells. Using transgenic lines overexpressing specific lectin genes or incorporation of the purified lectin into an artificial diet, it was shown that an important group of these vacuolar lectins is involved in plant defense against pathogens or predators (Peumans and Van Damme, 1995; Van Damme et al., 1998). However, during the last decade, evidence has been accumulating that plants also synthesize lectins in minute amounts in response to some specific stress factors and changing environmental conditions. Several plant lectins have been discovered that, unlike the previously characterized “vacuolar” lectins, are not synthesized in the endoplasmic reticulum (ER) but on free ribosomes in the cytosol. Further localization studies have shown that this group of lectins locates to the nuclear and/or cytoplasmic compartment of the plant cell. According to their subcellular localization, these lectins were called nucleocytoplasmic plant lectins (Lannoo and Van Damme, 2010). Since the expression of several nucleocytoplasmic lectins was shown to be inducible by (a)biotic stress factors (Claes et al., 1990; Van Damme, 2008; Fouquaert et al., 2009; Vandenborre et al., 2009), these lectins are believed to be involved in stress signaling (Van Damme et al., 2004, Van Damme, 2008). However, until now, this hypothesis has awaited confirmation, since no putative receptors have been identified for nucleocytoplasmic plant lectins (Lannoo and Van Damme, 2010).
The tobacco (Nicotiana tabacum) agglutinin (Nictaba) was one of the first representatives of this novel group of nucleocytoplasmic plant lectins. In 2002, Nictaba was purified from tobacco leaves (cv Samsun NN) pretreated with jasmonic acid methyl ester (Chen et al., 2002). The lectin is not expressed under normal physiological conditions but accumulates in leaf parenchyma cells after treatment with jasmonic acid methyl ester, jasmonic acid, and its precursor 12-oxophytodienoic acid. It was shown that hydroxylation, sulfation, or glucosylation of jasmonic acid results in compounds that are unable to trigger Nictaba synthesis. Since no other plant hormone tested was able to induce lectin accumulation, Nictaba can be considered a genuine jasmonate-inducible protein (Lannoo et al., 2007; Vandenborre et al., 2009). Recently, it was also demonstrated that Nictaba synthesis is up-regulated by insect herbivory, in particular caterpillar feeding. Interestingly, the accumulation of Nictaba is not only confined to the leaf subjected to jasmonate treatment or insect herbivory but can also be observed in other leaves of the tobacco plant, indicating a systemic response (Vandenborre et al., 2009). However, no accumulation of jasmonic acid or its derivatives was observed in systemic leaves, suggesting the involvement of an unknown signaling system that works downstream of the jasmonate pathway.
Although the tobacco lectin was originally reported in tobacco cv Samsun NN, it was shown later that the lectin sequence is also present in other tobacco species and cultivars. Nictaba accumulated in nine out of 19 tobacco species tested, including all cultivars from tobacco (Lannoo et al., 2006a).
Cloning of the Nictaba coding sequence from tobacco cv Samsun NN revealed that it contains a basic tetra-Lys nuclear localization sequence (102-LysLysLysLys-105). Using immunohistochemistry and confocal microscopy with a GFP-Nictaba fusion protein, it was confirmed that Nictaba resides in the nucleocytoplasmic compartment of the plant cell and is virtually absent from the nucleolus (Chen et al., 2002; Lannoo et al., 2006b; Vandenborre et al., 2009). Using hapten inhibition assays and glycan array technology, it was demonstrated that Nictaba exhibits specificity for GlcNAc oligomers but also interacts with high-Man and complex N-glycans (Lannoo et al., 2006b).
Hitherto, the molecular function of Nictaba has remained unknown. In this paper, we used a proteomics approach to isolate and identify Nictaba-interacting proteins in the nucleus and the cytoplasm of tobacco cv Xanthi cells using lectin affinity chromatography as well as pull-down assays. Both approaches revealed that Nictaba primarily associates with core histone proteins. Binding of Nictaba to these nucleosomal proteins was confirmed in an affinity chromatography experiment with a calf thymus histone preparation. Proteins could be eluted from the lectin column with GlcNAc monomers. Moreover, western-blot analysis using an anti-O-GlcNAc antibody and lectin blotting with the GlcNAc-specific lectin wheat germ agglutinin (WGA) suggest that these histone proteins interact with Nictaba through GlcNAc modifications.
RESULTS AND DISCUSSION
Purification of Nuclei from Tobacco cv Xanthi Cultured Cells
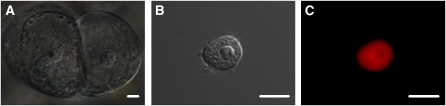
Intact nuclei were isolated from cultured cv Xanthi cells (according to protocol 2 below). Differential interference phase contrast microscopy as well as fluorescence microscopy revealed that the structure of the isolated nuclei was intact and that the nuclear preparation was almost devoid of any cellular debris. The identity of the isolated nuclei was confirmed by staining with 4′,6-diamidino-2-phenylindole or propidium iodide (Fig. 1). Fields of purified nuclei with corresponding propidium iodide staining are shown in Supplemental Figure S1.
Figure 1.
Microscopical analysis of tobacco cells and nuclei-enriched fractions. A, Light microscopy of cv Xanthi cultured cells. B and C, Nuclei extracted from cv Xanthi cells visualized under transmission light (B) and stained with propidium iodide (C). Bars = 10 μm. [See online article for color version of this figure.]
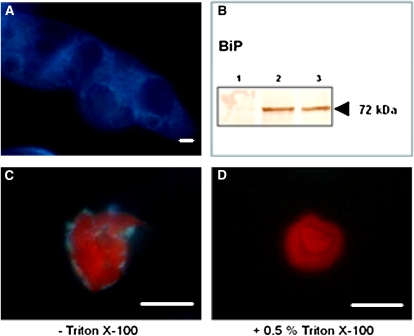
Since addition of Triton X-100 to the nuclear isolation buffer was shown to be sufficient for lysis of ER structures and removal of ER remnants associated with the outer nuclear envelope (Watson and Thompson, 1986), a comparative analysis was conducted in which nuclei were purified in the presence or absence of 0.5% Triton X-100. Using the ER-specific stain ER-Tracker, it was observed that nuclei extracted in the absence of Triton X-100 were to a greater extent associated with ER remnants, compared with nuclei purified in the presence of 0.5% Triton X-100 (Fig. 2).
Figure 2.
Removal of ER remnants associated with the outer nuclear envelope by addition of 0.5% Triton X-100 in the extraction buffer. A, ER of cv Xanthi cells stained with ER-Tracker Blue-White DPX. B, The ER-associated chaperone BiP is detected in an unfractionated protein extract but not in a nuclear protein extract: lane 1, 30 μg of nuclear protein; lanes 2 and 3, 50 and 30 μg of unfractionated protein, respectively. C and D, cv Xanthi nuclei extracted in the presence of Triton X-100 (D) are not stained for ER in comparison with nuclei extracted without Triton X-100 (C). Bars = 10 μm.
The contamination rate of nuclear-enriched fractions was estimated by measuring the activity of marker enzymes. Enzymatic assays revealed that the nuclear protein extracted from tobacco cells was largely depleted from proteins residing in other subcellular compartments. The activity of the ER-localized cytochrome c reductase in the nuclear protein extract was only 0.37% compared with the total amount of unfractionated protein that was extracted from an equal amount of cells. No activity was detected in the nuclear protein fraction for the cytosolic Glc-6-P dehydrogenase, the Golgi-localized IDPase, and the plasma membrane ATPase. Moreover, the ER-localized chaperone protein BiP, which was clearly observed in the unfractionated protein fraction from tobacco cells, was absent from the nuclear protein extract (Fig. 2B).
Nictaba Interacts with a Variety of Nucleocytoplasmic Proteins from Tobacco
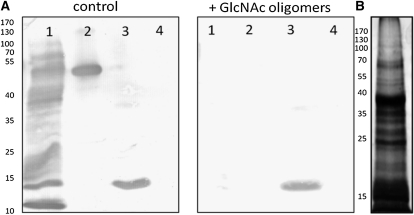
Lectin blotting of nuclear proteins revealed the interaction of Nictaba with several proteins, the most abundant ones having molecular masses ranging between 10 and 15 kD, 35 and 40 kD, and 70 and 100 kD (Fig. 3). In this experiment, carboxypeptidase Y and peanut (Arachis hypogaea) lectin (PNA) were used as positive (glycosylated) and negative (nonglycosylated) controls for lectin binding, respectively. Preincubation of Nictaba with a mixture of GlcNAc oligomers clearly inhibited the binding of this lectin to the nuclear proteins.
Figure 3.
A, Interaction of Nictaba with nuclear proteins from tobacco in the absence (left panel) or presence of GlcNAc oligomers (right panel). Lane 1, 30 μg of nuclear protein; lane 2, 0.1 μg of carboxypeptidase Y; lane 3, 1 μg of Nictaba; lane 4, 0.5 μg of PNA. B, SDS-PAGE and silver staining of 30 μg of total nuclear protein. The positions and sizes (kD) of the marker proteins are indicated on the left.
Identification of Nictaba-Interacting Proteins from Tobacco cv Xanthi Nuclei Using Lectin Affinity Chromatography
To specifically look for interacting partners for the tobacco lectin in the nuclear compartment of the plant cell, tobacco nuclei were purified and nuclear protein was extracted using a method that allowed the purification of milligram amounts of nuclear protein (protocol 1). However, the pitfall of this method is the contamination of the nuclear extract with small amounts of cell wall proteins. Interacting partners for Nictaba in the nuclear protein fraction were identified using affinity chromatography on a column with covalently coupled Nictaba. Only a small fraction of the nuclear protein (purified according to protocol 1) was retained on the column, representing 1.2% of the total protein loaded on the column. Analysis of the eluted protein by SDS-PAGE revealed polypeptides with molecular masses ranging from 10 to 100 kD (Supplemental Fig. S2).
Proteins eluted from the Nictaba column were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS; Table I; Supplemental Table S1). Quantification of eluted proteins was performed by calculating the exponentially modified protein abundancy index (emPAI). This label-free protein quantification method has previously been shown to correctly estimate the abundance of proteins identified by MS (Ishihama et al., 2005). The most abundant proteins eluting from the Nictaba column were the histone proteins. In particular, core histone proteins H2A, H2B, H3, and H4 (for formation of the octameric nucleosome core particle) are highly represented in the eluted fractions and are more abundant than the linker histones (H1). The emPAI values for all proteins in the Nictaba-eluted fraction were compared with those of proteins identified in the total nuclear extract that was applied on the column and allowed us to conclude that histone proteins are clearly enriched in the protein fraction bound by Nictaba (Supplemental Tables S1 and S2).
Table I. Nictaba-interacting proteins identified in a cv Xanthi nuclear extract using lectin affinity chromatography.
| National Center for Biotechnology Information Gene Identifier No. | Protein Description | emPAI |
| 117307343 | Histone H4 | 9 |
| 156720295 | Histone H3.1 | 5.8124 |
| 27529852 | Histone H2A | 3.6419 |
| 158053020 | Histone H2B | 2.5942 |
| 10241929 | Elicitor-inducible protein | 1.1543 |
| 4097575 | NTFP1 | 1.1543 |
| 1169476 | Elongation factor 1-α (EF-1-α), vitronectin-like adhesion protein 1, PVN1 | 0.8168 |
| 3759184 | Phi-1 | 0.7191 |
| 90265703 | Tobacco fibrillarin homolog | 0.5399 |
| 4027901 | α-Expansin precursor | 0.3896 |
| 1498168 | Endoxyloglucan transferase-related protein | 0.311 |
| 4027891 | α-Expansin precursor | 0.2915 |
| 1255951 | PS60 | 0.2915 |
| 10799832 | Ribosomal protein L11-like | 0.2328 |
| 27597229 | Purple acid phosphatase | 0.1787 |
| 14031051 | Peroxidase | 0.166 |
| 2493321 | l-Ascorbate oxidase precursor (ascorbase; ASO) | 0.1601 |
| 6048277 | Pectin methylesterase | 0.1548 |
| 5381253 | Peroxidase | 0.145 |
| 2541876 | CND41, chloroplast nucleoid DNA-binding protein | 0.1054 |
| 16903349 | Endo-β-1,4-glucanase precursor | 0.0857 |
Next to these nucleosomal proteins, a stress-inducible protein (elicitor-inducible protein), a chaperone protein (NTFP1), and some proteins involved in translation and ribosome biogenesis (elongation factor 1α, fibrillarin homolog, and ribosomal protein L11-like) were identified as Nictaba-binding proteins, albeit at a much lower abundance. In addition, a few proteins that presumably locate to the extracellular compartment (e.g. phi-1, α-expansin precursor, endoxyloglucan transferase-related protein, PS60, peroxidase, etc.) were identified by MS (Supplemental Table S1).
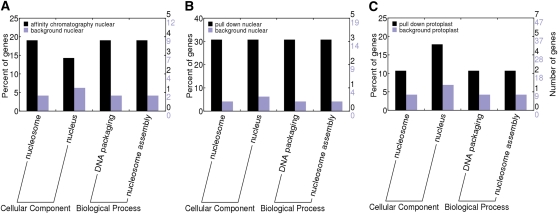
Amino acid sequences of the identified proteins were also analyzed using the InterProScan tool (Zdobnov and Apweiler, 2001) to functionally annotate Nictaba-interacting proteins. Afterward, the Web Gene Ontology Annotation Plotting (WEGO) tool (Ye et al., 2006) was used to identify the cellular localization and the biological processes of eluted proteins that were enriched when compared with a total tobacco cv Xanthi nuclear extract (further designated as “background”). It is clear from Figure 4A that biological processes such as DNA packaging and nucleosome assembly, both processes obviously associated with histone proteins, are highly represented among the Nictaba-interacting proteins when compared with the background.
Figure 4.
InterProScan scanning and WEGO plotting of Nictaba-interacting proteins identified by affinity chromatography (A) and pull-down assay (B) in a nuclear protein extract or identified by pull-down assay in an unfractionated protoplast extract (C). Proteins interacting with the lectin were compared with a total tobacco background containing all nuclear or protoplast proteins. [See online article for color version of this figure.]
Identification of Nictaba-Interacting Proteins from Tobacco cv Xanthi Nuclei Using Pull-Down Assays
Since MS analysis and identification of Nictaba-binding proteins retained on the tobacco lectin column revealed small amounts of proteins that locate to the plant cell wall, a more rigid fractionation was done to obtain highly pure tobacco nuclei (according to protocol 2). After enzymatic removal of the cell wall of tobacco cells, nuclei were fractionated from the protoplasts using a 25% to 36% iodixanol gradient. As a consequence, the yield of protein was much lower (125 mg versus 25 mg from a 100-g cell weight for protocols 1 and 2, respectively). This nuclear protein fraction was used in pull-down assays with a recombinant His-tagged Nictaba. The nuclear protein extract from cv Xanthi cells was used as the prey protein source to pull down Nictaba-interacting partners. Table II gives an overview of the released proteins identified by LC-MS/MS analysis (using an LTQ OrbiTRAP XL mass spectrometer) together with their emPAI values. Analyses with this protein preparation retrieved very few extracellular proteins, confirming the superior purity of this nuclear preparation from tobacco cells (Supplemental Table S3).
Table II. Nictaba-interacting proteins identified in a tobacco nuclear extract using a pull-down assay.
| National Center for Biotechnology Information Gene Identifier No. | Protein Description | emPAI |
| 110738359 | Histone H4-like protein | 2.1623 |
| 110736078 | Histone H2A.F/Z | 1.1543 |
| 10177235 | Histone H2B-like protein | 0.9953 |
| 117168143 | At5g65350 (putative histone H3-like 5) | 0.4679 |
| 132270 | Rubber elongation factor protein (REF; allergen Hev b 1) | 0.2589 |
| 33333422 | Maturase K | 0.1787 |
| 115446065 | Os02g0466400 | 0.1105 |
| 10177535 | Unnamed protein product | 0.0927 |
| 62701659 | F-box domain, putative | 0.0827 |
| 115459208 | Endoglucanase 12 | 0.0772 |
| 15226197 | Leu-rich repeat transmembrane protein kinase, putative | 0.0624 |
| 3687301 | Subtilisin-like protease | 0.0607 |
| 115461811 | Os05g0122600 | 0.0563 |
| 3970757 | SBT4E protein | 0.0563 |
The MS results from the pull-down assay are in line with those obtained by lectin affinity chromatography in that the most abundant Nictaba-interacting proteins are core histone proteins. No linker histone protein was identified. Comparison with a background list containing all proteins in the nuclear protein fraction (Supplemental Table S2) revealed that these nucleosomal proteins are specifically enriched in the pull-down assay. InterProScan scanning of identified protein sequences and WEGO plotting again revealed that biological processes typically associated with histone proteins were highly enriched among the Nictaba-interacting proteins when compared with a background extract (Fig. 4B).
Proteins identified in the elution fraction at a lower abundance (emPAI < 0.3) include a rubber (Hevea brasiliensis) elongation factor, a protein involved in posttranscriptional processing (maturase K), a protein involved in ribosome biogenesis (unnamed protein product), and an F-box protein with unknown function.
Identification of Nictaba-Interacting Proteins from Tobacco cv Xanthi in an Unfractionated Protoplast Extract Using Pull-Down Assays
To provide additional proof that Nictaba interacts with histone proteins in tobacco cells, a similar pull-down experiment was performed using an unfractionated protoplast extract as the source of prey protein. From the results shown in Table III, it is clear that histone proteins H2A.F/Z, H4, and H2B are present at relatively high abundance in the protein fraction interacting with Nictaba (Table III; Supplemental Table S4), demonstrating that Nictaba interacts with histone proteins, even in a protein extract not specifically enriched for nuclear proteins. In addition, this experiment also allowed us to identify putative cytoplasmic proteins that can interact with Nictaba. The most abundant protein pulled down from this extract corresponds to the putative heat shock protein PS1, a chaperone protein involved in the stress response. Furthermore, two hypothetical proteins with an unknown function (28927693 and 12323759), a protein with transcription factor activity (147785120), and the rubber elongation factor were detected among the Nictaba-interacting proteins at a relatively high abundance (emPAI > 0.25). The specificity of the interaction with Nictaba was verified by comparison with a background list of proteins identified in an unfractionated protoplast extract (Supplemental Table S5). Analysis of the list of identified Nictaba-binding proteins using the InterProScan tool revealed that Nictaba-binding proteins in protoplast extracts are involved in histone-associated processes (Fig. 4C), though to a lower extent compared with the experiments performed with nuclear proteins.
Table III. Nictaba-interacting proteins identified in a cv Xanthi unfractionated protoplast extract using a pull-down assay.
| National Center for Biotechnology Information Gene Identifier No. | Protein Description | emPAI |
| 109892469 | Putative heat shock protein PS1 | 9 |
| 28927693 | Hypothetical protein | 1.1543 |
| 110736078 | Histone H2A.F/Z | 0.4679 |
| 110738359 | Histone H4 | 0.4679 |
| 12323759 | Hypothetical protein | 0.2915 |
| 132270 | Rubber elongation factor protein (REF; allergen Hev b 1) | 0.2589 |
| 10177235 | Histone H2B-like protein | 0.2589 |
| 147785120 | Hypothetical protein | 0.2589 |
| 14423552 | Unknown protein | 0.2328 |
| 124301100 | At2g01610 | 0.2328 |
| 9279614 | Unnamed protein product | 0.166 |
| 115485275 | Xanthine/uracil/vitamin C permease family protein, putative, expressed | 0.166 |
| 469457 | Polygalacturonase inhibitor protein | 0.1548 |
| 14009657 | Secreted acid phosphatase | 0.1366 |
| 119004 | Glucan endo-1,3-β-glucosidase, basic vacuolar isoform | 0.1288 |
| 11133446 | Malate dehydrogenase, cytoplasmic 2 | 0.122 |
| 1002531 | Actin-4 | 0.1105 |
| 110741992 | Putative Fru bisphosphate aldolase | 0.1105 |
| 1009712 | Calreticulin | 0.0889 |
| 117165712 | Calreticulin-1 | 0.0889 |
| 11762222 | Adenosylhomocysteinase 1 | 0.0857 |
| 115449495 | Os02g0812600 | 0.0827 |
| 12644189 | Chaperonin CPN60, mitochondrial | 0.07 |
| 115481628 | Putative ribosomal protein S23 (S12) | 0.07 |
| 15226197 | Leu-rich repeat transmembrane protein kinase, putative | 0.0624 |
| 108862990 | 5-Methyltetrahydropteroyltriglutamate-homo-Cys methyltransferase, putative, expressed | 0.0578 |
| 33235471 | Lipoxygenase | 0.0428 |
| 62733821 | Retrotransposon protein, putative | 0.0366 |
| 116057152 | TNF receptor-associated factor (ISS) | 0.0266 |
GlcNAc-Dependent Interaction of Calf Histone Proteins with Nictaba and WGA
Since histone proteins are known to be the most conserved proteins in eukaryotes, the interaction of histone proteins from calf thymus with Nictaba was investigated. A total nonfractionated preparation of purified histone proteins was analyzed on the Nictaba-Sepharose 4B column. Proteins bound to the lectin column were eluted and analyzed by SDS-PAGE (Fig. 5). Protein elution could be established by adding a buffer with high pH. Alternatively, histone proteins were eluted from the Nictaba column with 1 m GlcNAc (Fig. 5). Lower concentrations of GlcNAc (0.1–0.5 m) did not result in desorption of bound histone proteins. Similarly, 1 m Glc did not destroy the interaction of the bound histone proteins to Nictaba (Supplemental Fig. S3A).
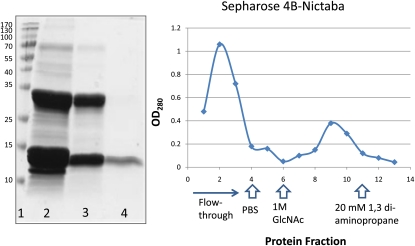
Figure 5.
Interaction of histone proteins with Nictaba-Sepharose 4B. The left panel shows affinity chromatography of histone proteins from calf thymus on Nictaba-Sepharose 4B. Lane 1, Molecular mass markers (Fermentas); lane 2, 50 μg of whole nonfractionated histone proteins from calf thymus; lane 3, flow through of the Nictaba column; lane 4, histone proteins eluted from the Nictaba column. The right panel shows the absorbance profile (OD280) of a Nictaba-Sepharose 4B column loaded with 10 mg of calf thymus histone proteins and eluted with 1 m GlcNAc and 20 mm 1,3-diaminopropane. [See online article for color version of this figure.]
As shown in Figure 5, only a small fraction of the total histone preparation was retained on the lectin column, representing approximately 16% of the total protein loaded on the column. N-terminal sequencing of the polypeptide with a molecular mass of approximately 12 kD yielded a 12-amino acid sequence (PEPAKSAPAPKK), which allowed us to unambiguously identify this polypeptide as histone H2B (100% sequence identity).
To exclude the possibility that histone proteins interact aspecifically with the Sepharose 4B matrix, the calf histone extract was also analyzed on a Sepharose 4B matrix (without any lectin coupled to it). It was observed that virtually no histone proteins were retained on the Sepharose 4B matrix (Supplemental Fig. S3B).
To investigate if histone proteins also interact with the WGA, a lectin known to specifically recognize GlcNAc and O-GlcNAc, a similar affinity chromatography experiment with calf histone proteins was also performed on a WGA column. Analysis of the histone proteins eluted from this column revealed that histone 2B as well as histone 3 interact with WGA (Supplemental Fig. S3C).
Lectin-Interacting Histone Proteins Are Recognized by CTD110.6 Antibody
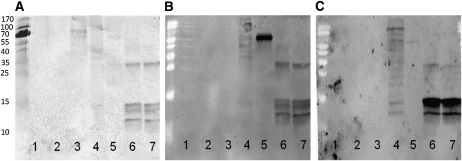
Histone proteins bound to the Nictaba or WGA column were analyzed by SDS-PAGE and reacted on a blot with fluorescein isothiocyanate (FITC)-labeled WGA and Nictaba as well as with the CTD110.6 antibody, known to specifically detect the O-GlcNAc modification on Ser/Thr residues of some nucleocytoplasmic animal proteins (Fig. 6). Nuclear proteins extracted from human ECV cells, as well as 5 ng of bovine serum albumin (BSA) chemically coupled to a GlcNAc residue (BSA-aminophenyl-GlcNAc), were used as positive controls. In addition, 100 ng of carboxypeptidase Y, a protein known to be glycosylated by high-Man glycans, was used as a positive control for the Nictaba-FITC blot, whereas 5 μg of PNA (an unglycosylated lectin) and 5 ng of BSA-aminophenyl were used as negative controls. The anti-O-GlcNAc antibody showed good interaction with BSA-GlcNAc and both calf thymus histone preparations retained on the Nictaba and WGA columns. Whereas the interaction of CTD110.6 with ECV proteins was very weak, these nuclear proteins reacted with Nictaba-FITC, but especially with WGA-FITC. Both lectins also showed good reactivity toward the histone fraction retained on both lectin columns. As expected, only Nictaba-FITC reacted with carboxypeptidase Y, since the lectin is known to recognize the GlcNAc2 core of high-Man N-glycans.
Figure 6.
Interaction of calf thymus histone proteins eluted from a Nictaba or WGA column with CTD110.6 antibody (A), Nictaba-FITC (B), and WGA-FITC (C). Lane 1, 5 ng of BSA-aminophenyl; lane 2, 5 μg of PNA; lane 3, 5 ng of BSA-aminophenyl-GlcNAc; lane 4, 10 μg of nuclear fractionated protein from a human transformed endothelial cell line (ECV 304); lane 5, 100 ng of carboxypeptidase Y; lane 6, 3 μg of histone protein eluted from a Nictaba-Sepharose column; lane 7, 3 μg of histone protein eluted from a WGA-Sepharose column.
MS Analysis of Nictaba- and WGA-Interacting Calf Histone Proteins Indicates Potential O-GlcNAcylation and Similarity in Substrate Binding
Calf histone proteins eluted from Nictaba and WGA-Sepharose 4B columns were analyzed by MS following digestion and reverse phase (RP)-HPLC separation. A detailed comparison of targets bound by both WGA and Nictaba revealed that over 75% of all identified proteins were bound to both lectin columns. Next, we performed a mild β-elimination using butylamine onto the eluted proteins. This step converts all O-GlcNAc carrying Ser and Thr residues and adds a mass marker of 55 D onto the modified amino acids. Following β-elimination, proteins were digested and separated by RP-HPLC and the samples were analyzed on a LTQ OrbiTRAP XL mass spectrometer. Here, we could identify several peptide sequences that hinted toward O-GlcNAc modification. Notably, Ser-65 from sequences NH2-AMGIMNSFVNDIFER-COOH and NH2-AMGNMNSFVNDIFER-COOH from histone H2B were prominent in both analyses. In addition, also the Thr-80 from NH2-EIAQDFNTDLR-COOH and Thr-101 in NH2-ALVQNDTLLQVK-COOH that could be mapped to histone H3.3 and H1, respectively, were identified as O-GlcNAc-modified peptides among the WGA-interacting proteins.
Our results suggest O-GlcNAcylation of histone H2B on Ser-65. This result complements the observations of Sakabe et al. (2010), who also mapped an O-GlcNAc site on histone H2B at position Ser-36. These authors suggested that probably other unidentified O-GlcNAc sites were present on this protein. The O-GlcNAc modification of histone H3.3 and H1 on Thr-80 and Thr-101, respectively, is yet undescribed. Since β-elimination not only removes the O-GlcNAc modification from Ser and Thr amino acids but also phosphate, the possibility that the detected O-GlcNAc sites are in fact phosphorylation sites had to be excluded. Therefore, we analyzed Nictaba- and WGA-interacting histone proteins that were not subjected to β-elimination on a LTQ OrbiTRAP XL mass spectrometer. No phosphorylated peptides were identified, suggesting that the modified Ser and Thr residues are genuine O-GlcNAcylation sites.
A peptide list of purified proteins and potential O-GlcNAc-modified sites is presented in Supplemental Table S6. Our results show that both Nictaba and WGA bind similar histone targets and that several of the potential O-GlcNAc-modified amino acids are present in both histone preparations retained on the lectin columns.
CONCLUSION
In the past decade, several plant lectins were discovered that reside in the nucleus and the cytoplasm of plant cells. Since the expression of most of these lectins is up-regulated by various (a)biotic stress factors, it was hypothesized that these lectins fulfill a role in cellular signaling events in response to stress conditions (Lannoo and Van Damme, 2010). Hitherto, the physiological role of none of these proteins was investigated. This study describes, to our knowledge for the first time, the identification of binding partners for a nucleocytoplasmic lectin from tobacco.
Using a combination of enzyme assays, western-blot analyses, and microscopical studies, it was shown that the preparation of tobacco nuclei was essentially pure. Nevertheless, the results from the MS analysis revealed small amounts of proteins that presumably locate to the plant cell wall. It is known that purification of nuclei is very cumbersome, and contamination of tobacco nuclei with debris corresponding to cell wall fragments has been reported before (Dahan et al., 2009). The extracellular compartment of plant cells contains many heavily glycosylated proteins (Knox, 1995; Jose-Estanyol and Puigdomenech, 2000; Léonard et al., 2002). Taking into account the carbohydrate-binding properties of Nictaba as determined by glycan array analyses (Lannoo et al., 2006b), it is not surprising that these proteins are retained on the lectin column. Therefore, a more rigid purification and fractionation of tobacco cells was performed. Our results show that enzymatic removal of the cell wall prior to purification of nuclei results in nuclear protein fractions with high purity.
Two independent approaches (lectin affinity chromatography and pull-down assays) identified histone proteins as the most abundant nuclear Nictaba-interacting proteins. Moreover, Nictaba also pulled down histone proteins from an unfractionated protoplast extract, strongly emphasizing the specificity of the interaction.
The affinity of Nictaba for histone H2B was confirmed by lectin affinity chromatography with a nonfractionated histone preparation from calf thymus. In addition, the interaction between Nictaba and histone H2B was proven to be GlcNAc dependent, since histone H2B was eluted with GlcNAc but not with other sugars. Furthermore, all these results were confirmed by a parallel experiment where histone proteins were chromatographed on a WGA column. Blotting experiments confirmed that lectin-bound histone proteins clearly interacted with the FITC-labeled lectins WGA and Nictaba but also with an anti-O-GlcNAc antibody. All these data suggest the presence of O-GlcNAc-modified proteins among the lectin-binding partners, as confirmed by MS analyses after mild β-elimination on lectin-bound histone proteins. Furthermore, it should also be noted that the binding of Nictaba to blotted plant nuclear proteins such as histone H2B was strongly inhibited by preincubating the lectin with GlcNAc oligomers.
Because histone proteins are one of the most abundant proteins in the cell, the possibility remains that the Nictaba-interacting histone proteins are the result of aspecific protein binding to the affinity matrix. However, this seems unlikely for several reasons. First, many other proteins that are abundantly present in a background extract are not found among the Nictaba-interacting proteins. When one compares the lists of Nictaba-interacting proteins and the proteins identified in the background extracts, the protein composition has clearly been changed, suggesting a specific enrichment of certain proteins by interaction with Nictaba. InterProScan scanning of Nictaba-interacting and background protein sequences also suggests the enrichment of nucleosomal proteins. Second, emPAI calculation demonstrates that histone proteins are by far the most abundant proteins among the Nictaba-interacting proteins, while they are not in the background extract. Third, calf histone proteins are specifically removed from a Nictaba-Sepharose 4B column by GlcNAc but not by other sugars. This latter observation suggests that bound proteins are removed from the column by competition with GlcNAc for the sugar-binding site. Finally, when a histone extract was chromatographed on the Sepharose 4B matrix, no proteins were bound. In the future, the interaction of Nictaba with histone proteins will have to be confirmed by other biochemical, genetic, and microscopical techniques. Negative control experiments with a Nictaba mutant protein affected in its carbohydrate-binding activity (Schouppe et al., 2010) could also be useful to prove the interaction with histone proteins.
Recently, it has been reported that O-GlcNAc can dynamically cycle on core histone proteins (H2A, H2B, H4) and therefore can be considered a part of the “histone code” (Sakabe et al., 2010). The O-GlcNAc modification is an abundant carbohydrate modification on many animal proteins involved in important cellular processes. Moreover, O-GlcNAc modification is dynamic and can change upon an induction stimulus like stress (Butkinaree et al., 2010). Furthermore, extensive cross talk between O-GlcNAcylation and phosphorylation has been reported and plays an important role in multiple signaling pathways for cellular regulation (Hu et al., 2010; Zeidan and Hart, 2010).
O-GlcNAc modification of plant proteins was reported by Heese-Peck et al. (1995). These authors also suggested that, in contrast to animal proteins that carry a single O-GlcNAc, nuclear plant proteins carry oligosaccharides with more than five GlcNAc residues. Unfortunately, until now the latter observation was not confirmed by other research groups. Although the presence of O-GlcNAc on plant proteins is evident, the function and importance of O-GlcNAc signaling in plants is largely undefined. At present, two different O-GlcNAc transferases, one of presumed prokaryotic origin and another of eukaryotic origin with changing expression levels in response to different conditions, have been identified unambiguously, indicating the presence of this modification on nucleocytoplasmic plant proteins (Olszewski et al., 2010). Recently, Xing et al. (2009) reported a jacalin-related lectin called VER2 in wheat (Triticum aestivum). VER2 exhibits specificity toward GlcNAc and Gal, and its expression is induced predominantly at potential nuclear structures in shoot tips and young leaves and weakly in cytoplasm in response to vernalization. It was shown that an O-GlcNAc-modified protein coimmunoprecipitated with VER2 in vernalized wheat plants but not in devernalized materials. Furthermore, evidence was provided to show that O-GlcNAc signaling and phosphorylation are involved in the vernalization response in wheat, indicating the involvement of O-GlcNAc protein modification in response to environmental stresses in plants.
At present, the tobacco lectin is one of the few nucleocytoplasmic plant lectins that have been studied in detail. It should be mentioned, however, that proteins homologous to the tobacco lectin can be retrieved from sequence databases of most higher plants (Lannoo et al., 2008), indicating that Nictaba homologs are widespread in Viridiplantae. In addition, the Nictaba domain was also identified as part of different types of chimeric proteins, suggesting that this domain served different purposes during evolution.
Our results suggest that Nictaba binds to several O-GlcNAcylated plant proteins in the nuclear compartment, in particular histone proteins. These results are in agreement with previous observations that showed the specific interaction of Nictaba with GlcNAc oligomers (Lannoo et al., 2006b). Future studies will concentrate on the ultrastructural colocalization of Nictaba and histone proteins in the plant cell and the functional implications resulting from this interaction. In particular, the interaction between Nictaba and histone proteins in relation to different plant stresses will be investigated. It is known that plants respond and adapt to both biotic and abiotic stresses in order to survive. Molecular and genomic studies have revealed the coordination of gene expression and chromatin regulation in response to environmental stresses. Furthermore, it was shown that several histone modifications are dramatically altered under stress conditions. In addition, chromatin-related proteins such as histone modification enzymes, histone proteins, and components of the chromatin-remodeling complex influence gene regulation as a result of stress responses (Chinnusamy and Zhu, 2009; Kim et al., 2010). In light of these findings, it is tempting to speculate that the interaction of Nictaba with histones reflects the ability of the lectin to alter chromatin conformation and folding and, as such, alter gene expression.
MATERIALS AND METHODS
Cell Cultures
Tobacco (Nicotiana tabacum ‘Xanthi’) cells were grown on a rotary shaker (150 rpm at 25°C) in 250-mL Erlenmeyer flasks in the dark. Cell suspensions were maintained in 100 mL of medium containing 3% saccharose, 0.44% Linsmaïer and Skoog salts (Duchefa), 0.02% Gln, 0.165 mg L−1 2,4-dichlorophenoxyacetic acid, 0.5 mg L−1 folic acid, 0.1 mg L−1 kinetin, 2 mg L−1 Gly, 2 mg L−1 biotin, 0.1 mg L−1 thiamine, 0.5 mg L−1 pyridoxin, 5 mg L−1 nicotinic acid, and 3 mg L−1 Ca2+ panthotenic acid, pH 5.5. Cells were subcultured weekly by transferring 10 mL of cell suspension into 90 mL of fresh medium.
Preparation of Nuclear Extract for Affinity Chromatography (Protocol 1)
A rather crude nuclear extract adapted from Dahan et al. (2009) was used for lectin affinity chromatography. The extract was prepared from cv Xanthi cells diluted with an equal volume of fresh medium on the 7th d of culture. On day 8, cells were harvested by filtration and frozen in liquid nitrogen. Frozen cells were ground with mortar and pestle to obtain a fine powder, which was solubilized in NB buffer (50 mm Tris-MES, pH 7.5, 2 mm orthovanadate, 20 mm, sodium fluoride, 100 mm β-glycerophosphate, 20 mm dithiothreitol, 10 mm EDTA, 10 mm EGTA, and 0.5% Triton X-100). Nuclei were filtered through a 31-μm mesh and collected by centrifugation (500g, 10 min) before loading on a 25% iodixanol layer (Sigma-Aldrich). After centrifugation for 30 min at 3,000g, nuclei were collected below the iodixanol layer and washed with NB buffer. Nuclei were stored at −80°C until use.
Preparation of Nuclei for Pull-Down Assay (Protocol 2)
Highly pure nuclei were prepared from protoplasts using a protocol adapted from Calikowski and Meier (2006) and Xiong et al. (2004). Protoplasts were prepared from 2- to 5-d-old cv Xanthi cells. Therefore, cells were collected by centrifugation (5 min, 500g) and plasmolyzed for 1 min in 0.4 m mannitol. Subsequently, plasmolyzed cells were incubated with enzyme buffer (42.5 mm MES, 10 mm CaCl2, 0.3 m mannitol, 2% cellulase RS [Duchefa], and 0.2% Pectolyase Y [Duchefa], pH 5.5; 100 mL of buffer for a 100-mL cell culture) for 2 h with gentle swirling. The protoplast suspension was filtered through a 100-μm cloth, and 5 mL was loaded on a 6% Ficoll-400 layer (Sigma-Aldrich) in a 15-mL glass tube. After centrifugation for 30 min at 3,000g, purified protoplasts were recovered from the interface between the water and the Ficoll layer. Afterward, protoplasts were washed twice with protoplast buffer (25 mm Tris, 25 mm MES, 0.3 m mannitol, and 25 mm CaCl2, pH 5.6).
Xanthi protoplasts were suspended in NIB buffer (0.5 m hexylene glycol, 20 mm KCl, 20 mm HEPES, 5 mm EDTA, and 0.5% Triton X-100, pH 7.4) and passed six times through a 0.45- × 12-mm needle (Terumo) to completely lyse the cells. This solution was then filtered through a 31-μm mesh to remove unlysed protoplasts. To further purify the nuclei, 2 mL of lysed protoplast solution was loaded on a 25%/36% iodixanol gradient (2 mL of each concentration, diluted in NIB buffer without Triton X-100, OptiPrep Density Gradient Medium [Sigma-Aldrich]) in a 15-mL glass tube. Gradients were centrifuged for 30 min at 3,000g, and colorless nuclei were isolated from the 25%/36% interface. Purified nuclei were washed twice with NIB buffer without Triton X-100 to remove iodixanol and solvent remnants.
Extraction of Nuclear Proteins
For each 1 mL of nuclear suspension obtained from cv Xanthi cells, 1 mL of NLB1 buffer (20 mm KCl, 20 mm HEPES, 5 mm EDTA, 1 m NaCl, and 1 m Mg2Cl2, pH 7.4) was added. This solution was mixed gently at 4°C and 20 rpm for 30 min using a Stuart rotator type SB3. After centrifugation for 30 min at 13,000g and 4°C, the supernatant containing the soluble nuclear proteins was concentrated using an Amicon Ultracel PL-10 centrifugal device (molecular mass cutoff of 10,000 D; Millipore).
Microscopy
Nucleus integrity was checked by means of differential interference phase contrast microscopy and fluorescence microscopy. For fluorescent detection of nuclei, dilutions of purified nuclei were stained with 10 ng mL−1 4′,6-diamidino-2-phenylindole (Invitrogen), 10 ng mL−1 propidium iodide (Invitrogen), and/or 100 nm ER-Tracker Blue-White DPX (Invitrogen) and pipetted onto glass-bottom dishes. Images were acquired through the appropriate filters with an automated Nikon TE2000E epifluorescence microscope (Nikon) equipped with a 40× Plan Fluor oil objective (numerical aperture 1.3) and a Nikon RGB camera.
Enzyme Assays
Cytochrome c reductase activity was assessed using the Cytochrome C Reductase Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Glc-6-P dehydrogenase activity was analyzed by mixing the nuclear protein with 650 μL of 100 mm triethanolamine/100 mm NaOH, 50 μL of 100 mm MgCl2, 50 μL of 35 mm Glc-6-P, and 25 μL of 11 mm NADP+ in a 1-mL cuvet and measuring the increase in A340 at room temperature. IDPase activity was assayed by mixing 90 μL of enzyme buffer [10 mm Mo7O24(NH4)6·4H2O, 0.1 m MgCl2, 2.5% Triton X-100, 25 mm Na2IDP, and 50 mm Tris-HCl] with 10 μL of nuclear protein at 37°C for several time intervals. The reaction was stopped and the increase of phosphate was visualized by adding Ames reagent containing 1.8% SDS and measuring the A820. ATPase activity was measured by mixing nuclear protein with 100 mm Tris-MES (pH 6.5) buffer containing 150 mm ATP, 1 m KCl, and 100 mm MgSO4 and incubation at 37°C for different time intervals. To inhibit the vacuolar and mitochondrial ATPases, 100 mm KNO3 and 10 mm NaN3 were added, respectively. The acid phosphatase was inhibited by the addition of 100 mm Na2MoO4 in the buffer. The enzyme reaction was stopped by the addition of Ames reagent (containing 1.8% SDS), and the increase of phosphate was monitored by measuring the A820.
Affinity Chromatography
Nictaba and WGA were coupled to Sepharose 4B using the divinylsulfone method (Pepper, 1994). Approximately 30 mg of a crude nuclear extract from cv Xanthi cells (obtained using protocol 1) was dialyzed against 50 mm Tris-MES (pH 7.5) containing 10 mm EDTA, 10 mm EGTA, and 200 mm NaCl and loaded on the Nictaba-Sepharose column (diameter 1 cm, height 2 cm) equilibrated with 50 mm Tris-MES, pH 7.5. The column was washed with 3 column volumes of the same buffer, and bound proteins were eluted with 20 mm 1,3-diaminopropane. Fractions were collected and analyzed by SDS-PAGE and MS.
A total histone type II preparation from calf thymus (Sigma-Aldrich) dissolved in phosphate-buffered saline (50 mm potassium phosphate, 150 mm NaCl, pH 7.2) was chromatographed on Nictaba- or WGA-Sepharose 4B. After washing with phosphate-buffered saline, the column was eluted with 20 mm 1,3-diaminopropane. Alternatively, 1 m GlcNAc was used to elute the column. Fractions were collected and analyzed by SDS-PAGE. Eluted polypeptides were subjected to N-terminal amino acid sequencing.
Pull-Down Assays
Pull-down assays were carried out with the His Protein Interaction Pull-Down Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Approximately 50 μg of recombinant His-tagged Nictaba protein (produced as described by Vandenborre et al. [2008]) was used as bait protein, while 150 and 300 μg of protein extracted from tobacco nuclei or protoplasts (obtained using protocol 2), respectively, were used as prey proteins.
β-Elimination of O-GlcNAc-Modified Proteins
Purified proteins (150 μg) were completely dried and redissolved in 500 μL of a 10% (v/v) butylamine (Sigma-Aldrich) solution in water. Next, β-elimination was carried out for 2 h at 50°C. Excess reagent was removed using a NAP-5 desalting column (Amersham Biosciences), whereby 500 μL was desalted in 1 mL of a 50 mm triethylbicarbonate solution (Sigma-Aldrich). Digestion of proteins and subsequent RP-HPLC separation of peptides are described below.
Preparation of Peptides for LC-MS/MS Analysis
Purified proteins were completely dried and redissolved in freshly prepared 50 mm ammonium bicarbonate buffer (pH 7.8). Prior to digestion, protein mixtures were boiled for 10 min at 95°C followed by cooling down on ice for 15 min. Sequencing-grade trypsin (Promega; Benelux) was added in a 1:100 (trypsin:substrate) ratio (w/w), and digestion was allowed overnight at 37°C. The sample was acidified with 10% acetic acid and loaded for RP-HPLC separation on a 2.1-mm i.d. × 150-mm 300SB-C18 column (Zorbax; Agilent Technologies) using the Agilent 1100 Series HPLC system. Following a 10-min wash with 10 mm ammonium acetate (pH 5.5) in water:acetonitrile (98:2, v/v), both Baker HPLC analyzed (Mallinckrodt Baker), a linear gradient to 10 mm ammonium acetate (pH 5.5) in water:acetonitrile (30:70, v/v) was applied over 100 min at a constant flow rate of 80 μL min−1. Eluting peptides were collected in 48 fractions between 20 and 80 min, and fractions separated by 15 min were pooled and vacuum dried until further analysis.
MS Analysis
Sample Analysis on the Esquire HCT Mass Spectrometer
The dried fractions were redissolved in 100 μL of 2.5% acetonitrile. Eight microliters of this peptide mixture was applied for nano-LC-MS/MS analysis on an Ultimate (Dionex) in-line connected to an Esquire HCT mass spectrometer (Bruker). The sample was first trapped on a trapping column (PepMap C18 column, 0.3 mm i.d. × 5 mm; Dionex). After back flushing from the trapping column, the sample was loaded on a 75-μm i.d. × 150-mm RP column (PepMap C18; Dionex). The peptides were eluted with a linear gradient of 3% solvent B (0.1% formic acid in water:acetonitrile [3:7, v/v]) increase per minute at a constant flow rate of 0.2 μL min−1. Using data-dependent acquisition, multiple charged ions with intensities above threshold (adjusted for each sequence according to the noise level) were selected for fragmentation. During MS/MS analysis, a MS/MS fragmentation amplitude of 0.7 V and a scan time of 40 ms were used.
Sample Analysis on the LTQ OrbiTRAP XL Mass Spectrometer
Dried fractions were redissolved in 100 μL of 2% acetonitrile, and 8 μL was used for LC-MS/MS analysis using an Ultimate 3000 HPLC system (Dionex) in-line connected to a LTQ OrbiTRAP XL mass spectrometer (Thermo Electron). Peptides were first trapped on a trapping column (PepMap C18 column, 0.3 mm i.d. × 5 mm [Dionex]), and following back flushing from the trapping column, the sample was loaded on a 75-μm i.d. × 150-mm RP column (PepMap C18; Dionex). Peptides were eluted with a linear gradient of 1.8% solvent B (0.05% formic acid in water:acetonitrile [2:8, v/v]) increase per minute at a constant flow rate of 300 nL min−1.
The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the six most abundant ion peaks per MS spectrum. Full-scan MS spectra were acquired at a target value of 1E6 with a resolution of 30,000. The six most intense ions were then isolated for fragmentation in the linear ion trap. In the LTQ, MS/MS scans were recorded in profile mode at a target value of 5,000. Peptides were fragmented after filling the ion trap with a maximum ion time of 10 ms and a maximum of 1E4 ion counts.
Protein Identification and Bioinformatics
Data Analysis (Esquire HCT Mass Spectrometer)
The fragmentation spectra were converted to mgf files using the Automation Engine software (version 3.2; Bruker) and were searched using the MASCOT database search engine (version 2.2.0; Matrix Science; http://www.matrixscience.com) against Swiss-Prot filtered for sequences from Viridiplantae. Peptide mass tolerance was set at 0.5 D and peptide fragment mass tolerance at 0.5 D, with the electrospray ionization ion trap as selected instrument for peptide fragmentation rules. Peptide charge is set to 1+, 2+, or 3+. Variable modifications were set to Met oxidation, pyro-Glu formation of N-terminal Gln, acetylation of the N terminus, deamidation of Gln or Asn. The enzyme is set on trypsin. Only peptides that were ranked 1 and scored above the threshold score set at 95% confidence were withheld.
Data Analysis (LTQ OrbiTRAP XL)
Data Analysis of Plant Proteins. MS/MS peak lists were searched with Mascot using the Mascot Daemon interface (version 2.2.0; Matrix Science). Spectra were searched against the Swiss-Prot database, and taxonomy was set to Viridiplantae. Variable modifications were set to Met oxidation, pyro-Glu formation of N-terminal Gln, acetylation of the protein’s N terminus, and deamidation of Gln and Asn. Mass tolerance of the precursor ions was set to ±10 ppm and of fragment ions to ±0.5 D. The peptide charge was set to 1+, 2+, or 3+, and one missed tryptic cleavage site was allowed. Also, Mascot’s C13 setting was to 1. Only peptides that were ranked 1 and scored above the identity threshold score set at 99% confidence were withheld.
Data Analysis of Histone Proteins. For the analysis of affinity-purified histone proteins, MS/MS peak lists were searched with Mascot using the Mascot Daemon interface (version 2.2.0; Matrix Science). Spectra were searched against the Swiss-Prot database, taxonomy was set to Mammalia, and enzyme was set to trypsin. Variable modifications were set to Met oxidation, pyro-Glu formation of N-terminal Gln, acetylation of the protein’s N terminus, and deamidation of Gln and Asn. For the non β-eliminated samples, we added phosphorylation of Ser and Thr as well as O-GlcNAc-modified Ser and Thr to the list of variable modifications. For the β-eliminated samples, we added butylamine-modified (+55 D) Ser and Thr to the list of variable modifications. No fixed parameters were used.
Mass tolerance of the precursor ions was set to ±10 ppm and of fragment ions to ±0.5 D. The peptide charge was set to 1+, 2+, or 3+, and one missed tryptic cleavage site was allowed. Also, Mascot’s C13 setting was to 1. Only peptides that were ranked 1 and scored above the identity threshold score set at 99% confidence were withheld.
emPAI Analysis
The emPAI index was calculated to estimate the abundance of the glycoproteins based on the number of identified tryptic peptides (Nesvizhskii et al., 2007; Vaudel et al., 2010). Protein sequences were searched for conserved functional profiles using InterProScan (Zdobnov and Apweiler, 2001; Ye et al., 2006) and plotted with the WEGO tool (Ye et al., 2006).
Analytical Methods
The total protein content was estimated using the Coomassie (Bradford) Protein Assay Kit (Thermo Fisher Scientific), based on the dye-binding procedure of Bradford (1976) or using the BCA Protein Assay Kit (Thermo Fisher Scientific). Proteins were analyzed by SDS-PAGE using 15% polyacrylamide gels under reducing conditions as described by Laemmli (1970). Proteins were visualized after staining with Coomassie Brilliant Blue R-250 or using the PageSilver Silver Staining Kit (Fermentas).
For western-blot analysis, samples separated by SDS-PAGE were electrotransferred to 0.45-μm polyvinylidene fluoride membranes (Biotrace PVDF; PALL, Gelman Laboratory). After blocking the membranes in Tris-buffered saline (TBS; 10 mm Tris, 150 mm NaCl, and 0.1% [v/v] Triton X-100, pH 7.6) containing 5% (w/v) BSA for 1 h at room temperature, blots were incubated for 1 h with the mouse CTD110.6 monoclonal antibody (Covance) directed against O-GlcNAc (diluted 1:1,000 in TBS buffer). The secondary antibody was a rabbit anti-mouse IgG labeled with horseradish peroxidase (Dako). This incubation was followed by a 1-h incubation with the peroxidase-antiperoxidase complex (Sigma-Aldrich). After several washings in TBS buffer, immunodetection was performed using a colorimetric assay with 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich) as a substrate (Graham and Karnovsky, 1966).
For lectin blots, membranes were incubated for 1 h with 1 μg mL−1 Nictaba, washed three times with TBS, and incubated for 1 h with a rabbit polyclonal antibody directed against Nictaba (diluted 1:80). The secondary antibody was a goat anti-rabbit IgG labeled with horseradish peroxidase (Sigma-Aldrich). This incubation was followed by three times with TBS and a 1-h incubation with the peroxidase-antiperoxidase complex (Sigma-Aldrich). Immunodetection was achieved by a colorimetric assay using 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich) as a substrate (Lannoo et al., 2006b). Inhibition of Nictaba binding was achieved by a 1-h preincubation of Nictaba with a mixture of GlcNAc oligomers (at a final concentration 10-fold higher than that required to completely inhibit the agglutination activity of a 100 μg mL−1 solution of Nictaba; Lannoo et al., 2006b). Alternatively, WGA and Nictaba were labeled with FITC using the EZ-Label FITC Protein Labeling Kit (Thermo Fisher Scientific). Blots were incubated with 1 μg mL−1 FITC-labeled lectin in the dark for 1 h, and bound proteins were visualized using the FLA-5100 imaging system (GE Healthcare). BSA-aminophenyl and BSA-aminophenyl-GlcNAc were gifts from Prof. N. Zachara.
N-terminal sequencing was carried out on affinity-purified protein fractions separated by SDS-PAGE and electroblotted on a ProBlot polyvinylidene difluoride membrane (Applied Biosystems). The membrane was stained with a 1:1 mix of Coomassie Brilliant Blue and ethanol, and the proteins of interest were excised from the blot. The N-terminal sequence was determined by Edman degradation performed on a model Procise 491cLC protein sequencer without alkylation of Cys residues (Applied Biosystems).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Fields of tobacco nuclei (purified according to protocol 2) stained with propidium iodide (A) and visualized with differential interference contrast (B); C shows an overlay of the two channels, and the inset in D shows a detailed view of the rectangular selection in C.
Supplemental Figure S2. SDS-PAGE analysis of nuclear proteins purified from cv Xanthi cells according to protocol 1 and retained on a Nictaba-Sepharose affinity column.
Supplemental Figure S3. Interaction of histone proteins with Nictaba-Sepharose 4B, WGA-Sepharose 4B, and Sepharose 4B.
Supplemental Table S1. Nictaba-interacting proteins identified in a cv Xanthi nuclear extract using lectin affinity chromatography.
Supplemental Table S2. Proteins identified in a cv Xanthi total nuclear extract.
Supplemental Table S3. Nictaba-interacting proteins identified in a tobacco nuclear extract using a pull-down assay.
Supplemental Table S4. Nictaba-interacting proteins identified in a tobacco protoplast extract using a pull-down assay.
Supplemental Table S5. Proteins identified in a cv Xanthi protoplast extract.
Supplemental Table S6. Nictaba- and WGA-interacting calf histone proteins and potential O-GlcNAc-modified sites.
Supplementary Material
Acknowledgments
We are grateful to Natasha Zachara (Department Biological Chemistry, John Hopkins University School of Medicine) for providing BSA-aminophenyl and BSA-aminophenyl-GlcNAc. We are indebted to Romina Termote-Verhalle and Kirsten Plas (Laboratory of Biochemistry and Glycobiology, Department of Molecular Biotechnology, Faculty of Bioscience Engineering, Ghent University) for excellent technical assistance.
References
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW. (2010) O-Linked β-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta 1800: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calikowski TT, Meier I. (2006) Isolation of nuclear proteins. Methods Mol Biol 323: 393–402 [DOI] [PubMed] [Google Scholar]
- Chen Y, Peumans WJ, Hause B, Bras J, Kumar M, Proost P, Barre A, Rougé P, Van Damme EJM. (2002) Jasmonic acid methyl ester induces the synthesis of a cytoplasmic/nuclear chito-oligosaccharide binding lectin in tobacco leaves. FASEB J 16: 905–907 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK. (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A. (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan J, Pichereaux C, Rossignol M, Blanc S, Wendehenne D, Pugin A, Bourque S. (2009) Activation of a nuclear-localized SIPK in tobacco cells challenged by cryptogein, an elicitor of plant defence reactions. Biochem J 418: 191–200 [DOI] [PubMed] [Google Scholar]
- Fouquaert E, Peumans WJ, Vandekerckhove TTM, Ongenaert M, Van Damme EJM. (2009) Proteins with an Euonymus lectin-like domain are ubiquitous in Embryophyta. BMC Plant Biol 9: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RC, Jr, Karnovsky MJ. (1966) The early stages of absorption of injected horseradish peroxidase in the proximal tubules of mouse kidney: ultrastructural cytochemistry by a new technique. J Histochem Cytochem 14: 291–302 [DOI] [PubMed] [Google Scholar]
- Heese-Peck A, Cole RN, Borkhsenious ON, Hart GW, Raikhel NV. (1995) Plant nuclear pore complex proteins are modified by novel oligosaccharides with terminal N-acetylglucosamine. Plant Cell 7: 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Shimoji S, Hart GW. (2010) Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett 584: 2526–2538 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Jose-Estanyol M, Puigdomenech P. (2000) Plant cell wall glycoproteins and their genes. Plant Physiol Biochem 38: 97–108 [Google Scholar]
- Kim JM, To TK, Nishioka T, Seki M. (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Knox JP. (1995) The extracellular matrix in higher plants. 4. Developmentally regulated proteoglycans and glycoproteins of the plant cell surface. FASEB J 9: 1004–1012 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lannoo N, Peumans WJ, Van Damme EJM. (2006a) The presence of jasmonate-inducible lectin genes in some but not all Nicotiana species explains a marked intragenus difference in plant responses to hormone treatment. J Exp Bot 57: 3145–3155 [DOI] [PubMed] [Google Scholar]
- Lannoo N, Peumans WJ, Pamel EV, Alvarez R, Xiong TC, Hause G, Mazars C, Van Damme EJM. (2006b) Localization and in vitro binding studies suggest that the cytoplasmic/nuclear tobacco lectin can interact in situ with high-mannose and complex N-glycans. FEBS Lett 580: 6329–6337 [DOI] [PubMed] [Google Scholar]
- Lannoo N, Peumans WJ, Van Damme EJM. (2008) Do F-box proteins with a C-terminal domain homologous with the tobacco lectin play a role in protein degradation in plants? Biochem Soc Trans 36: 843–847 [DOI] [PubMed] [Google Scholar]
- Lannoo N, Van Damme EJM. (2010) Nucleocytoplasmic plant lectins. Biochim Biophys Acta 1800: 190–201 [DOI] [PubMed] [Google Scholar]
- Lannoo N, Vandenborre G, Miersch O, Smagghe G, Wasternack C, Peumans WJ, Van Damme EJM. (2007) The jasmonate-induced expression of the Nicotiana tabacum leaf lectin. Plant Cell Physiol 48: 1207–1218 [DOI] [PubMed] [Google Scholar]
- Léonard R, Costa G, Darrambide E, Lhernould S, Fleurat-Lessard P, Carlué M, Gomord V, Faye L, Maftah A. (2002) The presence of Lewis a epitopes in Arabidopsis thaliana glycoconjugates depends on an active α4-fucosyltransferase gene. Glycobiology 12: 299–306 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Vitek O, Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods 4: 787–797 [DOI] [PubMed] [Google Scholar]
- Olszewski NE, West CM, Sassi SO, Hartweck LM. (2010) O-GlcNAc protein modification in plants: evolution and function. Biochim Biophys Acta 1800: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper DS. (1994) Some alternative coupling chemistries for affinity chromatography. Mol Biotechnol 2: 157–178 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJM. (1995) Lectins as plant defense proteins. Plant Physiol 109: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe K, Wang ZH, Hart GW. (2010) β-N-Acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA 107: 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouppe D, Rougé P, Lasanajak Y, Barre A, Smith DF, Proost P, Van Damme EJM. (2010) Mutational analysis of the carbohydrate binding activity of the tobacco lectin. Glycoconj J 27: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM. (2008) Plant lectins as part of the plant defence system against insects. Schaller A, , Induced Plant Resistance to Herbivory. Springer, Dordrecht, The Netherlands, pp 285–307 [Google Scholar]
- Van Damme EJM, Barre A, Rougé P, Peumans WJ. (2004) Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Peumans WJ, Barre A, Rougé P. (1998) Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit Rev Plant Sci 17: 575–692 [Google Scholar]
- Vandenborre G, Lannoo N, Smagghe G, Daniel E, Breite A, Soin T, Jacobsen L, Van Damme EJM. (2008) Cell-free expression and functionality analysis of the tobacco lectin. In Vitro Cell Dev Biol Anim 44: 228–235 [DOI] [PubMed] [Google Scholar]
- Vandenborre G, Miersch O, Hause B, Smagghe G, Wasternack C, Van Damme EJM. (2009) Spodoptera littoralis-induced lectin expression in tobacco. Plant Cell Physiol 50: 1142–1155 [DOI] [PubMed] [Google Scholar]
- Vaudel M, Sickmann A, Martens L. (2010) Peptide and protein quantification: a map of the minefield. Proteomics 10: 650–670 [DOI] [PubMed] [Google Scholar]
- Watson JC, Thompson WF. (1986) Purification and restriction endonuclease analysis of plant nuclear DNA. Methods Enzymol 118: 57–75 [Google Scholar]
- Xing LJ, Li J, Xu YY, Xu ZH, Chong K. (2009) Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS ONE 4: e4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong TC, Jauneau A, Ranjeva R, Mazars C. (2004) Isolated plant nuclei as mechanical and thermal sensors involved in calcium signalling. Plant J 40: 12–21 [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L.et al (2006) WEGO: a Web tool for plotting GO annotations. Nucleic Acids Res 34: W293– W297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. (2001) InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zeidan Q, Hart GW. (2010) The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J Cell Sci 123: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.