Abstract
Plants possess two myosin classes, VIII and XI. The myosins XI are implicated in organelle transport, filamentous actin organization, and cell and plant growth. Due to the large size of myosin gene families, knowledge of these molecular motors remains patchy. Using deep transcriptome sequencing and bioinformatics, we systematically investigated myosin genes in two model plants, Arabidopsis (Arabidopsis thaliana) and Brachypodium (Brachypodium distachyon). We improved myosin gene models and found that myosin genes undergo alternative splicing. We experimentally validated the gene models for Arabidopsis myosin XI-K, which plays the principal role in cell interior dynamics, as well as for its Brachypodium ortholog. We showed that the Arabidopsis gene dubbed HDK (for headless derivative of myosin XI-K), which emerged through a partial duplication of the XI-K gene, is developmentally regulated. A gene with similar architecture was also found in Brachypodium. Our analyses revealed two predominant patterns of myosin gene expression, namely pollen/stamen-specific and ubiquitous expression throughout the plant. We also found that several myosins XI can be rhythmically expressed. Phylogenetic reconstructions indicate that the last common ancestor of the angiosperms possessed two myosins VIII and five myosins XI, many of which underwent additional lineage-specific duplications.
One of the most prominent features of plant cell biology is extensive dynamics of the cell interior. This dynamics involves trafficking of organelles, including endoplasmic reticulum (ER), mitochondria, peroxisomes, Golgi stacks, and endomembrane vesicles, collectively called cytoplasmic streaming (Shimmen and Yokota, 2004). Using chemical inhibitors, it has been shown that the organelle trafficking relies primarily on the actomyosin motility system (Lee and Liu, 2004; Sparkes, 2010). Plant myosins are traditionally partitioned into three classes: algal class XIII myosins and two classes of flowering plant myosins, VIII and XI (Bezanilla et al., 2003; Foth et al., 2006). However, recent sequencing of the several complete genomes of the green algae, mosses, dicots, and monocots provided the necessary data for a much deeper insight into myosin evolution and classification (Avisar et al., 2008b). It was found that the flowering plants generally possess large families of myosin genes. For instance, Arabidopsis (Arabidopsis thaliana) has 13 class XI myosin motors compared with only two in the moss Physcomitrella patens (Reddy and Day, 2001; Vidali et al., 2010). The myosins XI are the fastest known processive motors (Tominaga et al., 2003; Shimmen and Yokota, 2004). However, the biological significance of the organelle trafficking and other myosin-dependent processes in plants remained poorly understood, in part because the pharmaceutical approach is unsuitable for identification of the functions of myosins in plant development.
Recent progress in plant myosin research (Sparkes, 2010) is due to the use of RNA interference and dominant negative inhibition approaches (Avisar et al., 2008a, 2008b, 2009; Sparkes et al., 2008; Natesan et al., 2009; Sattarzadeh et al., 2009) as well as the power of gene knockout technology (Ojangu et al., 2007; Peremyslov et al., 2008, 2010; Prokhnevsky et al., 2008; Ueda et al., 2010). The first genome-wide characterization of the myosins XI in Arabidopsis yielded a surprising outcome: none of the 13 myosin gene knockouts had a discernible developmental phenotype under optimal growth conditions (Peremyslov et al., 2008). However, a closer analysis of the mutant plants revealed reduced root hair growth in the plants with inactivated myosins XI-K and XI-2 (Ojangu et al., 2007; Peremyslov et al., 2008). These same myosins have been shown to contribute to the transport of Golgi stacks, peroxisomes, and mitochondria (Peremyslov et al., 2008). Taken together, these results suggested that the functions of myosins in plants are redundant and that the analysis of multiple knockouts is needed to obtain an adequate functional map of the entire set of plant myosins.
The following characterization of the double knockout mutants revealed that myosins XI-1 and XI-B also contribute to organelle transport and root hair growth, respectively. Furthermore, it was found that simultaneous inactivation of the pair of closely related myosin paralogs, XI-K and XI-1, caused a moderate reduction in plant stature (Prokhnevsky et al., 2008). The most recent work on the triple and quadruple myosin XI gene knockouts highlighted critical contributions of the highly expressed myosins XI-K, XI-1, XI-2, and XI-I to both diffuse and polarized cell expansion as well as to plant growth and development (Peremyslov et al., 2010). Inactivation of these myosins resulted in stunted plants, delayed flowering, and dramatic reductions in cell sizes, up to 10-fold in the case of root hairs. In addition, quadruple knockouts had virtually immobile organelles and exhibited cell type-specific changes in the architecture of F-actin bundles (Peremyslov et al., 2010). The myosins XI-K, XI-1, and XI-2 were also shown to be responsible for the bulk ER flow along thick F-actin bundles in the elongated cells of cotyledonal petioles (Ueda et al., 2010). Interestingly, a recent study of the two myosins XI in the moss P. patens revealed functions in polarized cell growth and F-actin organization that are analogous to those of Arabidopsis myosins XI (Vidali et al., 2010).
Parallel studies using dominant negative inhibition generally concurred with the conclusions of the gene knockout analyses and, in addition, implicated myosins XI-C, XI-E, and XI-I in the transport of Golgi stacks and mitochondria (Sparkes et al., 2008; Avisar et al., 2009). Both dominant negative inhibition and RNA interference were used to show that myosins XI contribute to the dynamics of the chloroplast extensions called stromules (Natesan et al., 2009; Sattarzadeh et al., 2009).
With the rapid growth of interest in plant myosin motors, it also became clear that this field would benefit from a more systematic, genome-wide approach. Because myosins are relatively large proteins, the gene maps based on existing bioinformatics predictions (e.g. The Arabidopsis Information Resource [TAIR] 8 gene models) are complex and not entirely reliable. This problem is further exacerbated by the rapidly growing number of plant genome sequences with different qualities and degrees of completeness. Although there are multiple genome-wide microarray databases of plant gene expression profiles, including those of myosins, these data are scattered and not always easily accessible.
In this work, we employed bioinformatics and data from deep transcriptome sequencing (RNA-seq) of Arabidopsis and the model grass Brachypodium distachyon (referred to as Brachypodium in the rest of this paper) to investigate the genome-wide patterns of splicing, expression, and phylogeny of the plant myosins. We amend myosin gene maps and provide the data in an easy-to-use format. We combined RNA-seq data with our and other publicly available microarray analyses to deduce the tissue-specific and temporal patterns of myosin gene expression. A novel, myosin XI-K-derived gene of Arabidopsis that is expressed preferentially in the emerging vascular tissue is described. We also constructed a detailed phylogenetic tree of plant myosins that sheds new light on myosin evolution and classification. Our analysis provides a broad framework for future experimental designs aimed at the functional genomics of plant myosins and mechanisms through which these molecular motors contribute to cell growth and plant development.
RESULTS AND DISCUSSION
Myosin Evolution from Algae to Mosses to Angiosperms
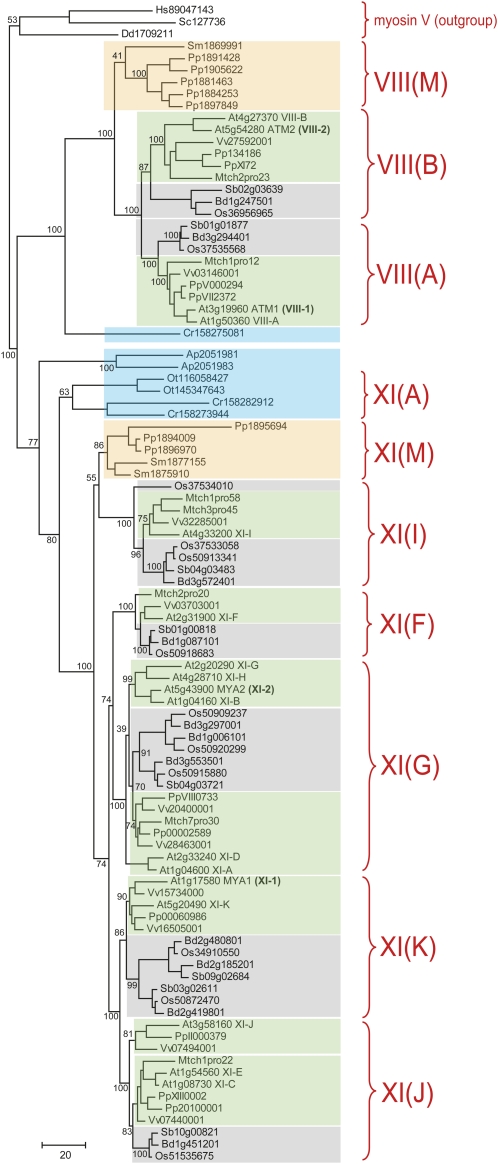
With several new plant genomes available (e.g. the lycopsid Selaginella moellendorffii, the moss P. patens, the dicot Medicago truncatula, and the monocots Brachypodium and Sorghum bicolor), we are now in a position to validate the classification of myosins proposed previously (Bezanilla et al., 2003; Avisar et al., 2008b) and potentially gain new insights into the evolution of myosins among diverse land plants and algae. A maximum-likelihood tree of class VIII, XI, and XIII myosins was constructed using three sequences from myosin class V as an outgroup (Fig. 1). The topology of the resulting tree shows that the ancestors of land plants, the green algae, possess both class VIII and class XI myosins (blue boxes in Fig. 1). Previously, the two algal myosins from Acetabularia peniculus that cluster with class XI myosins were classified as a separate class XIII (Foth et al., 2006), but our phylogenetic analysis shows that there is no need for this additional class. Together with other previously unclassified algal myosins, the A. peniculus myosins form evolutionary lineages that clearly fall into class XI (Fig. 1).
Figure 1.
Phylogenetic tree of the motor domains of plant myosins. Each terminal node of the tree is labeled by the two-letter abbreviation of the corresponding species name and the unique identifier. For Arabidopsis, the myosin identifiers proposed in this work are also indicated. The myosin clusters are highlighted according to corresponding plant taxa as follows: algae in blue, mosses in light brown, dicots in green, and monocots in gray. Abbreviations are as follows: Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Dd, Dictyostelium discoideum; Ap, Acetabularia peniculus; Cr, Chlamydomonas reinhardtii; Ot, Ostreococcus tauri; Pp, Physcomitrella patens, Sm, Selaginella moellendorffii; At, Arabidopsis thaliana; Mt, Medicago truncatula; Pt, Populus trichocarpa; Vv, Vitis vinifera; Sb, Sorghum bicolor; Bd, Brachypodium distachyon; Os, Oryza sativa. For selected major branches, bootstrap support values (percentage) are shown. Braces at the right designate corresponding subfamilies of the class VIII and XI myosins.
Mosses (P. patens) and lycopsids (S. moellendorffii) possess the same two classes of myosins, which, as expected, are closer to the flowering plant clade than to the corresponding algal myosins (brown boxes in Fig. 1). Counterintuitively, moss myosins XI in the group XI (M) cluster with the flowering plant group XI (I) rather than outside the flowering plant clade. Because this branch is weakly supported by bootstrap analysis, it likely represents a long-branch attraction artifact that could be corrected when more moss myosin sequences become available. Both P. patens and S. moellendorffii also exhibit conspicuous lineage-specific expansions of paralogous myosin VIII gene families, and the functional implications of these expansions remain to be experimentally addressed.
Importantly, with the analysis of additional available genomes, we confirmed all the groups detected previously for angiosperms, namely, groups A and B for class VIII and groups I, F, G, K, and J for class XI (Fig. 1; Avisar et al., 2008b). A few cases where representatives of one or another group were missing in certain genomes could be attributed to the state of sequence assembly and the annotation state of the corresponding genomes (we did not attempt to assemble myosin genes from unannotated genome sequences). For instance, apparent orthologs of Arabidopsis myosins XI-I and XI-F are present in Populus trichocarpa but lack the motor domain that was used for tree reconstruction.
Taken together, the results of the phylogenetic analysis of plant myosins indicate that the origin of the flowering plants was associated with a burst of duplication in class XI and a single duplication in class VIII; most likely, these duplications were followed by subfunctionalization of the paralogous myosins. Subsequent, additional functional specialization in different groups of plants probably resulted from lineage-specific duplications. More detailed classification based on independent duplication events within dicot and monot branches is starting to emerge; for example, there are three paralogous clades within the group XI (K) myosins in monocots compared with only two in dicots. Only the myosins in group XI (F) have not been duplicated in any of the plant lineages analyzed so far.
By and large, the available experimental data confirm the proposed evolutionary scenario for plant myosins. A gradual subfunctionalization with a degree of redundancy is particularly clear within the recently characterized pairs of closely related paralogs that emerged via relatively recent gene duplications, namely, myosins XI-K/XI-1 and XI-2/XI-B. The former pair shares functions in organelle transport, whereas only myosin XI-K is also required for polarized root hair growth. In the latter pair, both myosins are required for root hair growth, but only XI-2 is involved in organelle transport (Prokhnevsky et al., 2008; Peremyslov et al., 2010). On the other hand, specific functions of the myosins within groups XI (I), XI (F), and XI (J) remain unknown; inactivation of these myosins via insertional T-DNA mutagenesis produced no discernible phenotypes in the vegetative Arabidopsis plants (Peremyslov et al., 2008). The case of the entire myosin class VIII is even more striking: the functions of these myosins remain obscure. Future sequencing of the fern and gymnosperm genomes will further refine the evolutionary scenario presented here, but dramatic changes seem unlikely.
Myosin Nomenclature
So far, there is no universally accepted classification of the plant myosins. The originally described Arabidopsis myosin genes designated MYA1/MYA2 and ATM1/ATM2 (Knight and Kendrick-Jones, 1993; Kinkema et al., 1994) belong to classes XI and VIII, respectively. The genome-wide description of Arabidopsis myosins retained these designations and named additional myosins as XIA to XIK and VIIIA/B (Reddy and Day, 2001). Here and in our previous publications, we adopted a nomenclature that attempts to be more systematic and phylogenetically relevant than the currently used designations. We preserved the designations proposed by Reddy and Day (2001) but added a hyphen to separate the Roman numeral from the following capital letter (i.e. XI-A to XI-K). We also renamed MYA1 and MYA2 as XI-1 and XI-2 and renamed ATM1 and ATM2 as VIII-1 and VIII-2. In accord with the existence of five phylogenetic lineages of myosins XI and two lineages of myosins VIII in higher plants (Fig. 1), we subdivide Arabidopsis myosins into seven groups (Table I).
Table I. Nomenclature of the Arabidopsis class XI and class VIII myosins.
| Evolutionary Group | Gene Identifier | Proposed Name | Other Name |
| XI(F) | AT2G31900 | XI-F | XIF |
| XI(G) | AT5G43900 | XI-2 | MYA2 |
| AT1G04160 | XI-B | XIB | |
| AT2G20290 | XI-G | XIG | |
| AT4G28710 | XI-H | XIH | |
| AT1G04600 | XI-A | XIA | |
| AT2G33240 | XI-D | XID | |
| XI(I) | AT4G33200 | XI-I | XII |
| XI(J) | AT1G08730 | XI-C | XIC |
| AT1G54560 | XI-E | XIE | |
| AT3G58160 | XI-J | XIJ | |
| XI(K) | AT5G20490 | XI-K | XIK |
| AT1G17580 | XI-1 | MYA1 | |
| VIII(A) | AT3G19960 | VIII-1 | ATM1 |
| AT1G50360 | VIII-A | VIIIA | |
| VIII(B) | AT5G54280 | VIII-2 | ATM2 |
| AT4G27370 | VIII-B | VIIIB |
A broader adoption of this nomenclature poses formidable challenges, primarily due to the variability in the numbers and phylogenetic patterns of the myosin genes among the plant species (Fig. 1). However, we propose that the myosins of flowering plant species other than Arabidopsis also are assigned to the seven groups shown in Figure 1. Such an assignment will provide phylogenetic reference and will be useful to guide experimental analysis. Here, we adopt this approach for the Brachypodium myosins.
Gene Models and Alternative Splicing of the Myosin Genes in Arabidopsis and Brachypodium
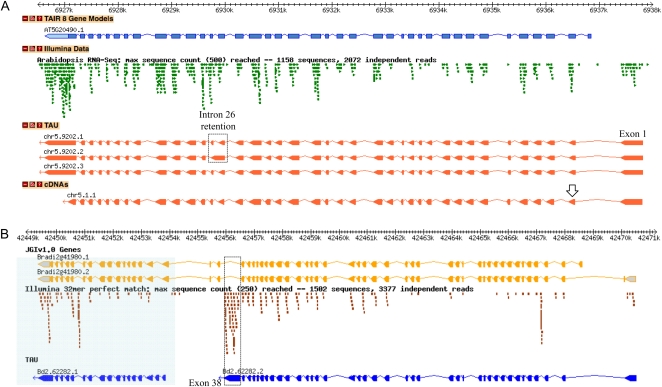
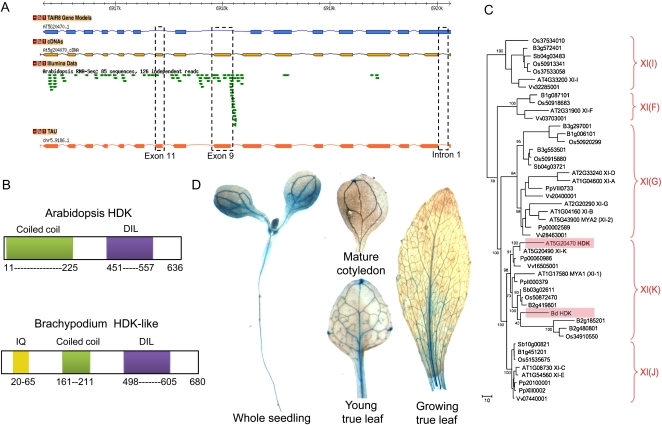
Due to the large sizes of myosin XI genes and the complexity of myosin mRNA splicing patterns, currently available myosin gene models (e.g. at TAIR [http://www.arabidopsis.org/]) are not entirely reliable, as pointed out previously (Ojangu et al., 2007). To refine and optimize the myosin gene models for the model dicot plant Arabidopsis and the model monocot plant Brachypodium, transcript evidence generated using ultra-high-throughput transcriptome sequencing (Fox et al., 2009) was coupled with all publicly available myosin transcript data to generate revised gene models using a reference-guided assembly approach. Briefly, public Illumina RNA-seq transcriptome data and public EST data downloaded from GenBank were combined with existing gene predictions using the Transcription-unit Assembly Utility (TAU) algorithm (Filichkin et al., 2010; http://mocklerlab-tools.cgrb.oregonstate.edu/). Based on this approach, we revised the gene models for 17 Arabidopsis and 11 Brachypodium class XI and class VIII myosins. Figure 2 shows the examples of reannotated gene models for the genes encoding Arabidopsis myosin XI-K (Fig. 2A; AT5G20490) and its apparent Brachypodium ortholog (Fig. 2B; Bradi2g41980). Myosin XI-K attracted much attention thanks to its prominent roles in organelle and ER transport, F-actin organization, as well as cell and organ growth in Arabidopsis (Peremyslov et al., 2010; Ueda et al., 2010). It will be interesting to determine if similar roles are played by the ortholog of this myosin in Brachypodium (Bradi2g41980).
Figure 2.
Transcript evidence and gene models for Arabidopsis and Brachypodium myosins. Gbrowse genome viewer screenshots depicting Arabidopsis and Brachypodium myosin genes, supporting transcript evidence, and inferred gene models are shown. A, Arabidopsis XI-K (AT5G20490). A box and an arrow mark alternative splicing events. B, Brachypodium XI-K ortholog (Bradi2g41980). A box denotes novel exon 38; the area corresponding to a separate transcript, Bd2.62282.1, is shaded. The TAIR 8 Gene Models track represents the annotated Arabidopsis gene models (http://www.arabidopsis.org). The JGIv1.0 Genes track shows the annotated Brachypodium gene models provided by the Brachypodium Genome Initiative (2010; http://www.brachypodium.org). The cDNAs track represents the transcript structures supported by cloned cDNAs. The Illumina Data track represents the raw Illumina RNA-seq transcriptome data aligned to the genome. The TAU track shows the gene models assembled from available empirical transcript data using the TAU algorithm.
In the case of the Arabidopsis XI-K gene, the Illumina RNA-seq transcript evidence (Filichkin et al., 2010) supported inclusion of a novel first exon that was not previously annotated and indicated that the annotated first exon was incorrect (Fig. 2A). In addition, the RNA-seq data revealed a novel intron retention event in annotated intron 26 (boxed in Fig. 2A) and an alternative acceptor site that changes the start position for the second exon (arrow in Fig. 2A). These data were combined to generate one preferentially expressed (chr5.9202.1) and two new alternatively spliced (chr5.9202.2 and chr5.9202.3) gene models for the Arabidopsis XI-K gene, as shown in the TAU track of Figure 2A.
To further evaluate the revised, TAU-derived XI-K gene models, we isolated RNA from the Arabidopsis rosette leaves, reverse transcribed, PCR amplified, and sequenced the entire coding region of the XI-K mRNA. The resulting consensus sequence fully corresponded to the TAU model chr5.9202.1. Furthermore, we performed 5′ RNA ligase-mediated (RLM)-RACE to determine the correct transcription initiation site(s) and to sequence the adjacent cDNA region. The resulting PCR products were cloned, and 12 independent clones were sequenced. Eleven seemingly random transcription initiation sites between nucleotides 6,937,789 and 6,936,808 on chromosome 5 were identified, with the first cDNA nucleotide being a purine in all clones (eight clones started with A and four started with G). All 12 clones showed identical nucleotide sequence downstream from nucleotide 6,936,808 that corresponded closely to the 5′ terminus of the first exon in TAU models. A putative translation initiation AUG codon of the XI-K coding sequence was found at nucleotide 6,937,379, followed by an 834-nucleotide-long intron and 129-nucleotide-long exon that had been considered part of an intron according to the TAIR gene model. Therefore, direct sequencing of the XI-K cDNA has fully confirmed the TAU model chr5.9202.1 (Fig. 2A, compare tracks cDNAs and TAU) and revealed that the myosin XI-K mRNA population possesses 5′-untranslatable regions of variable length exceeding 570 nucleotides. It seems reasonable to suggest that such an unusually long untranslatable region plays a role in the regulation of myosin XI-K expression.
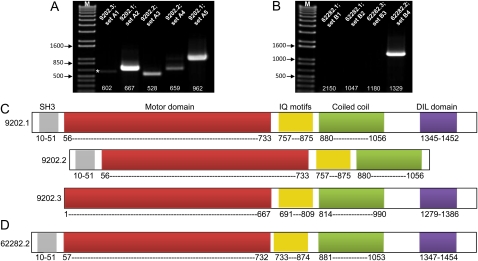
To determine if the alternatively spliced variants chr5.9202.2 and chr5.9202.3 are indeed expressed in Arabidopsis, we interrogated the corresponding areas of the conceptually transcribed mRNA variants using primer sets specific to each of the variants. The results of reverse transcription (RT)-PCR analysis supported occurrence of the intron 26 retention in the chr5.9202.2 variant as well as of an alternative acceptor site in the chr5.9202.3 variant (Fig. 3A). Interestingly, the former splicing event was present in substantial quantities, whereas the latter was barely detectable (asterisk in Fig. 3A). As expected, the “canonical” splicing variant chr5.9202.1 was expressed predominantly (Fig. 3A).
Figure 3.
Splicing variants of the Arabidopsis myosin XI-K gene transcripts and of the Brachypodium XI-K ortholog: experimental validation (A and B) and protein domain structures (C and D). A, Gel analysis of the RT-PCR products validates splicing isoforms of XI-K transcript in Arabidopsis. M, DNA size markers; A1 to A5, primer sets used to interrogate each splicing site (Supplemental Table S1); cDNA, the major isoform identified by cDNA sequencing; 92021.1 and 92021.2, isoforms corresponding to TAU models in Figure 2A. The asterisk marks a faint band corresponding to isoform 9202.1. The PCR product sizes are shown at the bottom. B, Similar analysis validates the splicing isoform Bd2.6228.2 of the XI-K ortholog (Bradi2g41980) in Brachypodium. The isoforms shown in Figure 2B were interrogated using primer sets B1 to B4 (Supplemental Table S1). C and D, Domain structures of the protein isoforms corresponding to validated splicing variants for Arabidopsis XI-K (C) and its Brachypodium ortholog (D). The domain borders are shown at the bottom.
The predicted domain architectures of the three variants of Arabidopsis myosin XI-K are shown in Figure 3C. Of these, the dominant variant chr5.9202.1 possesses all domains typically found in class XI and V myosins: the N-terminal Src homology 3 (SH3) domain and a motor/head domain as well as the C-terminal tail region including IQ motifs, a coiled-coil domain, and a DIL domain. The alternatively spliced chr5.9202.2 and chr5.9202.3 transcripts encode proteins that lack the DIL and SH3 domains, respectively (Fig. 3C).
We conclude that the previously accepted gene model for XI-K was incorrect, thus affecting experiments aimed at cloning, expressing, and tagging of the XI-K gene or cDNA. In particular, the 5′-terminal sequence of the XI-K cDNA reported by Ojangu et al. (2007) does not correspond to our validated model; apparently, it represents an additional minor transcription variant. Although we did not detect this variant among the 12 sequenced RLM-RACE clones (see above), we were able to amplify it at very low efficiency using oligonucleotide primers designed by the authors (data not shown).
A second example of reannotation illustrates the use of empirical transcript data to revise the gene models for the Brachypodium myosin XI from the XI (K) group encoded by the Bradi2g41980 gene (Figs. 1 and 2B). Conceptual translation of the mRNAs corresponding to two existing gene models (Fig. 2B, track JGIv1.0) yielded unusually large proteins with duplication of the tail region unseen in any previously characterized myosins XI or V (data not shown). The public Illumina RNA-seq data (Brachypodium Genome Initiative, 2010) confidently refuted the Joint Genome Institute model Bradi2g41980.1 and supported two additional gene models (Fig. 2B, track TAU; data not shown). As shown in the Figure 2B Illumina RNA-seq track, a large number of RNA-seq reads aligned within the annotated intron 38, consistent with an alternative 3′-terminal exon 38 and 3′-bias expected for RNA-seq data derived from oligo(dT)-primed RT products (boxed in Fig. 2B).
Each of the initial TAU models was interrogated using RT-PCR and four variant-specific primer sets. The results shown in Figure 3B unequivocally supported only one of these models (Bd2.62282.2 in the TAU track of Fig. 2B). As could be expected, conceptual translation of the Bd2.62282.2 transcript yielded a full-size myosin XI possessing all domains also found in its apparent Arabidopsis ortholog XI-K (Fig. 3D). Strikingly, it was demonstrated that the transcript encoding an additional tail region (shaded in Fig. 2B) was indeed expressed in Brachypodium, but as a separate mRNA (Bd2.62282.1 in the TAU track of Fig. 2B; see also Fig. 3B).
Thus, combined empirical transcript evidence enabled modification of the myosin gene models in Arabidopsis and Brachypodium, including the identification of new alternative splicing variants. The next challenge is to investigate the functional relevance of the predicted myosin variants and to determine if, similar to mammalian myosins V (Wu et al., 2002; Roland et al., 2009), alternative splicing is involved in functional regulation of the plant myosins XI.
We developed the complete, genome-wide sets of the amended gene models for all Arabidopsis and Brachypodium myosins XI and VIII to facilitate further experimental inquiry into functional genomics of the plant myosins. The detailed information on these models, including amino acid sequences of myosins corresponding to each splicing variant, can be found at http://myosins.cgrb.oregonstate.edu.
Patterns of Myosin Gene Expression
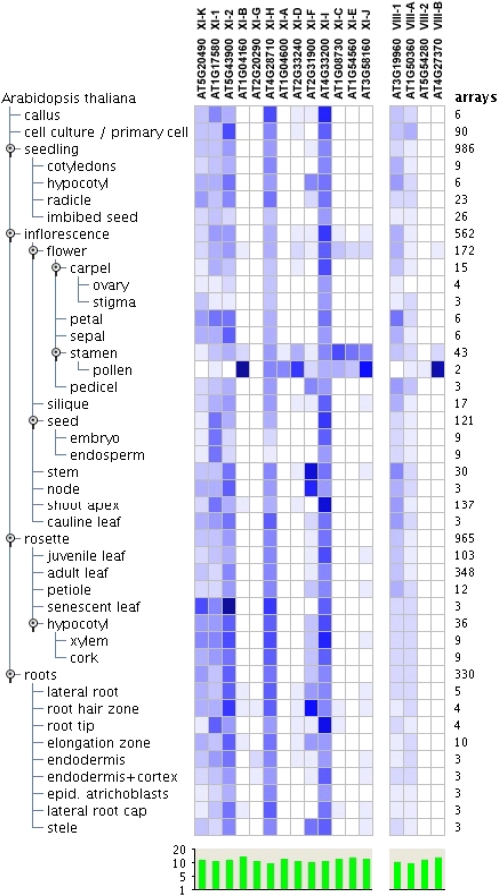
To investigate the patterns of myosin gene expression in Arabidopsis, we mined public Affymetrix DNA microarray data using the Genevestigator suite of tools (https://www.genevestigator.com; Hruz et al., 2008). The resulting expression heat map for the individual anatomical categories and distinct developmental stages that combined all available data is shown in Figure 4. This heat map revealed that six myosin XI genes, XI-K, XI-1, XI-2, XI-H, XI-I, and XI-F, are transcribed at varying but substantial levels in distinct organs and tissues. The expression pattern of the XI-G gene was difficult to evaluate due to low levels of the microarray signals (Fig. 4). The remaining six myosin XI genes, XI-A, XI-B, XI-C, XI-D, XI-E, and XI-J, exhibited particularly high expression levels in pollen and stamen (Fig. 4), suggesting a degree of tissue specialization and involvement in male reproductive function.
Figure 4.
Development- and anatomy-specific expression profiles of Arabidopsis myosins. Public Affymetrix DNA microarray data sets for Arabidopsis were mined to generate an expression heat map using the Genevestigator (https://www.genevestigator.com) Meta-Profile Analysis tool with default settings. Myosin genes are organized according to their class, and blue indicates that a gene is expressed specifically in a particular developmental stage or organ.
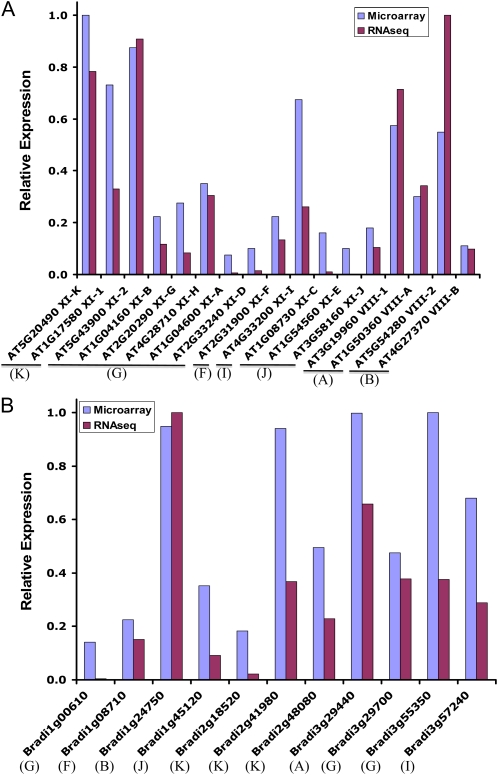
We also investigated global relative expression levels of Arabidopsis and Brachypodium myosins in whole plants. Initially, we compared the estimated average expression levels of Arabidopsis myosins measured by either Affymetrix DNA microarrays (https://www.genevestigator.com) or Illumina RNA-seq transcriptome analysis (Filichkin et al., 2010). In general, there was a substantial correlation between the expression levels estimated using these different technologies (Fig. 5A). However, whole plant comparisons also gave an additional insight into myosin expression patterns with potential functional relevance.
Figure 5.
Relative expression levels of Arabidopsis and Brachypodium myosins in whole plants. A, The microarray expression values for Arabidopsis seedlings were estimated using the Genevestigator Meta-Profile Analysis tool. The RNA-seq values were estimated from published data (Filichkin et al., 2010), using the RPKM (reads per kilobase transcript per million mapped reads) approach. B, The microarray expression values for Brachypodium seedlings grown in light-dark cycles and sampled over a diurnal time course were estimated from Brachypodium Affymetrix data (S.E. Fox and T.C. Mockler, unpublished data). The RNA-seq values were estimated from published RNA-seq data (Brachypodium Genome Initiative, 2010) using the RPKM approach.
According to their whole plant expression levels, Arabidopsis myosin XI genes could be roughly separated into three categories. The first category includes five genes (XI-K, XI-1, XI-2, XI-I, and XI-H) with highly abundant transcripts (Fig. 5A), suggesting broad roles in plant development and physiology. Indeed, data available for four of these myosins showed that they collectively provide important, albeit redundant, contributions to intracellular dynamics, cell expansion, and aerial organ growth (Peremyslov et al., 2010). The functions of the highly expressed myosin XI-H remain to be determined.
The second category includes four genes (XI-F, XI-G, XI-B, and XI-J) transcribed at intermediate levels (Fig. 5A), which could be interpreted as an indication of less central roles, perhaps limited to specialized cell types. The functions of the XI-F, XI-G, and XI-J myosin genes remain uncharacterized, whereas XI-B has been shown to provide important contributions to root hair elongation (Prokhnevsky et al., 2008; Peremyslov et al., 2010). On the other hand, the XI-B and XI-J genes are preferentially expressed in pollen (Fig. 4), implying a role in pollen tube growth. A potential dual function of XI-B (and, perhaps, also XI-J) is in line with the prominent mechanistic parallels in the tip growth of the root hairs and pollen tubes (Cole and Fowler, 2006; Lee and Yang, 2008). Comparative investigation of the myosin XI-B and XI-E functions in these cell types is a promising area for future research into the mechanisms of polarized cell elongation in plants.
The third category of Arabidopsis myosin XI genes includes XI-A, XI-D, XI-C, and XI-E, for which the transcription levels at the entire plant level are extremely low (Fig. 5A). Given that each of these genes is expressed almost exclusively in stamen and pollen (Fig. 4), specialized function in pollen formation and/or growth seems very likely. Indeed, preliminary experiments with the mutants in which genes XI-C and/or XI-E were inactivated suggested that these myosins are required for efficient pollen tube growth (R. Cole, J. Fowler, V.V. Peremyslov, and V.V. Dolja, unpublished data).
Among four myosin VIII genes in Arabidopsis, VIII-1, VIII-A, and VIII-2 are transcribed to relatively high levels, whereas VIII-B transcription is relatively low (Fig. 5A). Similar to myosins XI, it can be suggested that the former three myosins VIII function throughout the plant, whereas VIII-B is more specialized, in accord with its preferential expression in pollen (Fig. 4). It should be mentioned that the genes encoding myosins XI-G and VIII-2 show barely detectable expression in organ-specific Affymetrix assays (Fig. 4) but substantial expression levels in whole plants, as supported by both Affymetrix and RNA-seq data (Fig. 5A). This apparent discrepancy appears to be an artifact due to a relatively low sensitivity of the Affymetrix assays done with isolated tissues, a nearly uniform accumulation of corresponding transcripts throughout plants, or both.
A global expression analysis was also performed for Brachypodium by comparing Affymetrix DNA microarray data for whole seedlings (S.E. Fox and T.C. Mockler, unpublished data) with public Illumina RNA-seq data (Brachypodium Genome Initiative, 2010). Similar to Arabidopsis, the relative expression estimates made using the two platforms were well correlated (Fig. 5). Moreover, separation of the Brachypodium myosin genes into categories expressed to high, intermediate, and low levels revealed striking correspondence to the pattern seen in Arabidopsis. Indeed, Brachypodium possessed five highly expressed myosin genes XI, with two from groups XI (K) and XI (G) each and one of group XI (I) (Bradi2g41980, Bradi2g48080, Bradi3g55350, Bradi3g29700, and Bradi3g57240, respectively), exactly mimicking the distribution of five highly expressed Arabidopsis myosin XI genes (Fig. 5). This result suggests conservation of the high expression levels and, perhaps, broad functions in cell dynamics and plant growth among myosins from the evolutionarily related myosin XI groups (Fig. 1) in a monocot and dicot plants.
The two Brachypodium myosin genes XI transcribed to intermediate levels (Bradi1g08710 and Bradi1g45120; Fig. 4B) belonged to two out of three groups of such myosins, XI (F) and XI (J), in Arabidopsis. Finally, two Brachypodium myosin XI genes with low transcript abundance were Bradi1g00610 and Bradi2g18520 from groups XI (G) and XI (K) (Fig. 5B). By analogy to Arabidopsis, it could be expected that these latter genes and probably the group XI (J) gene from Brachypodium are pollen specific. It will be interesting to see if future analyses confirm the existence of the three pollen-specific myosins in Brachypodium compared with six such genes in Arabidopsis.
Diurnal Regulation of the Myosin Gene Expression
To determine whether the expression of plant myosins is regulated by light and temperature, we investigated the accumulation of myosin transcripts under different diurnal conditions. This has been done with all 17 Arabidopsis myosin genes and their apparent orthologs from rice (Oryza sativa subspecies japonica; a monocot); analogous data from Brachypodium are not yet available. The analysis of mRNA profiles using whole genome expression microarrays showed that the transcripts of a majority of myosin genes in both Arabidopsis and rice accumulated in a cyclical fashion under the diurnal conditions, suggesting that their expression can be driven by photocycles and/or thermocycles (Table II).
Table II. Cyclical accumulation of the myosin gene transcripts under diurnal conditions in Arabidopsis and rice.
N/A, Not applicable; this probe set is not presented at the DIURNAL portal database; N/D, a cyclical transcript expression is not detected (i.e. a correlation coefficient r between the data and the model is below an arbitrarily selected cutoff value of 0.8 or greater). Asterisks denote putative orthologs of the respective Arabidopsis myosin as determined by the best mutual BLAST match hit. Other rice gene identifiers represent putative homologs of Arabidopsis as predicted by the best BLAST hit. Both orthologs (shown in boldface) and homologs were determined by using the Orthomap tool (http://orthomap.cgrb.oregonstate.edu). Percentage of similarity with the respective Arabidopsis protein is shown in parentheses.
| Arabidopsis | Rice | ||
| Locus Identifier | Diurnal Profilea | Locus Identifier | Diurnal Profilea |
| At5g20490; XI-K | SD; LD; LDHC; LDHH | Os03g48140* (88%) | LDHC; LDHH |
| At1g17580; XI-1 | SD; LDHH; LLHC | Os03g48140 (84%) | LDHC; LDHH |
| At5g43900; XI-2 | LD | Os02g34080 (79%) | LLHH_LDHH |
| At1g04160; XI-B | LL-LDHC | Os02g34080* (79%) | LLHH_LDHH |
| At2g20290; XI-G | LLHC; LDHH | Os02g34080 (75%) | LLHH_LDHH |
| At4g28710; XI-H | LD; LL_LDHC | Os02g34080 (85%) | LLHH_LDHH |
| At1g04600; XI-A | LD; LLHC | Os02g34080 (57%) | LLHH_LDHH |
| At2g33240; XI-D | N/D | Os02g34080 (62%) | LLHH_LDHH |
| At2g31900; XI-F | LD; LLHC | Os03g53660* (83%) | LDHC; LDHH |
| At4g33200; XI-I | LDHC | Os03g48140 (72%) | LDHC; LDHH |
| At1g08730; XI-C | N/D | Os06g29350 (90%) | LLHH_LDHH |
| At1g54560; XI-E | LDHC | Os06g29350* (90%) | LLHH_LDHH |
| At3g58160; XI-J | LLHC | Os06g29350 (78%) | LLHH_LDHH |
| At3g19960; VIII-1 | LLHC; LDHC; LDHH | Os10g34710* (86%) | LDHC; LDHH |
| At1g50360; VIII-A | N/A | Os10g34710 (85%) | LDHC; LDHH |
| At5g54280; VIII-2 | LDHC | Os07g37560* (73%) | LDHC; LDHH |
| At4g27370; VIII-B | LL_LDHC | Os07g37560 (68%) | LDHC; LDHH |
Transcript is rhythmically expressed under the following diurnal conditions: LDHH, 12 h of light (L)-12 h of dark (D) and continuous temperature (HH); LDHC, 12 h of light (L)-12 h of dark (D) and high/low physiological temperature (HC); LLHC, continuous light (LL) for 24 h and high/low physiological temperature (HC); SD, short day (8 h of light); LD, long day (16 h of light). Circadian (or free-running) conditions are indicated by LL_ or LLHH_ (i.e. a continuous light LL_ and/or temperature LLHH_ that followed the respective diurnal condition [i.e. _LDHC]). Details of the underlying experimental conditions and microarray data sets are available at the DIURNAL portal at http://diurnal.cgrb.oregonstate.edu.
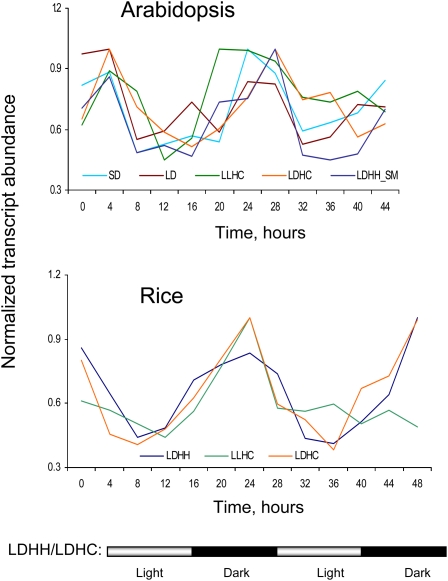
Interestingly, expression levels of myosins XI that showed strong cycling behavior peaked at a similar phase in Arabidopsis and rice, as illustrated for the Arabidopsis XI-K gene and its apparent rice ortholog in Figure 6. This phase corresponded to dawn and/or morning hours, suggesting that many myosin genes are transcribed actively in anticipation of the photosynthetic activity during the light period. In contrast, only a minority of the tested myosins showed rhythmic expression under continuous light/dark/temperature conditions, suggesting that their cycling is regulated by the circadian clock (Table II). Among 17 Arabidopsis myosin genes, such circadian expression behavior was detected only for XI-B, XI-H, and VIII-B.
Figure 6.
Cyclical regulation of the transcript abundance for the Arabidopsis myosin XI-K gene and its putative ortholog in rice (locus identifier Loc_Os03g48140). Diurnal conditions were as follows: LDHH, 12 h of light (L)-12 h of dark (D) and continuous temperature (HH); LDHC, 12 h of light (L)-12 h of dark (D) and high/low physiological temperature (HC); LLHC, continuous light (LL) for 24 h and high/low physiological temperature (HC); SD, short day (8 h); LD, long day (16 h). SM, According to the Smith et al. (2004) protocol (http://diurnal.cgrb.oregonstate.edu/diurnal_details.html).
Previously, regulation of plant myosin gene expression in a photoperiod-sensitive manner was reported for the rice myosin XI gene OSMYOXIB, which is required for normal pollen development (Jiang et al., 2007). Our data show that many class XI family myosins are diurnally regulated and that the accumulation of their transcripts is driven by photocycles and/or thermocycles. Because none of the tested class XI myosins has been implicated in chloroplast transport or positioning (Avisar et al., 2008b; Suetsugu et al., 2010), myosins are unlikely to directly regulate chloroplast performance during the photoperiod. Instead, an overall myosin-driven intracellular dynamics could play a role in more uniform photoassimilate redistribution and elevation of a metabolic status in the cells of light-exposed aerial organs, as recently proposed (Peremyslov et al., 2010).
A Novel, Developmentally Regulated Arabidopsis Gene That Encodes a Headless Variant of Myosin XI-K
As discussed above, gene duplication is the predominant mode of myosin gene family evolution in flowering plants, with many relatively recent events occurring in a lineage-specific manner. An example of this process is seen in myosin group XI (K), where a single duplication in the Arabidopsis lineage yielded genes XI-K and XI-1 with overlapping functions (Prokhnevsky et al., 2008). However, in addition to the full copy of the XI-K gene (AT5G20490), in the same chromosome 5 there is its “shadow” that emerged via partial tandem gene duplication (ATG5G20470). Because RNA-seq data indicated that this gene is transcribed (Fig. 7A, Illumina data track), we were interested in potential functions of the encoded protein.
Figure 7.
Expression, evolution, and regulation of the HDK-like genes in Arabidopsis and Brachypodium. A, Gene models of Arabidopsis HDK (AT5G20470). B, Conserved protein domains in the Arabidopsis HDK and Brachypodium HDK-like proteins. C, Phylogenetic tree of the tail fragments of plant myosins including HDK-like proteins (shaded). Designations and abbreviations are the same as in Figure 1. D, The expression of a GUS reporter driven by a putative HDK gene promoter is observed primarily in young seedlings and the vasculature of young leaves as visualized by in situ staining.
To validate the RNA-seq data, we reverse transcribed and PCR amplified the predicted cDNA corresponding to the ATG5G20470 gene transcript using RNA isolated from young Arabidopsis seedlings. Sequencing of the resulting PCR product revealed a single 1,911-nucleotide-long open reading frame (ORF) and supported modification of the TAIR gene model to include a new first intron within the annotated first exon, novel exons within the annotated introns 7 and 8, as well as modified boundaries for several exons (Fig. 7A, tracks cDNAs and TAU).
Reconstruction of the domain architecture of the product of the conceptually translated ORF using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de) revealed an N-terminal coiled-coil domain and a DIL domain (Fig. 7B, top). Because the predicted product completely lacked the motor head domain, we named it HDK for headless derivative of myosin XI-K. The two conserved domains of HDK were most closely related to those of myosin XI-K, with a 72% amino acid sequence identity. The phylogenetic tree for the C-terminal region of myosins corresponding mostly to the cargo-binding subdomains fully supports this observation (Fig. 7C). Thus, the HDK gene most likely evolved via a relatively recent partial duplication of the XI-K gene.
The high similarity between the amino acid sequences of HDK and myosin XI-K suggested that these proteins bind similar cargoes. However, HDK cannot transport the bound cargoes due to the absence of a motor domain. Therefore, it could be expected that the expression of HDK would interfere with XI-K-dependent cargo transport, by analogy to the commonly used experimental approach in which ectopic expression of headless myosins is employed for dominant negative inhibition of myosin functions (Avisar et al., 2008a, 2008b, 2009; Sparkes et al., 2008, 2009; Sattarzadeh et al., 2009).
Given the intriguing possibility that HDK is involved in regulating myosin XI-K, we investigated the expression pattern and the potential functions of HDK in Arabidopsis. An expression cassette was engineered in which a putative promoter region of the HDK gene (a 1-kb-long genomic DNA fragment starting upstream from the HDK initiation codon) designated PrHDK was fused to the ORF encoding bacterial GUS, a sensitive enzymatic reporter of gene expression. The resulting PrHDK::GUS cassette was transformed into Arabidopsis and used to assay HDK promoter activity in growing plants.
Conspicuously, it was found that the expression of the HDK gene is regulated in a development- and tissue-specific manner. Indeed, substantial GUS activity was observed throughout the roots and young cotyledons of the PrHDK::GUS transgenic plants, with the vascular tissues showing the strongest in situ GUS staining (Fig. 7D). In the mature cotyledons, reporter protein expression was limited to the vascular bundles. A similar expression pattern was seen in the young true leaves. With leaf growth, GUS activity gradually declined, virtually disappearing in the mature leaves (Fig. 7D).
To gain insight into the potential HDK function in plant growth and development, we obtained mutant Arabidopsis lines SALK_04694 and SALK_085824 with insertions four nucleotides upstream of the ATG start codon and in the fifth exon of the HDK-encoded ORF, respectively, and generated the corresponding homozygous lines. Characterization of these lines using RT-PCR showed undetectable levels of HDK mRNA expression, confirming the insertional inactivation of the HDK gene. However, the HDK-deficient plants showed no discernible phenotypes compared with the phenotype of wild-type plants (data not shown). Given that xi-k knockouts also showed no obvious developmental phenotype (Peremyslov et al., 2008), this is not an unexpected outcome. If, as proposed, the HDK function is to modulate XI-K activity via dominant negative inhibition, the effect of such modulation should not exceed the (undetectable) phenotypic effect of the XI-K inactivation.
Because the phenotypes of the myosin-deficient (Peremyslov et al., 2008) or HDK-deficient (this work) plants were investigated under optimal growth conditions, it remains possible that these proteins play more prominent roles in stress or pathogen responses. Even though inactivation of HDK does not affect plant performance in limited-scale experiments, it seems likely that the mutant plants are less fit than the wild-type plants. This possibility can be addressed in large-scale, multigeneration competition experiments under natural environment conditions.
As we have shown above (Fig. 2B, shaded box), Brachypodium also possesses and expresses a gene that apparently emerged via partial duplication of the myosin XI gene. Indeed, conceptual translation of the Bd2.62282.1 transcript yielded a single ORF encoding an HDK-like protein with an IQ motif, a truncated coiled-coil domain, and a complete DIL domain (Fig. 7B, bottom). Phylogenetic analysis showed that this protein was most closely related to the monocot myosins within the XI (K) group (Fig. 7C), suggesting that the corresponding gene has emerged via partial duplication of one of the genes in this lineage. Therefore, we concluded that the Arabidopsis HDK and Brachypodium HDK-like genes have emerged independently, suggestive of a broader importance of partial gene duplication. Additional candidate HDK-like genes were also detected in the rice and sorghum genomes (data not shown), although their expression remains to be validated experimentally.
CONCLUSION
In this work, we provide a unified view of the multigene family of key molecular motors, myosins, in plants. Using newly sequenced genomes of algae, mosses, and angiosperms, we develop a scenario of myosin evolution according to which all green plants possess two classes of myosins, VIII and XI, rather than three classes, as previously accepted. Furthermore, we show that angiosperms possess two groups of myosins VIII and five groups of myosins XI; genes in each of these groups undergo active lineage-specific duplication, most likely accompanied by subfunctionalization (Fig. 1). We use phylogenetic classification to propose a modified approach to myosin nomenclature (Table I) and to guide research into functional genomics of plant myosins.
We show that, in addition to gene proliferation via duplication, plant myosin genes also exhibit an additional level of complexity due to alternative splicing of their corresponding transcripts. Dominated by intron retention, alternative splicing has a potential to yield myosin variants with distinct functions. Using empirical transcript data and improved gene model-building algorithms, we amended gene models for all known Arabidopsis and Brachypodium myosins (http://myosins.cgrb.oregonstate.edu). Using direct cDNA sequencing and mapping of the 5′ termini, we validated the revised gene models for the Arabidopsis myosin XI-K that plays major roles in multiple myosin-dependent processes and for its ortholog in Brachypodium (Figs. 2 and 3).
Existence of the myosin XI splicing isoforms and HDK-like genes in Arabidopsis and Brachypodium highlights a possibility of regulatory functions executed via selective binding of myosin partners. Indeed, full-size myosins bind both F-actin and the cargo. In contrast, the DIL-less isoforms are likely to bind only F-actin, whereas the HDK-like proteins can only bind cargo, thus modulating the corresponding activities of the full-size myosins.
From comparative analysis of the available DNA microarray and RNA-seq data, we conclude that there are two principal modes of myosin gene expression in Arabidopsis (Figs. 4 and 5). One mode involves ubiquitous expression throughout the vegetative plant, whereas the second mode pertains to myosin genes that are preferentially expressed in male reproductive cells of pollen and stamen (Fig. 4). In terms of the expression levels, myosin genes could be roughly classified into abundant, intermediate, and low expression categories (Fig. 5). We also show that many myosin genes in Arabidopsis and rice are diurnally regulated (Fig. 6; Table II), implicating myosins in photoperiod-related aspects of plant physiology.
We characterize a novel Arabidopsis gene, HDK, that evolved via partial duplication of the XI-K gene and is preferentially expressed in young cotyledons and developing vascular tissue (Fig. 7). Although preliminary analysis of the hdk gene knockout lines did not reveal an obvious phenotype, we suggest that HDK might function as an accessory modulator of the myosin XI-K function in early development. This notion is supported by the expression of the analogous, HDK-like protein in Brachypodium (Figs. 2, 3, and 7). Using phylogenetic analysis, we show that the HDK genes of Arabidopsis and Brachypodium most likely evolved independently. Together with the experimental results described above, these findings suggest that partial duplication of myosin genes converged to produce truncated myosin forms. Searches in protein databases suggest that HDK-like proteins are broadly represented in plant genomes, an observation that is compatible with the functional relevance of these headless myosin derivatives.
Taken together, the experimental and computational results of this work provide a detailed picture of plant myosin gene evolution, splicing, and expression that constitutes a genome-wide framework for the future investigation of myosin gene function and regulation in angiosperms. The emerging field of plant myosin genomics yields an increasingly refined depiction of an evolutionarily dynamic, functionally diversified, and partially redundant multigene family. The evolution of the myosin family is characterized by ongoing processes of gene duplication followed by functional specialization, in particular, via the acquisition of developmental and tissue-specific patterns of gene regulation.
MATERIALS AND METHODS
Phylogenetic Analysis
Phylogenetic analysis was performed using the MOLPHY software to build maximum-likelihood unrooted trees (Adachi et al., 2000) on the basis of multiple alignments constructed using the MUSCLE program (Edgar, 2004). Poorly aligned regions were removed manually; the final alignments used for the phylogenetic reconstructions included 620 positions for the motor domain tree and 348 positions for the HDK tree. The MOLPHY program was also used to compute the resampling estimated log likelihood bootstrap probabilities (Adachi et al., 2000).
Myosin Gene Models, Expression, and Regulation
The Arabidopsis (Arabidopsis thaliana) genome sequence, annotation, and annotated sequence features were downloaded from TAIR (ftp://ftp.arabidopsis.org/home/tair/Sequences/). The Brachypodium (Brachypodium distachyon) genome sequence, annotation, and annotated sequence features were obtained from BrachyBase (http://www.brachybase.org). Arabidopsis and Brachypodium RNA-seq transcriptome data were downloaded from the National Center for Biotechnology Information Short Read Archive, accession numbers SRA009031 and SRA010177, respectively. Arabidopsis EST and cDNA sequences were obtained from GenBank, and Brachypodium ESTs were downloaded from BrachyBase. Arabidopsis microarray data were obtained from and mined using the Genevestigator suite of tools (https://www.genevestigator.com). RNA-seq reads were mapped to the Arabidopsis and Brachypodium genome assemblies using HashMatch (Filichkin et al., 2010) and supersplat (Bryant et al., 2010). Long-read Sanger and 454 ESTs were mapped to the Arabidopsis and Brachypodium genome assemblies using BLAT (Kent, 2002). The combined alignments of RNA-seq data, ESTs, and cDNAs were used to construct empirical transcription unit assemblies using TAU (http://mocklerlab-tools.cgrb.oregonstate.edu), which rapidly and accurately assembles transcript data into alternatively spliced (when applicable) transcript models by reference-guided assembly. The TAU transcript models and their supporting transcript data were loaded into a Gbrowse genome viewer for visualization.
The 5′ RLM-RACE to determine the transcription initiation site of the XI-K (At5g20490) mRNA and to sequence the downstream mRNA region was done using total RNA from young rosette leaves of Arabidopsis var Columbia-0 isolated with the Qiagen Plant RNA Prep kit and the First Choice RLM-RACE kit (Ambion) according to the manufacturers’ protocols. The following two nested PCRs were done using primers supplied by the manufacturer in combination with two gene-specific primers (5′-CACAGTTGTCAAACTGAAGCTCGAC-3′ and 5′-GTTGCCAAAAGCTTCAAGAACT-3′). A control reaction where the tobacco (Nicotiana tabacum) acid pyrophosphatase treatment was omitted was included to ensure that no truncated transcripts lacking the 5′ cap were amplified. PCR products were digested with BamHI and HindIII and cloned into pGEM3Zf(+) (Promega); 12 independent clones were sequenced.
To validate the mRNA splicing isoforms predicted by RNA-seq (Fig. 2), RNA samples prepared from leaves of Arabidopsis or Brachypodium were analyzed by RT-PCR. cDNA was synthesized using random hexamer primers. Isoform-specific PCR oligonucleotides were designed to span the novel junctions and used in combination with compatible reverse primers spanning constitutive exon-exon junctions to amplify the predicted splicing products (Supplemental Table S1). To confirm the identity of PCR products, the resulting DNA fragments were sequenced and aligned against the genomic sequences of the corresponding genes.
To investigate the cyclic regulation of myosin gene expression, plants were entrained for at least 7 d under the following diurnal (driven) conditions: LDHH, 12 h of light (L)-12 h of dark (D) and continuous temperature (HH); LDHC, 12 h of light (L)-12 h of dark (D) and high/low physiological temperature (HC); LLHC, continuous light (LL) for 24 h and high/low physiological temperature (HC); SD and LD, short-day (8 h) and long-day (16 h) conditions were used for Arabidopsis only. Leaf tissues were collected at 4-h intervals, and RNA was isolated as described previously (Filichkin et al., 2007).
Analyses of HDK Gene Expression and Regulation
Total RNA purified from young Arabidopsis seedlings was used to amplify putative cDNA encoded by the previously uncharacterized HDK (At5g20470) gene. RT was reformed using random primers; the following PCR was done using oligonucleotides complementary to the regions flanking the predicted HDK ORF (5′-GTCTTTTCTGCTTTTACATGCTC-3′ and 5′-TGCAACAACACTCAAACCAGC-3′). The PCR product was sequenced directly. The 1,048-nucleotide-long sequence upstream of the HDK ORF start codon (nucleotides 6,921,168–6,920,120 on chromosome 5) harboring its putative promoter was PCR amplified using oligonucleotides 5′-TATAGAATTCTCGTCACACGCCTTTGCC-3′ and 5′-TATAGGATCCAAGTAGAACCATGTCCTGA-3′ and cloned into a promoterless expression cassette consisting of the GUS coding region followed by the nopaline synthase poly(A) signal in pCAMBIA-1300 binary vector (http://www.cambia.org/daisy/cambia/585.html). The resulting plasmid was transformed to Agrobacterium tumefaciens strain GV3101. Arabidopsis plants were transformed by the floral dipping method, and the transformants were selected on hygromycin-containing medium. Whole seedlings or detached leaves were collected at different developmental stages, and in situ GUS activity was detected by incubating the samples in a 5-bromo-4-chloro-3-indolyl β-glucuronide-containing standard histochemical solution overnight at 37°C.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HQ427882, HQ427883, HQ427884, and HQ427885.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used for interrogation of the splice sites in the transcripts of the Arabidopsis XI-K gene and the Brachypodium Bradi2g41980 gene.
Supplementary Material
Acknowledgments
We thank Rex Cole and John Fowler for sharing unpublished data and Ellie Kurth for excellent laboratory and greenhouse management.
References
- Adachi J, Waddell PJ, Martin W, Hasegawa M. (2000) Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA. J Mol Evol 50: 348–358 [DOI] [PubMed] [Google Scholar]
- Avisar D, Abu-Abied M, Belausov E, Sadot E, Hawes C, Sparkes IA. (2009) A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol 150: 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Dolja VV. (2008a) Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J Virol 82: 2836–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. (2008b) Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol 146: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Horton AC, Sevener HC, Quatrano RS. (2003) Phylogenetic analysis of new plant myosin sequences. J Mol Evol 57: 229–239 [DOI] [PubMed] [Google Scholar]
- Brachypodium Genome Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Bryant DWJ, Jr, Shen R, Priest HD, Wong WK, Mockler TC. (2010) Supersplat: spliced RNA-seq alignment. Bioinformatics 26: 1500–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RA, Fowler JE. (2006) Polarized growth: maintaining focus on the tip. Curr Opin Plant Biol 9: 579–588 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, DiFazio SP, Brunner AM, Davis JM, Yang ZK, Kalluri UC, Arias RS, Etherington E, Tuskan GA, Strauss SH. (2007) Efficiency of gene silencing in Arabidopsis: direct inverted repeats vs. transitive RNAi vectors. Plant Biotechnol J 5: 615–626 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. (2006) New insights into myosin evolution and classification. Proc Natl Acad Sci USA 103: 3681–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox S, Filichkin S, Mockler TC. (2009) Applications of ultra high throughput sequencing in plants. Methods Mol Biol 553: 79–108 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Cai M, Ramachandran S. (2007) ORYZA SATIVA MYOSIN XI B controls pollen development by photoperiod-sensitive protein localizations. Dev Biol 304: 579–592 [DOI] [PubMed] [Google Scholar]
- Kent WJ. (2002) BLAT: the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Wang H, Schiefelbein J. (1994) Molecular analysis of the myosin gene family in Arabidopsis thaliana. Plant Mol Biol 26: 1139–1153 [DOI] [PubMed] [Google Scholar]
- Knight AE, Kendrick-Jones J. (1993) A myosin-like protein from a higher plant. J Mol Biol 231: 148–154 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Yang Z. (2008) Tip growth: signaling in the apical dome. Curr Opin Plant Biol 11: 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Liu B. (2004) Cytoskeletal motors in Arabidopsis: sixty-one kinesins and seventeen myosins. Plant Physiol 136: 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC. (2009) Myosin XI is required for actin-associated movement of plastid stromules. Mol Plant 2: 1262–1272 [DOI] [PubMed] [Google Scholar]
- Ojangu EL, Järve K, Paves H, Truve E. (2007) Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma 230: 193–202 [DOI] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. (2008) Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol 146: 1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, Dolja VV. (2010) Class XI myosins are required for development, cell expansion, and F-actin organization in Arabidopsis. Plant Cell 22: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky AI, Peremyslov VV, Dolja VV. (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA 105: 19744–19749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS. (2001) Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol 2: RESEARCH0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland JT, Lapierre LA, Goldenring JR. (2009) Alternative splicing in class V myosins determines association with Rab10. J Biol Chem 284: 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattarzadeh A, Krahmer J, Germain AD, Hanson MR. (2009) A myosin XI tail domain homologous to the yeast myosin vacuole-binding domain interacts with plastids and stromules in Nicotiana benthamiana. Mol Plant 2: 1351–1358 [DOI] [PubMed] [Google Scholar]
- Shimmen T, Yokota E. (2004) Cytoplasmic streaming in plants. Curr Opin Cell Biol 16: 68–72 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM. (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, Griffing L. (2009) Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21: 3937–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA. (2010) Motoring around the plant cell: insights from plant myosins. Biochem Soc Trans 38: 833–838 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Teanby NA, Hawes C. (2008) Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J Exp Bot 59: 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Dolja VV, Wada M. (2010) Why have chloroplasts developed a unique motility system? Plant Signal Behav 5: 1190–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Kojima H, Yokota E, Orii H, Nakamori R, Katayama E, Anson M, Shimmen T, Oiwa K. (2003) Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. EMBO J 22: 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I. (2010) Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Burkart GM, Augustine RC, Kerdavid E, Tüzel E, Bezanilla M. (2010) Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA., III (2002) Identification of an organelle receptor for myosin-Va. Nat Cell Biol 4: 271–278 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.