Abstract
Auxin/indole-3-acetic acid (Aux/IAA) proteins function as repressors of auxin response gene expression when auxin concentrations in a cell are low. At elevated auxin concentrations, these repressors are destroyed via the ubiquitin-proteasome pathway, resulting in derepression/activation of auxin response genes. Most Aux/IAA repressors contain four conserved domains, with one of these being an active, portable repression domain (domain I) and a second being an auxin-dependent instability domain (domain II). Here, we have analyzed the effects of amino acid substitutions in the repression domain of selected Aux/IAA proteins. We show that stabilized versions of Aux/IAA proteins with amino acid substitutions in domain I display contrasting phenotypes when expressed in transformed Arabidopsis (Arabidopsis thaliana) plants. An alanine-for-leucine substitution in the LxLxL (where L is leucine and x is another amino acid) repression domain of IAA3, IAA6, or IAA19 confers enhanced auxin response gene expression and “high-auxin” phenotypes when expressed from the 35S or IAA19 promoter (as tested with IAA19) in transformed Arabidopsis plants. In marked contrast, a single alanine-for-leucine substitution in domain I of IAA12 or IAA17 confers repression of auxin response genes and “low-auxin” phenotypes. These results point to intrinsic differences in the repression domain(s) of IAA proteins and suggest that some IAA proteins have stronger or more complex repression domains than others.
Auxin/indole-3-acetic acid (Aux/IAA, or IAA) proteins are, in general, short-lived, nuclear proteins that play a key role in regulating the expression of auxin response genes (for review, see Hagen and Guilfoyle, 2002; Liscum and Reed, 2002). Many of the 29 genes that encode these proteins in Arabidopsis (Arabidopsis thaliana) are induced by auxin themselves (Abel et al., 1995). The Arabidopsis genes encode proteins ranging in size from about 18 to 36 kD that, for the most part, contain four conserved domains (i.e. referred to as domains I, II, III, and IV). Of some 20 Aux/IAA proteins that have been tested to date, including 16 from Arabidopsis, all functioned as transcriptional repressors when expressed from effector plasmids in protoplast transfection assays (Ulmasov et al., 1997; Tiwari et al., 2001; Bargmann and Birnbaum, 2009). Consistent with the transfection results, targeted or constitutive expression of stabilized versions of IAA proteins in Arabidopsis plants resulted in suppression of auxin response genes and auxin-responsive growth and developmental responses (Fukaki et al., 2005; Kim et al., 2006; Nakamura et al., 2006; De Smet et al., 2007; Sato and Yamamoto, 2008; Ku et al., 2009; Li et al., 2009).
Protoplast transfection experiments identified domain I of Aux/IAA proteins as an active, portable repression domain with a conserved LxLxL (i.e. where L is Leu and x is any of several different amino acids) motif similar to the so-called ethylene response factor-associated amphiphilic repression (EAR) repression domain (Tiwari et al., 2004). Supporting evidence for domain I being a repression domain came from experiments that demonstrated this domain’s interaction with the TOPLESS (TPL) corepressor (Szemenyei et al., 2008). Domain II confers instability to the Aux/IAA proteins (Worley et al., 2000; Dreher et al., 2006), and this domain contains a degron that interacts with the Transport Inhibitor Response1 (TIR1) auxin receptor and related auxin-binding F-box receptors in an auxin-dependent manner (for review, see Mockaitis and Estelle, 2008). Domains III and IV of Aux/IAA proteins constitute a protein-protein interaction domain that facilitates homotypic and heterotypic interactions among Aux/IAA proteins and Auxin Response Factors (ARFs), which contain a C-terminal domain related to domains III and IV in Aux/IAA proteins (Kim et al., 1997; Ulmasov et al., 1997).
We have previously reported that mutations in domain II of Aux/IAA proteins, including IAA3/SHORT HYPOCOTYL2 (SHY2), IAA17/AUXIN RESISTANT3 (AXR3), and IAA19/MASSUGU2 (MSG2), increased their capacity to repress auxin-responsive reporter genes in protoplast transfection assays, which, as demonstrated with IAA17, correlated with their increased stability (Tiwari et al., 2001). For simplicity, IAA3/SHY2, IAA17/AXR3, and IAA19/MSG2 are referred to as IAA3, IAA17, and IAA19 throughout this article. Mutations in both domains I and II of IAA17 (i.e. IAA17mImII, where mI and mII refer to mutated domain I and mutated domain II, respectively) resulted in reduced capacity of the protein to repress the expression of an auxin-responsive reporter gene in transfected protoplasts, and deletion of domain I, as tested with IAA19, resulted in complete loss of repression (Tiwari et al., 2001). Subsequent studies showed that while IAA17mImII still repressed an auxin-responsive reporter gene in transfected protoplasts, an N-terminal fusion of the herpes simplex virus Viral Protein16 (VP16) activation domain onto IAA17mImII (i.e. VP16-IAA17mImII) resulted in constitutive activation of an auxin-responsive reporter gene (Tiwari et al., 2003). Furthermore, the wild-type domain I of IAA17 was shown to be an active, portable repression domain that could alleviate activation by VP16, while a domain I mutant of IAA17 could not (Tiwari et al., 2004).
Recently, Li et al. (2009) showed that when the cauliflower mosaic virus (CaMV) 35S promoter was used to drive the expression of IAA17mImII and VP16-IAA17mImII in stably transformed Arabidopsis plants, auxin responses, including auxin-responsive gene expression, were constitutively repressed in 35S: IAA17mImII lines and constitutively activated in 35S:VP16-IAA17mImII lines. It was further shown that these responses occurred in an auxin-independent manner (i.e. free IAA hormone concentrations were not altered in the transgenic lines compared with the wild type). These results supported earlier observations made with transfected protoplasts where IAA17 functioned as a transcriptional repressor of auxin response gene expression that could be converted to a transcriptional activator by fusing a VP16 activation domain onto an IAA17 protein with a compromised repression domain.
In the experiments described here, we analyzed how Ala substitutions for Leu in the repression domain (i.e. LxLxL) of selected Aux/IAA proteins affected their ability to repress auxin response gene expression in transformed Arabidopsis plants. These experiments revealed that a single Ala substitution for the first Leu in the LxLxL motif of IAA19, IAA6 (a sister pair of IAA19), or IAA3 (a more distantly related IAA protein to IAA19) resulted in constitutive activation of the Direct Repeat5 (DR5):GUS reporter gene (and other auxin response genes) and “high-auxin” phenotypes. In marked contrast, an identical Ala substitution in the repression domain of IAA17 and a single Ala substitution in IAA12/BODENLOS (referred to as IAA12 throughout this article; a more distantly related IAA protein to IAA17) resulted in constitutive repression of the DR5 reporter gene (and other auxin response genes) and “low-auxin” phenotypes. Additional Ala substitutions in the repression domain of IAA12 resulted in activation of auxin response genes and “high-auxin” phenotypes, but this was not observed with IAA17. These studies point to intrinsic differences in IAA proteins, some of which are located outside of the conserved domains I, II, III, and IV. We discuss the implications of these results in terms of a current model for auxin-regulated gene expression.
RESULTS
Constitutive Expression of Closely Related IAA Repressors with Identical Amino Acid Substitutions in Domain I Results in Contrasting Phenotypes
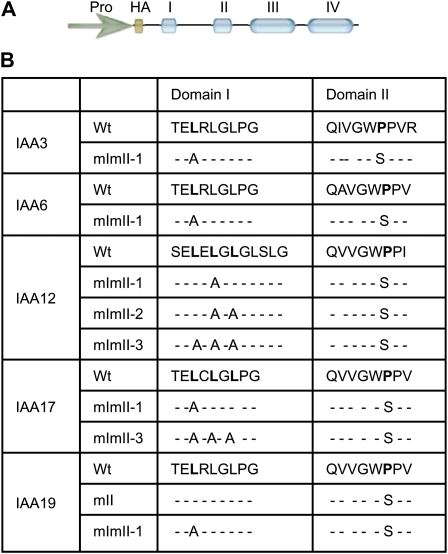
To evaluate the effects of mutations in the repression domain of Arabidopsis IAA proteins, we examined five different IAA proteins (i.e. IAA3, IAA6, IAA12, IAA17, and IAA19) that fall into distinct clades on a phylogenetic tree (Remington et al., 2004). We substituted an Ala for the first Leu in the LxLxL motif of domain I in IAA3, IAA6, IAA17, and IAA19 (Fig. 1). IAA12 differs from the other four IAA proteins examined in having an LxLxLxLxL motif in domain I and is discussed separately below. Each IAA protein also contained a Ser substitution for the first Pro in the GWPP motif of domain II to increase its stability compared with wild-type proteins (Worley et al., 2000; Gray et al., 2001; Ouellet et al., 2001; Ramos et al., 2001). The CaMV 35S promoter was used to drive expression of the mutant IAA proteins in stably transformed Arabidopsis ecotype Columbia (Col-0) plants, and transformed lines are referred to as 35S:IAAmImII with the specific IAA protein indicated (e.g. 35S:IAA19mImII-1). Transgenic lines were classified as having wild-type, “very-low-auxin,” “low-auxin,” or “high-auxin” phenotypes. Plants with “very-low-auxin” phenotypes had defective organs, and most did not survive beyond the young seedling stage. “Low-” and “high-auxin” phenotypes refer to transgenic plants that resembled plants that expressed 35S:IAA17mImII and 35S:VP16-IAA17mImII transgenes, respectively (Li et al., 2009).
Figure 1.
IAA transgene constructs and summary of amino acid substitutions in domain I and domain II of IAA proteins. A, Generalized diagram of IAAmImII constructs used for transforming Arabidopsis. Pro indicates either the CaMV 35S or IAA19 promoter. Conserved domains I through IV are diagrammed as blue ovals, and nonconserved regions are represented as the line. Each IAA protein contained a HA epitope tag at the N terminus. B, Summary table of amino acid sequences of wild-type (Wt) and mutated versions of domains I and II (mImII) for each IAA protein used in transforming Arabidopsis.
We had initially observed that 35S:IAAmII lines (i.e. IAA17mII and IAA19mII lines with a wild-type domain I) had “very-low-auxin” phenotypes like those shown for 35S:IAA19mII in Supplemental Figure S1. These “very-low-auxin” phenotypes would be expected for plants that express IAA proteins that are more stable (through an introduced mutation in conserved domain II) and have a wild-type domain I. The “very-low-auxin” phenotypes included seedlings with one cotyledon, triple cotyledons, defective primary roots, and strongly reduced DR5:GUS reporter gene expression, whether seedlings were treated with exogenous auxin or not. T1 seedlings that did survive had a dwarfish stature and produced few viable seeds. T2 plants were wild type in appearance (including DR5:GUS expression being identical to wild-type plants), indicating that the 35S:IAA17mII and 35S:IAA19mII lines were unstable. Instability with 35S:IAA17mII lines had been reported previously by Li et al. (2009).
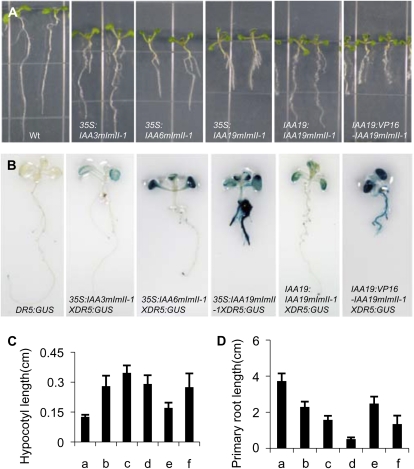
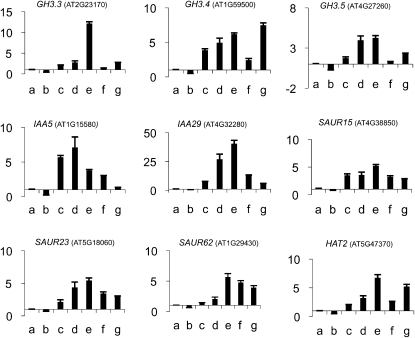
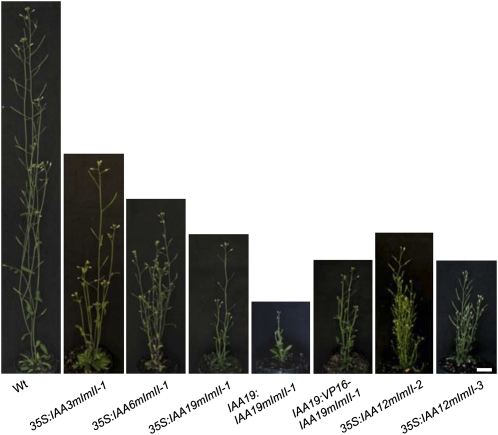
In contrast to the 35S:IAAmII lines, 35S:IAAmImII lines were relatively stable up to at least the T5 generation, making it possible to compare phenotypes in seedlings and adult plants expressing the different IAAmImII proteins. To determine if IAA proteins with an identical Ala substitution for the first Leu in domain I behaved like those previously reported for IAA17mImII (Li et al., 2009), we analyzed plants expressing 35S:IAA3mImII-1, 35S:IAA6mImII-1, and 35S:IAA19mImII-1 transgenes (Fig. 1B; Supplemental Fig. S2). IAA6 and IAA19 are close relatives phylogenetically (i.e. sister pairs) but are more distantly related to IAA3 and IAA17 (Remington et al., 2004). In marked contrast to plant lines constitutively expressing IAA19mII or our original IAA17mImII construct (Li et al., 2009), lines expressing IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1 had “high-auxin” phenotypes, including seedlings with short, highly branched roots, long hypocotyls and petioles, epinastic cotyledons and leaves, and enhanced DR5:GUS reporter gene expression (Fig. 2). To determine if a set of well-characterized natural auxin response genes behaved similarly to the DR5 reporter gene in the transformed Arabidopsis plants, we used quantitative reverse transcription (qRT)-PCR to measure the relative expression of three Gretchen Hagen3 genes, two IAA genes, three SMALL AUXIN-UP RNA genes, and the HOMEOBOX FROM ARABIDOPSIS THALIANA2 gene. Expression of these natural auxin response genes was elevated similar to DR5:GUS in these transgenic seedlings (Fig. 3, compare columns a with columns c–e). These “high-auxin” phenotype seedlings resembled those previously reported for plants expressing a 35S:VP16-IAA17mImII transgene (Li et al., 2009). When compared at the same age (e.g. 4 weeks old), 35S:IAA3mImII-1, 35S:IAA6mImII-1, and 35S:IAA19mImII-1 plants were shorter than wild-type plants (Fig. 4). The adult plants displayed a zigzag floral shoot phenotype, resembling that previously observed with 35S:VP16-IAA17mImII transgenic lines (Li et al., 2009).
Figure 2.
Phenotypes of wild-type and transformed seedlings expressing 35S:IAA3mImII-1, 35S:IAA6mImII-1, 35S:IAA19mImII-1, IAA19:IAA19mImII-1, and IAA19:VP16-IAA19mImII-1 transgenes. A, Photographs of 7-d-old seedlings. Two representative seedlings are shown for the wild type (Wt) and each IAA transformant. B, Histochemical GUS staining of 7-d-old wild-type and transformed seedlings. Seedlings were stained for 15 h at 37°C. C, Hypocotyl length for 7-d-old wild-type and transformed seedlings. Error bars indicate sd (n ≥ 15, ANOVA test, P < 0.05). D, Primary root length for 7-d-old wild-type and transformed seedlings. Error bars indicate sd (n ≥ 15, ANOVA test, P < 0.05). Lanes are as follows: a, the wild type; b, 35S:IAA3mImII-1; c, 35S:IAA6mImII-1; d, 35S:IAA19mImII-1; e, IAA19:IAA19mImII-1; f, IAA19:VP16-IAA19mImII-1.
Figure 3.
qRT-PCR analysis of natural auxin response gene expression in wild-type seedlings and seedlings expressing IAAmImII constructs. Levels of gene expression are shown relative to the wild type. Lanes are as follows: a, the wild type; b, 35S:IAA17mImII-1; c, 35S:IAA3mImII-1; d, 35S:IAA6mImII-1; e, 35S:IAA19mImII-1; f, IAA19:IAA19mImII-1; g, 35S:IAA12mImII-2.
Figure 4.
Phenotypes of wild-type (Wt) and transformed adult plants expressing 35S:IAA3mImII-1, 35S:IAA6mImII-1, 35S:IAA19mImII-1, IAA19:IAA19mImII-1, IAA19:VP16-IAA19mImII-1, 35S:IAA12mImII-2, and 35S:IAA12mImII-3. Plants were approximately 4 weeks old. Bar = 2 cm.
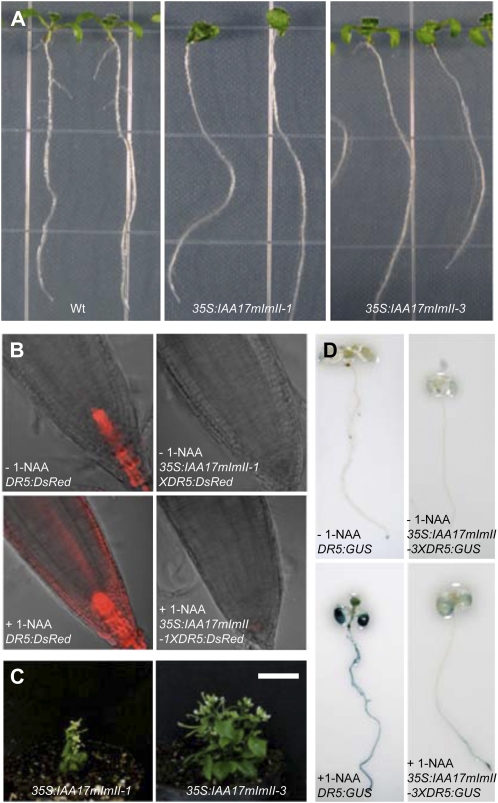
Because the 35S:IAA17mImII transgene that we previously tested had two amino acid substitutions in domain I (i.e. Val and Arg substituted for Glu and the first Leu in the TELCLGLPG motif; Li et al., 2009), we retested IAA17 with an identical Ala substitution for the first Leu in the LxLxL motif as that tested in IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1 (i.e. referred to as IAA17mImII-1; Fig. 1B). Plants expressing the 35S:IAA17mImII-1 transgene (Supplemental Fig. S2) had similar “low-auxin” phenotypes (Fig. 5) to those previously reported for 35S:IAA17mImII (Li et al., 2009). These “low-auxin” phenotypes, however, were less severe than the “very-low-auxin” phenotypes observed with 35S:IAA17mII or 35S:IAA19mII T1 seedlings. The “low-auxin” phenotypes included seedlings with agravitropic roots, reduced numbers of lateral roots, short hypocotyls and petioles, and strongly reduced DR5:DsRed expression in root tips with or without exposure to exogenous auxin (Fig. 5, A and B). Expression of the natural auxin response genes was down-regulated, similar to the DR5 reporter gene (Fig. 3, compare columns a and b). Adult plants had a dwarf stature with a compact inflorescence, short, wrinkled siliques, and reduced fecundity (Fig. 5C).
Figure 5.
Phenotypes of wild-type and transformed seedlings expressing 35S:IAA17mImII-1 and 35S:IAA17mImII-3 transgenes. A, Photographs of 7-d-old seedlings. Two representative seedlings are shown for the wild type (Wt), 35S:IAA17mImII-1, and 35S:IAA17mImII-3. B, DsRed signal in DR5:DsRed and 35S:IAA17mImII-1 × DR5:DsRed 7-d-old seedlings. Seedlings were mock treated or treated with 10 μm 1-NAA for 15 h. C, Adult plants of 35S:IAA17mImII-1 and 35S:IAA17mImII-3. Bar = 2 cm. D, GUS staining in 7-d-old seedlings for DR5:GUS and 35S:IAA17mImII-3 × DR5:GUS. Seedlings were mock treated or treated with 10 μm 1-NAA for 15 h.
Constitutive Expression of Transgenes with Increasing Numbers of Leu-to-Ala Substitutions in Domain I of IAA12 Results in the Conversion of “Low- to High-Auxin” Phenotypes
IAA12 is a member of group B IAA proteins and is more distantly related to IAA3, IAA6, IAA17, and IAA19, which are members of group A, based on the phylogenetic analysis of Remington et al. (2004). Interestingly, IAA12 differs from IAA6, IAA17, IAA19, and most other IAA proteins in having an LxLxLxLxL motif in domain I rather than the more common LxLxL motif (Fig. 1B).
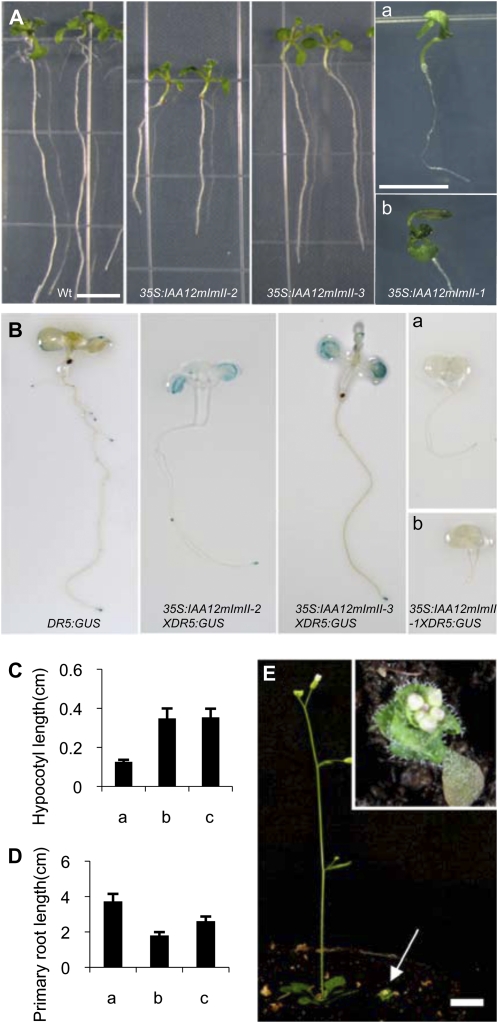
Expression of a 35S:IAA12mImII transgene (i.e. referred to as 35S:IAA12mImII-1) with substitution of an Ala for the second Leu in the SELELGLGLSLG motif of domain I resulted in plants with “very-low-auxin” phenotypes (i.e. like those observed for 35S:IAA19mII). These transgenic lines had phenotypes including seedlings with one cotyledon, fused cotyledons, no root or a short root, and little or no DR5:GUS activity (Fig. 6, A and B). Auxin treatment failed to induce the DR5:GUS reporter gene in these seedlings (Supplemental Fig. S3). Most of the 35S:IAA12mImII-1 lines did not survive, and those that did were extremely stunted (Fig. 6E). These results suggest that a single Leu-to-Ala substitution (i.e. at least for the second Leu in the LxLxLxLxL motif) in domain I of IAA12 is not sufficient to destroy the repression domain.
Figure 6.
Phenotypes of wild-type and transformed seedlings expressing 35S:IAA12mImII transgenes with a single, double, or triple mutation in domain I. A, Photographs of 7-d-old seedlings. Two representative seedlings are shown for the wild type (Wt) and 35S:IAA12mImII transformants. a and b show two 35S:IAA12mImII-1 primary transformants with fused cotyledons and stunted primary root, respectively. The wild type, 35S:IAA12mImII-2, and 35S:IAA12mImII-3 are at the same magnification. a and b for 35S:IAA12mImII-1 are at the same magnification. Bars = 0.5 cm. B, Histochemical GUS staining of 7-d-old wild-type and transformed seedlings. Seedlings were stained for 15 h at 37°C. a and b show two 35S:IAA12mImII-1 primary transformants with weaker GUS staining. C, Hypocotyl length for 7-d-old wild-type and transformed seedlings. Error bars indicate sd (n ≥ 15, ANOVA test, P < 0.05). Lanes are as follows: a, the wild type; b, 35S:IAA12mImII-2; c, 35S:IAA12mImII-3. D, Length of primary roots for 7-d-old wild-type and transformed seedlings. Error bars indicate sd (n ≥ 15, ANOVA test, P < 0.05). Lanes are as follows: a, the wild type; b, 35S:IAA12mImII-2; c, 35S:IAA12mImII-3. E, Photograph of a wild-type plant (left) and a T1 35S:IAA12mImII-1 transformant (arrow). Bar = 1 cm. The inset at right shows an enlargement of the T1 transformant.
In contrast to the 35S:IAA12mImII-1 lines, plants expressing IAA12 with Ala substitutions for the second and third Leu residues in the LxLxLxLxL motif (i.e. referred to as 35S:IAA12mImII-2) had “high-auxin” phenotypes. Seedlings had long hypocotyls and petioles with epinastic cotyledons and leaves, short roots, and enhanced DR5:GUS staining, especially in cotyledons (Fig. 6). In contrast to the “high-auxin” phenotypes described above for IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1, however, seedlings constitutively expressing IAA12mImII-2 had longer primary roots and fewer lateral roots. Natural auxin response genes were up-regulated in plants expressing 35S:IAA12mImII-2 compared with the wild type (Fig. 3, compare columns a and g). Adult plants expressing IAA12mImII-2 were bushier and shorter than wild-type plants (Fig. 4). Expression of a transgene with an additional Ala substitution in the first Leu in the SELELGLGLSLG motif (i.e. referred to as 35S:IAA12mImII-3) resulted in seedlings with similar enhanced “high-auxin” phenotypes (Figs. 4 and 6). These results with IAA12 suggest that two or three Ala substitutions for Leu residues in domain I of IAA12 alleviate the repression of stabilized IAA12 and result in increased auxin responses, including DR5:GUS and natural auxin response gene expression.
Constitutive Expression of Transgenes with Increasing Numbers of Leu-to-Ala Substitutions in Domain I of IAA17 Does Not Result in Conversion of “Low- to High-Auxin” Phenotypes
While the above results with IAA12 suggest that more than a single Leu-to-Ala substitution may be required to eliminate repression with an LxLxLxLxL motif, they do not explain why an identical Leu-to-Ala substitution with an LxLxL motif can eliminate repression with some IAA proteins (e.g. IAA19) but not others (e.g. IAA17). One possibility is that something about the LxLxL repression domain in IAA17 is intrinsically different from that in domain I of IAA19. If domain I of IAA17 was somehow a more potent repression domain than domain I in IAA19, a reasonable prediction would be that additional Leu-to-Ala substitutions in the LxLxL motif in domain I of IAA17 should eliminate repression (i.e. this would be similar to what was observed when additional Leu-to-Ala substitutions were introduced into the LxLxLxLxL motif of IAA12). To test this prediction, we substituted Ala residues for all three Leu residues in the LxLxL motif in domain I of IAA17 (IAA17mImII-3; Fig. 1B). These substitutions should eliminate interaction between the repression domain and the corepressor TPL, as reported for IAA12 (Szemenyei et al., 2008), and plants expressing a 35S:IAA17mImII-3 transgene might be predicted to have “high-auxin” phenotypes, as opposed to “low-auxin” phenotypes. In contrast to expectations, however, plants expressing the 35S:IAA17mImII-3 transgene had “low-auxin” phenotypes (Fig. 5A) only marginally less severe than phenotypes observed with plants expressing the 35S:IAA17mImII-1 transgene (i.e. IAA17 containing a single Leu-to-Ala substitution described above). These results argue against the LxLxL motif in domain I of IAA17 being the determinant that results in low-auxin phenotypes with plants expressing 35S:IAA17mImII-3 (or 35S:IAA17mImII-1) and suggest that some other determinant is responsible for the “low-auxin” phenotypes observed in plants expressing the 35S:IAA17mImII-1 and 35S:IAA17mImII-3 transgenes.
“High-Auxin” Phenotypes Do Not Result from a Cryptic Activation Domain in IAA Proteins
The “high-auxin” phenotypes displayed by 35S:IAA3mImII-1, 35S:IAA6mImII-1, and 35S:IAA19mImII-1 plants could theoretically result from a cryptic activation domain that becomes active upon destruction of the repression domain. Such a cryptic activation domain might substitute for a VP16 activation domain, as described for 35S:VP16-IAA17mImII plants (Li et al., 2009). To address whether IAA19 might contain a cryptic activation domain that could function if the repression domain becomes defective, effector genes encoding a yeast GALACTOSE4 DNA-binding domain (GD) fused to the VP16 activation domain alone, IAA19mImII-1, or VP16-IAA19mImII-1 were tested using a GAL4 UPSTREAM ACTIVATION SEQUENCE promoter:GUS (GAL4:GUS) reporter gene in transfected Arabidopsis protoplasts. The GAL4 system has been described previously by Tiwari et al. (2001, 2003, 2004). Supplemental Figure S4 shows that while transfection of effector constructs encoding GD-VP16 or GD-VP16-IAA19mImII-1 strongly induced expression of the GAL4:GUS reporter gene in an auxin-independent manner, transfection of effector constructs encoding only GD or GD-IAA19mImII-1 failed to induce reporter gene expression. These protoplast transfection experiments argue against the possibility that “high-auxin” phenotypes observed in Arabidopsis plants constitutively expressing IAA19mImII-1 result from a cryptic activation domain in IAA19. While we have not tested IAA3mImII-1 or IAA6mImII-1 in the protoplast system, we believe it is unlikely that this sister pair of IAA19 or the more distantly related IAA3 contains a cryptic activation domain that results in “high-auxin” phenotypes when IAA3mImII-1 or IAA6mImII-1 is constitutively expressed in Arabidopsis plants.
We also tested IAA12mImII-2 for a cryptic activation domain as described above for IAA19mImII-1 and found that expression of a GD-IAA12mImII-2 failed to induce expression of the GAL4:GUS reporter gene in transfected protoplasts (Supplemental Fig. S4). These results suggest that the “high-auxin” phenotypes do not appear to result from a cryptic activation domain in plants transformed with 35S:IAA19mImII-1 and 35S:IAA12mImII-2.
Activation of Auxin Signaling in Plants with the IAA19 Promoter Driving Expression of a Stabilized Version of IAA19 Containing a Domain I Mutation
In addition to using the strong 35S promoter, we used the natural IAA19 promoter to drive expression of the IAA19mImII-1 transgene in an Arabidopsis (Col-0) line containing the auxin-responsive DR5:GUS reporter gene (Fig. 1B; Supplemental Fig. S2). This experiment was conducted to rule out the possibility that the observations made above might be restricted to constructs containing the strong constitutive 35S promoter. With transgenic plants containing the IAA19 promoter driving expression of the GUS reporter gene, Tatematsu et al. (2004) had previously reported that weak GUS histochemical staining was observed in petioles, hypocotyls, and roots of light-grown seedlings. We confirmed these results on the expression pattern with the IAA19 promoter:GUS construct and documented the auxin inducibility of this construct in transformed Arabidopsis seedlings (Supplemental Fig. S5). Even with the relatively weak IAA19 promoter, IAA19:IAA19mImII-1 seedlings displayed “high-auxin” phenotypes with epinastic cotyledons, long hypocotyls, and enhanced DR5:GUS compared with wild-type seedlings (Fig. 2). Compared with the 35S promoter lines, the IAA19 promoter lines had longer and highly curled primary roots, shorter hypocotyls, and a reduction in DR5:GUS expression (Fig. 2). Expression of natural auxin response genes was also elevated in these seedlings compared with the wild type, but less so than that observed with 35S:IAA19mImII-1 seedlings (Fig. 3, compare columns a, e, and f). Adult plants transformed with IAA19:IAA19mImII-1 were smaller than both wild-type and 35S:IAA19mImII-1 plants (Fig. 4).
Because IAA19mImII-1 protein has no apparent cryptic activation domain, this protein must function to bring about “high-auxin” phenotypes without directly activating auxin response genes. To assess whether a well-characterized activation domain could enhance the “high-auxin” phenotypes observed in plants expressing IAA19:IAA19mImII-1, a VP16 activation domain was fused to the N terminus of IAA19mImII-1, and the fusion protein was expressed as an IAA19:VP16-IAA19mImII-1 transgene (Supplemental Fig. S2). Seedlings expressing the VP16 fusion protein had longer hypocotyls, shorter roots, more lateral roots, and increased DR5:GUS histochemical staining compared with plants expressing the IAA19mImII-1 protein (Fig. 2). These results suggest that the IAA19mImII-1 protein is not simply sequestering natural repressors (i.e. through IAA-IAA protein interactions) and preventing their ability to function on auxin response genes. Rather, the results with VP16-IAA19mImII-1 suggest that the IAA19mImII-1 protein is functioning through interactions with ARFs to derepress the expression of auxin response genes whether it possesses or lacks a functional activation domain (see “Discussion”).
DISCUSSION
Our results indicate that mutations in domain I of IAA proteins can have profoundly different consequences in terms of conferring auxin responses when stabilized IAA proteins are constitutively expressed in Arabidopsis plants. We have shown that (1) expression of IAA proteins with a defective repression domain can lead to derepression/activation of auxin response genes and subsequent “high-auxin” phenotypes; (2) depending on the IAA repressor expressed, an identical Ala substitution for the first Leu in the LxLxL motif of domain I can lead to constitutive repression of auxin response genes and “low-auxin” phenotypes or constitutive activation of auxin response genes and “high-auxin” phenotypes; (3) the LxLxLxLxL motif in domain I of IAA12 may represent a more extensive repression domain than the LxLxL motif found in most IAA proteins; and (4) selected IAA proteins may have more than a single repression domain.
How Can Expression of Some Mutant Forms of IAA Proteins Confer “High-Auxin” Phenotypes while Others Confer “Low-Auxin” Phenotypes?
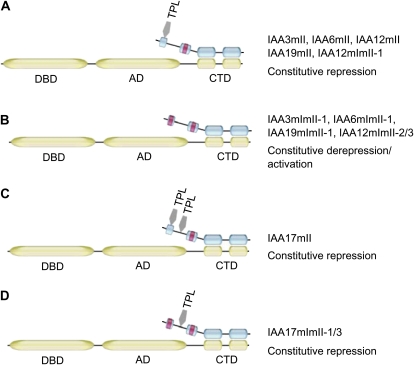
A stabilized repressor such as IAA3mII, IAA6mII, or IAAIAA19mII would be expected to strongly repress auxin response genes by interacting with TPL or a TPL-related corepressor through its domain I and with an ARF activator through its domains III and IV (Fig. 7A). The stabilized repressors would out-compete the natural short-lived IAA repressors for ARF targets and result in constitutive repression of auxin response gene expression and “low-auxin” phenotypes.
Figure 7.
Model for auxin response gene expression with different IAA domain I mutations. In this model, repression or activation of auxin response genes depends on whether a repression domain in an IAA protein functions or not (e.g. whether the repression domain interacts with a corepressor, TPL). A, IAA proteins such as IAA3, IAA6, IAA12, and IAA19 contain a single Leu-rich repression domain (domain I) that interacts with TPL to bring about the repression of auxin response genes. Mutations that destroy the degron in domain II (represented by the red box in domain II) permit auxin-independent, stable association of these IAA proteins with ARF activators, resulting in constitutive repression of auxin response genes (e.g. IAA3mII, IAA6mII, IAA12mII, and IAA19mII). Wild-type repressors like IAA3, IAA6, and IAA12 would also interact with ARF activators to repress auxin response genes, but this repression would be relieved by the destruction of IAA3, IAA6, and IAA12 when auxin levels are elevated. B, An Ala substitution (represented by the red box in domain I) for the first Leu in the LxLxL repression domain of IAA proteins such as IAA3, IAA6, and IAA19 impairs or prevents these IAA proteins from interacting with TPL, resulting in the loss of repression. Mutations in the degron of domain II would again promote auxin-independent, stable association of these IAA proteins with ARF activators (e.g. IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1). This stable association would prevent natural IAA repressors from interacting with ARF activators, which in turn would allow the ARF activation domain to function and result in constitutive activation of auxin response genes. IAA12 has an LxLxLxLxL motif in place of the more common LxLxL motif. Mutation of a single Leu residue in the LxLxLxLxL motif of domain I in IAA12 does not prevent repression, presumably because it can still interact with TPL. Thus, a stable form of IAA12 with a single Ala substitution in domain I (i.e. IAA12mImII-1) results in constitutive repression of auxin response genes (as in A). Additional Ala substitutions in the LxLxLxLxL motif of IAA12 impair the repression domain and result in constitutive activation of auxin response genes when IAA12 is stabilized by a mutation in domain II (e.g. IAA12mImII-2 and IAA12mImII-3). C, IAA proteins such as IAA17 contain a second repression motif located between domains I and II. This motif together with LxLxL in domain I results in repression of auxin response genes, and stabilization of IAA17 results in constitutive repression (e.g. IAA17mII). Wild-type repressors like IAA17 would also interact with ARF activators to repress auxin response genes, but this repression would be relieved by the destruction of IAA17 when auxin levels are elevated. D, An Ala substitution for the first Leu or additional Leu residues in the LxLxL motif of domain I in IAA17 prevents domain I interactions with TPL; however, a second LxLxL motif located between domains I and II in these IAA proteins still functions, and repression is maintained. Stabilization of the IAA17 protein with a nonfunctional repression domain in domain I results in constitutive repression of auxin response genes due to the second repression domain in these proteins (e.g. IAA17mImII-1 and IAA17mImII-3). DBD, DNA-binding domain; AD, activation domain; CTD, carboxyl terminal domain.
Our results with IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1 suggest that these three proteins have lost their capacity to repress auxin response genes, as might be expected for IAA repressors with a defective repression domain. Because the Leu residues in LxLxL or EAR repression motifs are important for repression (Hiratsu et al., 2004; Tiwari et al., 2004), Ala-for-Leu substitutions in the domain I LxLxL motif should impair repression by preventing the mutant IAA proteins from interacting with the TPL or TPL-related corepressor (Szemenyei et al., 2008). A defective repression domain, however, does not explain why expression of Aux/IAA proteins such as IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1 can lead to derepression/activation of auxin-responsive gene expression and “high-auxin” phenotypes. To explain the “high-auxin” phenotypes, we propose a model (Fig. 7) where the mutated domain I would lack repression activity (i.e. lack the ability to interact with TPL or a TPL-related corepressor) but the IAA protein would still interact through domains III and IV with ARF transcriptional activators located on auxin response genes. The stabilized (i.e. domain II mutant) IAA protein would out-compete the natural, unstable IAA repressors and prevent them from reaching their ARF targets. Because the repression domain is neutralized in the domain I mutant IAA proteins, the ARF activation domain would be allowed to function in a constitutive manner. This would be similar to the situation observed in protoplast transfection assays, where ARF activators lacking the C-terminal dimerization domain (i.e. which are unable to interact with IAA repressors) drive constitutive expression of auxin response genes (Tiwari et al., 2003; Wang et al., 2005).
How Might IAA17mImII-1 and IAA17mImII-3 Function as Repressors When the LxLxL Motif in Domain I Is Not Functional?
We have previously reported that constitutive expression of IAA17mII and IAA17mImII (i.e. with two amino acid substitutions in domain I) result in “low-auxin” phenotypes (Li et al., 2009). Here, we have shown that expression of IAA17mImII-1 results in constitutive repression of auxin response genes and “low-auxin” phenotypes even though the LxLxL motif in domain I contains the same Ala-for-Leu substitution as that in IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1. Furthermore, the “low-auxin” phenotypes are still observed with IAA17mImII-3, which has all three Leu residues substituted with Ala in the LxLxL motif of domain I. To explain this paradox, we propose that the domain I repression domain is neutralized in IAA17mImII-1 and IAA17mImII-3, but a second repression domain interacts with TPL or a TPL-related corepressor to bring about constitutive repression of auxin response genes (Fig. 7, C and D).
In fact, IAA17 contains a DLxLxL motif located between conserved domains I and II, and an identical or similar DLxLxL motif is found in IAA7, IAA14, and IAA16 (Supplemental Fig. S6). This consists of DLKLNL for IAA17, IAA4, and IAA16 and DLMLNL for IAA7. Identical or similar LxLxL motifs are found in a wide variety of IAA proteins from other plant species (Supplemental Fig. S6). Based upon a bioinformatics approach, the LxLxL motifs located between domains I and II have been predicted to be EAR-like repression domains in IAA7, IAA14, IAA16, and IAA17 proteins (Kagale et al., 2010). This second DLxLxL motif in IAA proteins is generally located at approximately 20 to 25 amino acids C terminal to the TELC/RLGLPG domain I motif. The second LxLxL motif is part of a more extensive conserved motif with the consensus sequence KRGFSETVDLKLNL. The KR represents part of a bipartite nuclear localization signal (Abel and Theologis, 1995). While most IAA proteins in Arabidopsis and other plants contain a KR motif between domains I and II, many fewer contain a second LxLxL motif. Nothing similar to this second LxLxL motif is found at any position C terminal to domain I in IAA3, IAA6, IAA12, IAA19, or any other Arabidopsis IAA protein than those listed in Supplemental Figure S6.
An acidic residue preceding the LxLxL motif may be important for repression, since a Glu (i.e. in domain I of IAA proteins) or Asp is commonly found adjacent to the first Leu in LxLxL repression domains (Hiratsu et al., 2004; Tiwari et al., 2004; Supplemental Fig. S6). Hiratsu et al. (2004) have reported that a hexapeptide EAR motif with an Asp at position 1 (i.e. DLxLxL) could function as a strong repression domain, whereas an LxLxL motif could not in protoplast transfection assays. Thus, the DLxLxL motif found between domains I and II in a class of phylogenetically related IAA proteins is predicted to function as an EAR-like repression domain. The presence of a second repression domain in IAA17 might also explain why a VP16 activation domain was required to relieve the repression of an IAA17 domain I mutant (Tiwari et al., 2003; Li et al., 2009).
Two previous genetic studies are consistent with IAA7 and IAA17 having two repression domains. The revertant axr2-1-r4 had a Leu-to-Phe substitution in the second L of the domain I LxLxL motif along with a Pro-to-Ser substitution in the GWPP domain II motif of the axr2-1/iaa7 mutant (Nagpal et al., 2000). The axr2-1-r4 mutant did not revert to a wild-type phenotype but had a phenotype intermediate between the wild type and axr2-1. Likewise, the revertant axr3-1R3 had a Leu-to-Phe substitution in the third L of the domain I LxLxL motif along with a Pro-to-Leu substitution in the GWPP domain II motif of the axr3-1/iaa17 mutant (Rouse et al., 1998). The axr3-1R3 mutant, like the axr2-1-r4 mutant, did not revert to the wild type and displayed an intermediate phenotype between the wild type and axr3-1. In these genetic studies, both revertants maintained “low-auxin” phenotypes that were less severe than those displayed by axr2-1 and axr3-1, suggesting that two repression domains may confer stronger repression than a single repression domain. The presence of two repression domains within an IAA repressor might provide a larger target and facilitate the recruitment of TPL or TPL-related corepressors.
Future experiments that test Ala substitutions for Leu in the LxLxL motif found in domain I simultaneously with equivalent substitutions in the LxLxL motif located between domains I and II should reveal whether this second LxLxL motif is a functional repression domain. If this is the case, it is expected (i.e. based on the model in Fig. 7) that constitutive expression of a stabilized IAA protein, like IAA17, with Ala-for-Leu substitutions in both LxLxL motifs, will result in “high-auxin” phenotypes, similar to those observed with IAA3mImII-1, IAA6mImII-1, and IAA19mImII-1.
The Repression Domain in IAA12 Contains an LxLxLxLxL Motif Compared with the LxLxL Motif Found in Most IAA Repressors
The LxLxLxLxL motif in domain I of IAA12 has been shown to function in repression by interacting with the corepressor TPL (Szemenyei et al., 2008). Ala substitutions for the first three Leu residues in this motif were shown to block the interactions between IAA12 and TPL. We found that an identical set of Ala substitutions in 35S:IAA12mImII-3 resulted in “high-auxin” phenotypes in Arabidopsis plants. Two Ala substitutions in 35S:IAA12mImII-2 also conferred “high-auxin” phenotypes, but a single Ala substitution in 35S:IAA12mImII-1 did not. It is possible that an LxLxLxLxL motif like that found in IAA12 might function as a stronger repression domain (e.g. by having a higher affinity or providing a larger target for TPL) than an LxLxL motif. Thus, a single Ala substitution for a Leu in an LxLxLxLxL motif would not be sufficient to inactivate the repression domain, but two or three Ala substitutions would neutralize the repression domain in IAA12 (Fig. 7, A and B). Results reported for the synthetic SRDX repression domain (i.e. LDLDLELRLGFA) are consistent with an LxLxLxLxL motif being a more potent repression domain than an LxLxL or an LxLxLxL repression domain when tested in Arabidopsis plants (Hiratsu et al., 2003).
IAA11 and IAA13 also contain an LxLxLxLxL motif, IAA10 contains an LxLxLxLxI motif, and IAA29 has an LxLxLxL motif. IAA proteins with LxLxLxL or LxLxLxLxL motifs are also found in the sequenced genomes of rice (Oryza sativa), sorghum (Sorghum bicolor), maize (Zea mays), and poplar (Populus trichocarpa; Jain et al., 2006; Kalluri et al., 2007; Wang et al., 2010a, 2010b) and in a wide variety of other plant species, indicating that these more extensive Leu-rich repression domains are conserved. Repression domains of different strengths (e.g. an LxLxL versus an LxLxLxLxL motif or one versus two LxLxL repression domains in the same IAA protein) may provide flexibility in regulating the expression of auxin response genes.
Do Natural Auxin Response Genes Lacking a Functional Repression Domain Play a Unique Role in Regulating Auxin Response Genes?
Arabidopsis itself contains a gene, IAA33, that lacks any obvious domain I or II. Furthermore, there are examples of predicted IAA proteins in a variety of seed plants that lack an obvious domain I or lack both domains I and II, and many of these are more closely related phylogenetically to IAA33 than to other Arabidopsis IAA proteins. For example, rice and poplar have several genes predicted to encode Aux/IAA proteins that lack one or both domains I and II. In poplar, PoptrIAA29.1, PoptrIAA29.2, and Poptr29.3 are reported to be missing domain I and PoptrIAA33.1, PoptrIAA33.2, and PoptrIAA34 appear to be missing both domains I and II (Kalluri et al., 2007). In rice, OsIAA4, OsIAA22, OsIAA27, OsIAA28, and OsIAA29 lack domain I or domains I and II (Jain et al., 2006). Likewise, sorghum (Wang et al., 2010a) and maize (Wang et al., 2010b) along with a variety of other plants have examples of Aux/IAA proteins that lack domain I or both domains I and II.
Whether natural Aux/IAA proteins that lack domain I or both domains I and II play any role in regulating auxin response genes is an open question; however, our results with IAA3mImII-1, IAA6mImII-1, IAA19mImII-1, IAA12mImII-2, and IAA12mImII-3 raise the possibility that Aux/IAA proteins lacking a functional domain I or domains I and II could regulate auxin response gene expression in a manner that differs from most Aux/IAA proteins. There is already evidence that Arabidopsis IAA proteins have diversified in how they regulate auxin response genes through differences in their stability (Dreher et al., 2006; Sato and Yamamoto, 2008). Perhaps this diversification might also include differences in how they regulate auxin response genes without a functional repression domain.
MATERIALS AND METHODS
Vector Constructs and Transgenic Lines
Domain I and domain II mutations in IAA3, IAA6, IAA12, IAA19, and IAA17 were obtained using the QuickChange site-directed mutagenesis kit (Stratagene). The hemagglutinin (HA) epitope tag was fused in frame to the N terminus of IAAmImII to create HA-IAAmImII, as described previously for IAA17 (Tiwari et al., 2001, 2003; Li et al., 2009). The 35S CaMV promoter or native IAA19 promoter was used to drive expression of the IAA transgenes as described previously (Tiwari et al., 2001, 2003). For the IAA19 promoter, a 1.9-kb fragment was amplified from wild-type Arabidopsis (Arabidopsis thaliana Col-0) with the primer pair IAA19 Pro Fw (5′-GTTGTCGACTCAAAACTCGAGTAGAAGCACATG-3′) and IAA19 Pro Rv (5′-CTCCATGGCTTGAACTTCTTTTTTTCCTCTCAC-3′). For DR5:DsRed, the same construct described by Ulmasov et al. (1997) was used, but instead of GUS, DsRed-Express was used. For GD constructs, the GAL4 DNA-binding domain was fused in frame with the N terminus of VP16, IAA12mImII-2, IAA19mImII-1, and VP16-IAA19mImII-1.
All constructs were cloned into the binary vector pPZP for transformation of Arabidopsis Col-0, and the cloning fidelity was confirmed by sequencing. Homozygous lines of DR5:DsRed and transgenic plants in the DR5:reporter background were obtained using Agrobacterium tumefaciens-mediated transformation by the floral dip method (Clough and Bent, 1998). Plants containing the DR5:GUS transgene have been described previously (Ulmasov et al., 1997). Seeds containing the IAA19 promoter:GUS transgene were kindly provided by Dr. Kotaro T. Yamamoto (Hokkaido University; Tatematsu et al., 2004). Five to 10 lines that overexpressed the individual single-copy transgenes were selected. A representative line for each construct was further analyzed. For 35S:IAA17mImII-3, T1 plants were analyzed.
Plant Growth and Treatments
Arabidopsis plants were grown in growth chambers at 22°C under continuous, cool-white fluorescent light. For experiments with seedlings, Arabidopsis seeds were sterilized and germinated on horizontal or vertical plates as described previously (Li et al., 2009). For auxin treatments, 7-d-old seedlings were removed from the agar plates and incubated with shaking in liquid medium containing or lacking 1-naphthalene acetic acid (1-NAA) for the times and at the concentrations indicated. After incubation, seedlings were stained for GUS, monitored for fluorescence (DsRed), or frozen in liquid nitrogen and stored at −70°C prior to RNA isolation. Primary root and hypocotyl lengths were measured using ImageJ (http://rsb.info.nih.gov/ij/) from photographs taken with a Canon digital camera.
Histochemical GUS Staining and Microscopy
GUS staining was carried out as described previously (Ulmasov et al., 1997). Seedlings were placed in GUS solution for 15 h at 25°C or 37°C and cleared in 70% ethanol. GUS staining of IAA19 promoter:GUS (Tatematsu et al., 2004) was monitored with a Leica stereomicroscope. DsRed in transgenic plants was analyzed using a Zeiss LSM 510 laser scanning confocal microscope at the University of Missouri Molecular Cytology Core.
Transient Expression Assays
Transient assays in protoplasts were carried out as described previously (Tiwari et al., 2001, 2003, 2004). In the GAL4 activity assays, 5 μg of effector DNA and 10 μg of reporter DNA were used for cotransfections. Assays were carried out in the presence or absence of 1 μM 1-NAA.
RNA Isolation and qRT-PCR Analysis
RNA was extracted using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s protocol. During RNA isolation, RNase-Free DNase (Qiagen) was used to eliminate any DNA contamination. First-strand cDNA was synthesized from 1 μg of total RNA using an Omniscript RT kit (Qiagen). One microliter of the resulting cDNA was subjected to qRT-PCR using SYBR Green Supermix in a CFX96 Real-Time System (Bio-Rad).
Efficiency of each primer pair was determined prior to qRT-PCR. qRT-PCR was carried out as described previously (Li et al., 2009). TUB5 (AT1G20010) and CBP20 (AT5G44200) were selected as internal references. In each PCR, three technical replicates were used. Data combined from two biological replicates are shown. The expression level of the natural gene in wild-type seedlings was set at 1. Oligonucleotide primers used for qRT-PCR are the same as described by Li et al. (2009) or otherwise listed in Supplemental Table S1.
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative data library under the following accession numbers: IAA3 (AT1G04240), IAA6 (AT1G52830), IAA12 (AT1G04550), IAA17 (AT1G04250), and IAA19 (AT3G15540).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. T1 transgenic seedlings expressing 35S:IAA19mII in the DR5:GUS background.
Supplemental Figure S2. qRT-PCR analysis of expression levels for IAA transgenes in Arabidopsis seedlings.
Supplemental Figure S3. T1 transgenic seedlings expressing 35S:IAA12mImII-1 in the DR5:GUS background.
Supplemental Figure S4. Activation domain assays in Arabidopsis protoplasts.
Supplemental Figure S5. Analysis of seedlings transformed with the IAA19:GUS reporter gene.
Supplemental Figure S6. Amino acid sequences between conserved domains I and II for IAA proteins that contain a second LxLxL motif.
Supplemental Table S1. Primer pairs used for qRT-PCR.
Supplementary Material
References
- Abel S, Nguyen MD, Theologis A. (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Abel S, Theologis A. (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8: 87–96 [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Birnbaum KD. (2009) Positive fluorescent selection permits precise, rapid, and in-depth overexpression analysis in plant protoplasts. Plant Physiol 149: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44: 382–395 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. (2004) Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun 321: 172–178 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6: 47–59 [DOI] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. (2010) Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri UC, Difazio SP, Brunner AM, Tuskan GA. (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT. (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku SJ, Park JY, Ha SB, Kim J. (2009) Overexpression of IAA1 with domain II mutation impairs cell elongation and cell division in inflorescences and leaves of Arabidopsis. J Plant Physiol 166: 548–553 [DOI] [PubMed] [Google Scholar]
- Li H, Cheng Y, Murphy A, Hagen G, Guilfoyle TJ. (2009) Constitutive repression and activation of auxin signaling in Arabidopsis. Plant Physiol 149: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M. (2006) Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J 46: 297–306 [DOI] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A. (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373 [DOI] [PubMed] [Google Scholar]
- Sato A, Yamamoto KT. (2008) Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant 133: 397–405 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang X-J, Hagen G, Guilfoyle TJ. (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Bai Y, Shen C, Wu Y, Zhang S, Jiang D, Guilfoyle TJ, Chen M, Qi Y. (2010a) Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics 10: 533–546 [DOI] [PubMed] [Google Scholar]
- Wang S, Tiwari SB, Hagen G, Guilfoyle TJ. (2005) AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17: 1979–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Deng D, Bian Y, Lv Y, Xie Q. (2010b) Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol Biol Rep 37: 3991–4001 [DOI] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21: 553–562 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.