Abstract
ZmPep1 is a bioactive peptide encoded by a previously uncharacterized maize (Zea mays) gene, ZmPROPEP1. ZmPROPEP1 was identified by sequence similarity as an ortholog of the Arabidopsis (Arabidopsis thaliana) AtPROPEP1 gene, which encodes the precursor protein of elicitor peptide 1 (AtPep1). Together with its receptors, AtPEPR1 and AtPEPR2, AtPep1 functions to activate and amplify innate immune responses in Arabidopsis and enhances resistance to both Pythium irregulare and Pseudomonas syringae. Candidate orthologs to the AtPROPEP1 gene have been identified from a variety of crop species; however, prior to this study, activities of the respective peptides encoded by these orthologs were unknown. Expression of the ZmPROPEP1 gene is induced by fungal infection and treatment with jasmonic acid or ZmPep1. ZmPep1 activates de novo synthesis of the hormones jasmonic acid and ethylene and induces the expression of genes encoding the defense proteins endochitinase A, PR-4, PRms, and SerPIN. ZmPep1 also stimulates the expression of Benzoxazineless1, a gene required for the biosynthesis of benzoxazinoid defenses, and the accumulation of 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside in leaves. To ascertain whether ZmPep1-induced defenses affect resistance, maize plants were pretreated with the peptide prior to infection with fungal pathogens. Based on cell death and lesion severity, ZmPep1 pretreatment was found to enhance resistance to both southern leaf blight and anthracnose stalk rot caused by Cochliobolis heterostrophus and Colletotrichum graminicola, respectively. We present evidence that peptides belonging to the Pep family have a conserved function across plant species as endogenous regulators of innate immunity and may have potential for enhancing disease resistance in crops.
Peptides regulate diverse processes pertaining to both development and defense in plants (Matsubayashi and Sakagami, 2006). Defensively, peptides can act as molecular messengers during plant interactions with other organisms, alerting the plant to potential attack and inducing defenses. Microbe-associated molecular patterns (MAMPs) are molecular fragments recognized by plants as indicators of potential invasion, and peptide MAMPs derived from microbial proteins, such as flg22, elf18, and Pep13, are bound by specific plant pattern-recognition receptors to elicit a cascade of downstream defense responses (Hahlbrock et al., 1995; Zipfel et al., 2004, 2006). Peptides also warn plants of attack by insect herbivores; the inceptin peptide is one such herbivory-associated molecular pattern (HAMP) that activates downstream defenses in response to herbivory (Schmelz et al., 2006; Mithöfer and Boland, 2008).
In addition to peptide MAMP elicitors that alert plants to the presence of invading organisms, there are several classes of endogenous plant peptides that regulate defenses, acting as internal elicitors (Ryan et al., 2007). Biotic stress resulting in cellular damage induces expression of the genes encoding endogenous peptide precursor proteins, and the activated peptides then contribute to defense through the amplification of plant responses. Systemin and hydroxyproline-systemin (HypSys) peptides function as endogenous regulators of defense against herbivores (Ryan and Pearce, 2003; Narváez-Vásquez et al., 2007). Signaling by these peptides promotes a myriad of antiherbivore responses, including the accumulation of proteinase inhibitor proteins and of other antinutritive proteins such as polyphenol oxidase, Thr deaminase, and arginase as well as systemic emission of volatiles (Pearce et al., 1991; Howe and Jander, 2008; Degenhardt et al., 2010). Other peptides are endogenous regulators of pathogen defense responses; recently, soybean (Glycine max) has been discovered to produce a unique peptide signal, GmSubPep, which activates the transcription of pathogen defense genes (Pearce et al., 2010). In Arabidopsis (Arabidopsis thaliana), elicitor peptide 1 (AtPep1) belongs to a family of peptides that interact with the PEPR receptors to regulate the expression of pathogen defense genes, including those encoding the PDF1.2 defensin and PR-1 (Huffaker et al., 2006; Yamaguchi et al., 2006, 2010).
While systemin and AtPep1 are endogenous defense signals as opposed to MAMP/HAMP exogenous elicitors and indicators of nonself, the signaling similarities shared by these peptide regulators closely resemble aspects of MAMP/HAMP-induced signaling (Ryan et al., 2007). AtPep family peptides and peptide MAMPs such as flg22 and elf18 activate similar downstream responses using many of the same molecular components (Ryan et al., 2007; Krol et al., 2010; Postel et al., 2010; Yamaguchi et al., 2010). Both flg22 and AtPeps bind specific Leu-rich repeat receptors, and both activate downstream defense genes through a myriad of downstream second messenger signals, which in addition to jasmonate and hydrogen peroxide are believed to include ethylene (ET), salicylate, and membrane depolarization (Yamaguchi et al., 2006; Huffaker and Ryan, 2007; Krol et al., 2010). The receptors for both flg22 and AtPep1 associate with an interacting receptor partner, BAK1, and likely activate cyclic nucleotide-gated calcium channels via receptor guanylyl cyclase activity (Ma et al., 2009; Postel et al., 2010). Treatment with flg22 up-regulates the transcription of genes encoding PROPEP family precursors and both PEPR receptors, and AtPep1 treatment induces the transcription of FLS2, the flg22 receptor (Zipfel et al., 2004; Ryan et al., 2007).
The breadth of responses regulated by endogenous peptides indicates their potential utility as a mechanism for manipulating resistance, a strategy that has been demonstrated through experiments with transgenic plant lines. Solanaceous plants constitutively expressing the genes encoding prosystemin or proHypSys accumulate herbivore defense proteins to much higher levels than wild-type plants and are more resistant to insect attack (Bergey et al., 1996; Ren and Lu, 2006). Similarly, Arabidopsis plants constitutively expressing the AtPROPEP1 precursor gene have higher basal expression levels of pathogen defense genes and demonstrate resistance to the necrotrophic pathogen Pythium irregulare (Huffaker et al., 2006). Direct application of peptide to plants is also an effective mechanism to manipulate defense; pretreatment of Arabidopsis plants with either flg22 or AtPep1 peptides prior to inoculation with the hemibiotrophic bacterial pathogen Pseudomonas syringae pv tomato DC3000 enhanced plant resistance (Zipfel et al., 2004; Yamaguchi et al., 2010).
Enhanced disease resistance obtained through peptide pretreatment or transgenic constitutive expression indicates that such methods could have potential use in the field, especially if the mechanisms are conserved across species. However, systemin is not active in nonsolanaceous plants, nor are AtPep peptides capable of signaling in other species (Ryan and Pearce, 2003; Yamaguchi et al., 2006). This species specificity has prevented the functional transfer of peptide-enhanced defense to diverse plant species. While a proHypSys ortholog has been identified in Ipomoea batatas, indicating that the systemin superfamily does exist in other species, homologs have not yet been identified in any other plant families (Chen et al., 2008). Whether this lack of identified systemin homologs is because related peptides evolved only in the Solanaceae or because the amino acid sequence of functional homologs has diverged to the point of being unrecognizable in other species is unknown.
Orthologs of AtPROPEP genes have been identified in other plant species through amino acid sequence comparisons. However, those orthologs share little direct sequence identity to AtPROPEP genes. This lack of sequence identity among species is unsurprising, as Arabidopsis peptides that bind the same receptor have precursor amino acid sequence identity between 12% and 47% (Yamaguchi et al., 2006). All Arabidopsis Pep family precursors do share homologous conserved domains, the combination of which has been used as a means of identification of orthologs in other species. First, all PROPEP family orthologs contain the predicted active peptide sequence at the C terminus of a larger precursor protein, a characteristic also shared by many animal peptide hormone precursors and by prosystemin (McGurl et al., 1992; Huffaker et al., 2006). None of the precursors has a traditional signal sequence for export through the secretory pathway, but each does encode an amphipathic helix motif that is potentially a site of protein-protein interactions (Rhoads and Friedberg, 1997; Huffaker et al., 2006). All predicted peptides are enriched in basic amino acids, and each precursor protein has several repeated EKE motifs, consisting of a high density of Asp/Glu residues interspersed with Lys/Arg (McGurl et al., 1992; Realini et al., 1994; Huffaker et al., 2006). None of the genes designated as AtPROPEP orthologs using the above criteria has been studied for functional homology, and it has been suggested that true AtPep1 homologs likely exist only in species closely related to Arabidopsis (Boller and Felix, 2009).
Our studies present evidence that the gene ortholog in maize (Zea mays), ZmPROPEP1, encodes a peptide, ZmPep1, which is an active signal regulating pathogen defense. The ZmPROPEP1 gene is expressed in response to jasmonic acid (JA) treatment and fungal infection. Treatment of leaves with ZmPep1 promotes production of the hormones JA and ET and induces the expression of genes encoding their biosynthetic enzymes, genes associated with pathogen defense, and the ZmPROPEP1 gene. ZmPep1 activates the biosynthesis of benzoxazinoid defenses and promotes the accumulation of 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc), a storage form of a highly reactive aglycone hydroxamic acid. Finally, pretreatment with ZmPep1 prior to infection enhances maize resistance to both the foliar pathogen Cochliobolis heterostrophus and the stalk rot pathogen Colletotrichum graminicola.
RESULTS
Maize Transcribes a Pathogen-Inducible Gene Orthologous to AtPROPEP1
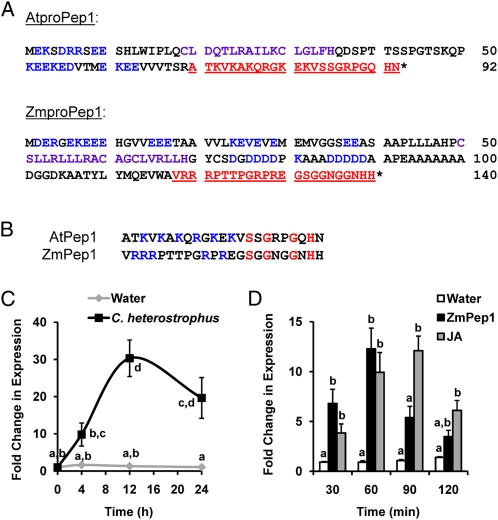
Using the AtPROPEP1 sequence to query National Center for Biotechnology Information maize nucleotide sequences, we identified ZmPROPEP1 as a potential homolog. While the amino acid identity between the two precursors is only 14%, both share the modular structural motifs characteristic of the PROPEP family (Fig. 1A). These motifs include the amphipathic helix motif that is potentially a site of protein-protein interactions, multiple EKE repeats, and location of the active peptides at the C terminus of both precursors. The native length of ZmPep1 is predicted to be 23 amino acids, as are both AtPep peptides that have been isolated biochemically (Huffaker et al., 2006; Pearce et al., 2008). Neither AtPROPEP1 nor ZmPROPEP1 has a conventional signal sequence for export through the secretory pathway, and both are predicted to localize to the cytosol.
Figure 1.
Comparison of the proteins encoded by the AtPROPEP1 and ZmPROPEP1 genes. A, Conserved precursor motifs are the EKE motif (blue), amphipathic helix motif (purple), and bioactive elicitor peptide (red, underlined). B, Comparison of conserved characteristics within the active AtPep1 and ZmPep1 peptides. Basic residues are blue, and identical amino acids are red. C, Average ± se (n = 3) induced ZmPROPEP1 gene expression in leaves by the fungal pathogen C. heterostrophus. D, Average ± se (n = 3) induced expression of the ZmPROPEP1 precursor gene in response to treatment of intact leaves with ZmPep1 or JA. In C, different letters (a–d) represent significant differences within the plot. In D, different letters (a and b) represent significant differences within each time point (all ANOVAs, P < 0.005; Tukey test corrections for multiple comparisons, P < 0.05).
The predicted peptide encoded by the ZmPROPEP1 gene has several conserved residues at the C-terminal end as compared with AtPep1 (Fig. 1B), including the Gly (Gly-17) shown to be essential for AtPep1 bioactivity (Pearce et al., 2008). Like AtPep1, the N-terminal end of ZmPep1 is enriched in basic residues and contains five Arg residues compared with the five Lys residues and one Arg in the N-terminal region of AtPep1 (Fig. 1B). The pI of both peptides is very high, 11.22 and 12.18 for AtPep1 and ZmPep1, respectively.
The maize genomic sequence encoding ZmPROPEP1 was cloned from both var Golden Queen (GQ), a commercially grown sweet corn, and var B73. As in the Arabidopsis AtPROPEP1 gene, both GQ and B73 genes contained a single short intron just upstream of the encoded peptide (Supplemental Fig. S1A). The cloned B73 sequence was found to be identical to database sequences, whereas the GQ gene encoded eight amino acid changes, none of which was in the predicted ZmPep1 peptide (Supplemental Fig. S1B). Several cDNAs encoding the ZmPROPEP1 precursor were amplified from young leaf tissue of 1-month-old GQ plants. Sequencing of six independent cDNA clones revealed that three had the intron alternatively spliced such that the transcripts encoded a precursor with five fewer amino acids (Supplemental Fig. S1B). This differential splicing could potentially contribute to the regulation of peptide processing, as the splice site is just upstream of the region encoding ZmPep1, where proteolytic activity likely would release the active peptide from the precursor.
To ascertain whether the ZmPROPEP1 gene responds to pathogen infection, ZmPROPEP1 transcript abundance was analyzed in intact plants that were infected with the fungus C. heterostrophus versus uninfected control plants. ZmPROPEP1 transcript levels increased in the fungus-infected plants (Fig. 1C). Expression of ZmPROPEP1 was also induced in intact leaves treated with either ZmPep1 peptide or JA but not in leaves treated with water (Fig. 1D).
The ZmPep1 Peptide Activates the Production of JA
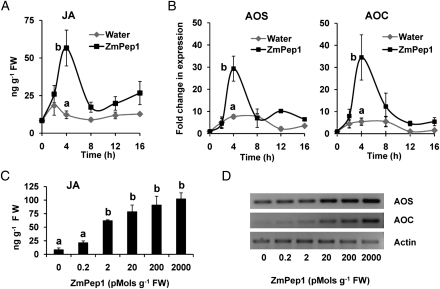
To confirm that ZmPep1 acts as a defense regulator, we quantified JA concentrations in excised leaves supplied with water or ZmPep1. After 4 h, ZmPep1 induced the accumulation of JA to levels 4.6-fold higher than that of control leaves supplied with water (Fig. 2A). To evaluate the dose dependence of ZmPep1 treatment and subsequent JA accumulation, leaves were treated with increasing concentrations of ZmPep1, ranging from 0.2 to 2,000 pmol g−1 fresh weight. After 4 h, JA levels in control leaves supplied with water averaged around half that of leaves supplied with the lowest concentration of ZmPep1 (Fig. 2C). Average JA levels increased with the application of increasing amounts of peptide, with a maximum 10 times that of water-supplied leaves (Fig. 2C). The concentration of ZmPep1 that induced half-maximal JA accumulation fell between 200 fmol and 2 pmol g−1 fresh weight. The vapor phase extraction method followed by gas chromatography (GC)-mass spectrometry (MS) analysis of JA allowed us to simultaneously measure salicylic acid, levels of which were not observed to change in our experiments.
Figure 2.
ZmPep1 induces JA accumulation and regulates the expression of related biosynthetic genes. A and B, Time-course analysis of JA levels (A) and AOS and AOC gene expression (B) in excised leaves supplied with water or ZmPep1 (2 nmol g−1 fresh weight). Relative transcript abundance levels were examined using semiquantitative PCR with actin as a control. C and D, Dose dependence of JA levels (C) and AOS and AOC gene expression (D) in response to ZmPep1 at 4 h. Each sample was a pool of two leaves (n = 3; ±se). At the time point of greatest mean change, different letters (a and b) represent significant differences (all ANOVAs, P <0.02; Tukey test corrections for multiple comparisons where applicable, P < 0.05). FW, Fresh weight.
In excised leaves, expression of the gene encoding allene oxide synthase (AOS) was wound inducible; however, leaves supplied with ZmPep1 exhibited a 3.8-fold greater induction of AOS transcript than did wounded leaves supplied with water (Fig. 2B). Expression of the allene oxide cyclase (AOC) gene was more specifically induced by ZmPep1 treatment. Compared with unwounded leaves at time zero, excised water-supplied leaves displayed modest 5-fold increases in transcript while ZmPep1-treated leaves exhibited a 30-fold induction (Fig. 2B). Maximal increases in AOC transcript abundance also occurred at 4 h. Similar to JA production, ZmPep1-induced expression of both the AOS and AOC genes was dose dependent. At 4 h, relative transcript levels of both genes showed increases in abundance starting at ZmPep1 applications of 20 pmol g−1 fresh weight (Fig. 2D).
ZmPep1 Induces ET Emission
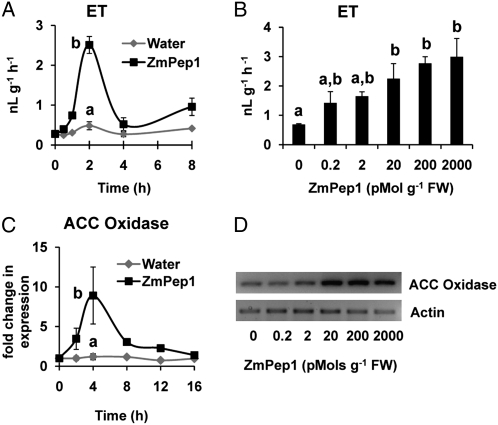
Given that ET commonly interacts with JA to regulate pathogen defenses, ET production was also investigated. After 2 h, ZmPep1-supplied leaves emitted a 5-fold increase in ET compared with water-supplied leaves (Fig. 3A). ZmPep1-induced ET production was dose dependent, and average emissions increased as the amount of peptide supplied to leaves increased (Fig. 3B). Expression of the gene encoding 1-aminocyclopropane-1-carboxylic acid oxidase (ACC Ox) also responded to ZmPep1 treatment. ZmPep1 induced an 8-fold increase in transcript levels above those detected in water-treated leaves, which showed no measurable change in ACC Ox expression (Fig. 3C). Similar to AOS and AOC genes, 20 pmol g−1 fresh weight ZmPep1 was observed to be the threshold level for effects on ACC Ox gene expression (Fig. 3D). While peak levels of ET emission occurred after 2 h of treatment, increased expression of the ACC Ox gene was greatest after 4 h. This implies that the early induction of ET in ZmPep1-treated leaves occurs either through activation of ACC Ox enzyme activity or translational activation rather than increases in transcription.
Figure 3.
ZmPep1 regulates the emission of ET and the expression of an ET biosynthesis gene. A, Time course of average (n = 4; ±se) induced ET emission in excised leaves supplied with water or with ZmPep1 (2 nmol g−1 fresh weight). B, Dose dependence of average (n = 4; ±se) ZmPep1-triggered ET emission at 2 h. C, Time course of average (n = 3; ±se) ACC Ox transcriptional changes in response to water or ZmPep1 (2 nmol g−1 fresh weight). D, Concentration effects of ZmPep1 on induced ACC Ox expression at 4 h. Relative transcript abundance levels were examined using semiquantitative PCR with actin as a control. Different letters (a and b) represent significant differences (all ANOVAs, P <0.05; Tukey test corrections for multiple comparisons where applicable, P < 0.05). FW, Fresh weight.
ZmPep1 Regulates the Expression of Pathogen Defense Genes
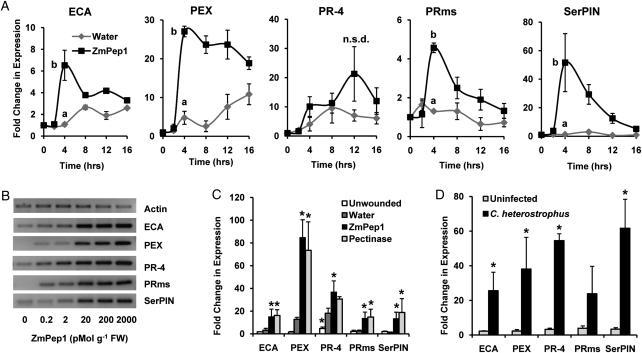
To examine defense processes associated with ZmPep1-activated production of JA and ET, we examined the expression of established defense marker genes (Doehlemann et al., 2008; Erb et al., 2009). Endochitinase A (ECA), pathogenesis-related 4 (PR-4), pathogenesis-related maize seed protein (PRms), and peroxidase (PEX) genes have been shown to be pathogen inducible in microarray experiments (Doehlemann et al., 2008), whereas SerPIN encodes a Bowman-Birk trypsin inhibitor that is strongly induced by JA treatment, elicitors, and biotic stresses (Erb et al., 2009).
Expression of all five genes was elevated in excised leaves that had been supplied with ZmPep1. Within 4 h, ECA transcript abundance increased 6-fold in ZmPep1-treated leaves as compared with unwounded control leaves (Fig. 4A). After longer treatment times, ECA transcripts were also modestly induced by wounding. PEX transcripts demonstrated a 25-fold increase in ZmPep1-treated leaves at 4 h, remaining elevated at 16 h. At 4 h, PEX was not strongly wound inducible, but at later time points, excision resulted in a gradual increase in transcription (Fig. 4A). Transcription of PR-4 was wound responsive in the excised leaves, and at early treatment times it was induced similarly by both water and ZmPep1 treatment. At 12 h, PR-4 transcripts accumulated to 2-fold higher levels in the ZmPep1-supplied leaves compared with water-supplied controls; however, this response was not statistically significant (Fig. 4A). Expression of the gene encoding PRms was modestly but consistently increased 4-fold higher than water-supplied or unwounded control leaves (Fig. 4A). At 4 h, ZmPep1 treatment resulted in the accumulation of transcript encoding SerPIN, to average levels 50-fold higher than those observed in either excised or unwounded 0-h control leaves (Fig. 4A).
Figure 4.
ZmPep1 activates defense gene expression. A, Average fold changes in transcript abundance over time in excised leaves treated with water or ZmPep1 (2 nmol g−1 fresh weight) versus untreated leaves. B, Dose dependence of ZmPep1-induced transcriptional changes in excised leaves at 4 h with actin amplification as a control. FW, Fresh weight. C, Local average changes in defense gene transcription in intact plants 4 h after application of water, 25 pmol of ZmPep1, or pectinase elicitor to a wound site. D, Local average changes in transcript abundance in intact plants 24 h after inoculation with 5 × 103 C. heterostrophus spores. For graphs in A, at the time point of greatest mean change, different letters (a and b) represent significant differences (all ANOVAs, P < 0.02; Tukey test corrections for multiple comparisons, P < 0.05). For graphs in C and D, asterisks represent significant differences from the water-treated control (P < 0.05). n.s.d. (not statistically different) indicates ANOVA P > 0.05. For all graphs, n = 3 (±se).
For each defense marker gene studied, the induced magnitude of change in transcript abundance was found to be dose dependent; excised leaves treated with ZmPep1 for 4 h displayed increased defense gene expression with increasing amounts of peptide application (Fig. 4B). Changes in transcriptional abundance of the gene encoding PRms were observed at the lowest ZmPep1 treatment level, 200 fmol g−1 fresh weight, while expression of PR-4 was clearly enhanced at 2 pmol g−1 fresh weight. Transcription of the ECA, PEX, and SerPIN genes was strongly induced in leaves supplied with 20 pmol g−1 fresh weight ZmPep1.
ZmPep1-induced gene expression was also observed in intact plants using 25 pmol of peptide solution applied to a small wound site (Fig. 4C). Transcript abundance of all five defense-associated genes was found to increase in the ZmPep1-treated leaves relative to wounded leaves treated with water, similar to the excised leaf assay. In intact plants, Rhizopus-derived pectinase elicitor also induced increased transcript abundance of each defense gene to comparable levels as ZmPep1 (Fig. 4C). Average expression of all five genes was also up-regulated in leaf tissue after 24 h of C. heterostrophus infection (Fig. 4D).
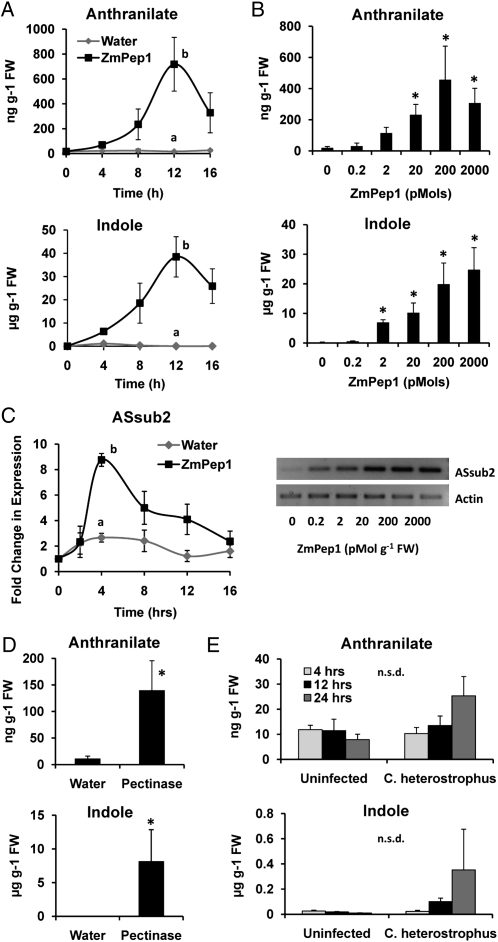
ZmPep1 Promotes Accumulation of the Defense Precursor Metabolites Anthranilate and Indole
To examine metabolites that fuel the biosynthesis of chemical defenses in maize, we examined benzoxazinoid hydroxamic acid-related precursors (Romero et al., 1995; Frey et al., 2009). ZmPep1 treatment increased the leaf concentrations of both anthranilate and indole after 12 h (Fig. 5A). Anthranilate increased from approximately 20 ng g−1 fresh weight in water-treated leaves to more than 700 ng g−1 fresh weight in peptide-treated leaves. Indole increased 30-fold in wounded leaves and more that 1,300-fold in ZmPep1-supplied leaves. Accumulation of anthranilate and indole in leaf tissue correlated to the amount of ZmPep1 used to treat the leaves (Fig. 5B). Peak induction was achieved at 200 pmol g−1 fresh weight.
Figure 5.
ZmPep1-induced defense-associated metabolites. A, Average anthranilate and indole accumulation in excised leaves. B, Dose dependence of ZmPep1-induced anthranilate and indole accumulation in excised leaves. C, Time course and dose dependence of ZmPep1-induced changes in expression of the ASsub2 gene. Transcript abundance was normalized via comparison with an actin control and expressed as fold change relative to an untreated leaf. For all experiments, unless otherwise indicated, ZmPep1 was supplied at 2 nmol g−1 fresh weight. Each sample was a pool of two leaves. For graphs in A and C, at the time point of greatest mean change, different letters (a and b) represent significant differences (all ANOVAs, P < 0.02; Tukey test corrections for multiple comparisons, P < 0.05). For graphs in B, asterisks represent significant differences from the water-treated control (P < 0.04). n.s.d. (not statistically different) indicates ANOVA P > 0.05. For all graphs, n = 3 (±se). FW, Fresh weight.
To determine whether the increase in anthranilate corresponded with transcriptional regulation of biosynthetic enzymes, expression of the gene encoding anthranilate synthase subunit 2 (ASsub2) was analyzed. Transcript abundance of ASsub2 increased in ZmPep1-treated leaves relative to water-treated leaves and was greatest after 4 h of treatment (Fig. 5C). Induced expression of the gene was dose dependent, and observable increases in transcript abundance were apparent at peptide concentrations as low as 200 fmol g−1 fresh weight (Fig. 5C). Accumulation of both anthranilate and indole occurred in leaves treated with a fungus-derived pectinase elicitor, with observed levels of both metabolites peaking at 12 h (Fig. 5D). Infection with C. heterostrophus only weakly influenced levels of anthranilate and indole in the leaves at the time points examined (Fig. 5E).
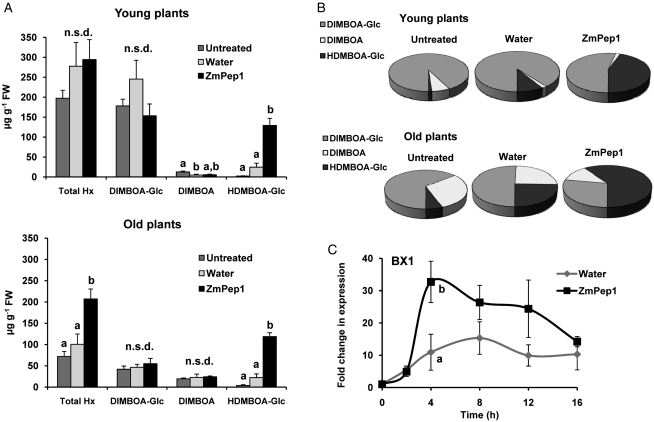
ZmPep1 Activates the Biosynthesis of Benzoxazinoid Defenses
Given the above patterns of induced anthranilate, a precursor to indole-derived benzoxazinoid hydroxamic acid defenses, the effect of ZmPep1 on benzoxazinoid metabolism was examined using intact plant assays (Romero et al., 1995; Frey et al., 2009). In both young plants and old plants, there was no significant difference in free 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA) or DIMBOA-Glc (Fig. 6A). However, HDMBOA-Glc responded modestly to wounding and strongly to peptide treatment.
Figure 6.
Effect of ZmPep1 treatment on benzoxazinoid defenses. A, Levels of benzoxazinoids in intact leaves after 24 h of treatment with water or 25 pmol of ZmPep1 (n = 4; ±se). FW, Fresh weight. B, Relative ratios of benzoxazinoids to one another in untreated plants versus water- and ZmPep1-treated plants. C, Time course of transcript accumulation of BX1 in leaves treated with water or ZmPep1. Relative transcript abundance was measured by semiquantitative reverse transcription-PCR with normalization to an actin control (n = 3; ±se). For all graphs in A, different letters (a and b) represent significant differences (all ANOVAs, P < 0.02; Tukey test corrections for multiple comparisons, P < 0.05). n.s.d. (not statistically different) indicates ANOVA P > 0.05. For the graph in C, at the time point of greatest mean change, different letters (a and b) represent significant differences (all ANOVAs, P < 0.05; Tukey test corrections for multiple comparisons, P < 0.05).
Young plants had a 3-fold higher basal total hydroxamic acid content than older plants (Fig. 6A). Total hydroxamic acid content in young plants was modestly increased by ZmPep1 treatment, but in older plants, the total hydroxamic acids more than doubled in response to ZmPep1, indicating that de novo hydroxamic acid synthesis was required (Fig. 6A). When the ratio of HDMBOA-Glc was compared with DIMBOA and DIMBOA-Glc in leaves, it accounted for an increased percentage of hydroxamic acid content in both young and old plants, but whereas DIMBOA-Glc predominated in young plants, HDMBOA-Glc became the predominant hydroxamic acid in old plants (Fig. 6B).
Production of indole by Benzoxazineless1 (BX1) is the first committed enzymatic reaction leading to benzoxazinoid synthesis. Expression of the gene encoding BX1 is responsive to biotic stresses, and modulation of BX1 expression is a mechanism regulating benzoxazinoid pathway activity (Frey et al., 2009; Niemeyer, 2009). To ascertain whether ZmPep1 might activate metabolic flux through the pathway by inducing BX1 gene expression, BX1 transcript abundance was analyzed in leaves. After 4 h of treatment with ZmPep1, BX1 transcripts accumulated to 30-fold higher levels than were found in time-zero control leaves (Fig. 6C). Excised leaves in water also had increased BX1 expression, but only 10-fold higher than that of time-zero control leaves.
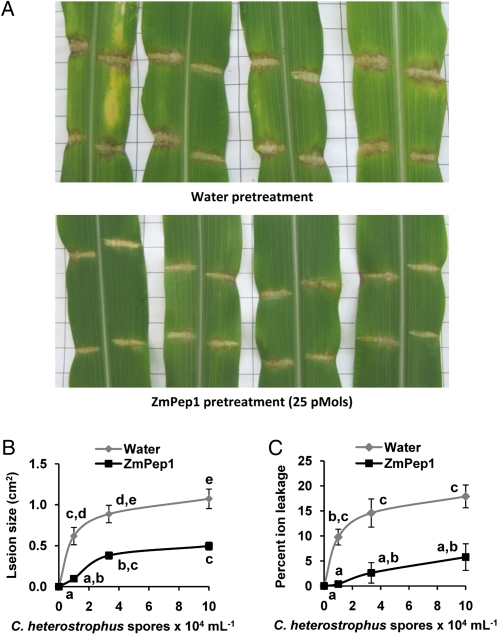
ZmPep1 Enhances Resistance to Southern Leaf Blight Disease
Because ZmPep1 activates the production of JA and ET, the expression of pathogen defense genes, and the accumulation of HDMBOA-Glc, we hypothesized that pretreatment of plants would improve plant disease resistance. To test this hypothesis, intact plants were treated with water or with ZmPep1 at 18 h prior to inoculation with C. heterostrophus, a fungal necrotroph that is the causative agent of southern leaf blight. Chlorotic lesions spread from the wound sites of infected leaves that had been pretreated only with water after 3 d (Fig. 7A). In the ZmPep1-pretreated leaves, lesions were contained at the edge of the wound site and had not spread. C. heterostrophus-induced lesion area in ZmPep1-treated leaves was less than half that of water-treated leaves even at high inoculation loads of C. heterostrophus (Fig. 7B).
Figure 7.
ZmPep1 pretreatment induces resistance to C. heterostrophus (southern leaf blight disease [SLB]). A, Lesions in leaves pretreated with water or with 25 pmol of ZmPep1 at 3 d after SLB infection. B, Average lesion area in plants pretreated with water or ZmPep1. C, Average SLB-induced cell death in samples from water- or ZmPep1-pretreated leaves as measured by the average percentage increase in ion leakage versus samples from uninfected leaves. For lesion analysis, n = 16 (±se); for ion leakage, samples were pools of four leaves, n = 4 (±se). For both graphs, different letters (a–e and a–c) represent significant differences (all ANOVAs, P < 0.001; Tukey test corrections for multiple comparisons, P < 0.05).
Leaves that had been pretreated with water had increased cell death, as estimated by ion leakage, relative to leaves that had been pretreated with ZmPep1 (Fig. 7C). As spore inoculation levels increased, subsequent average ion leakage from the infected leaves also increased. Across all inoculum levels, ZmPep1-pretreated leaves were more resistant to C. heterostrophus-induced cell death. At the lowest concentration of fungal inoculum applied, percentage ion leakage was 20-fold less in ZmPep1-pretreated leaves as compared with water controls.
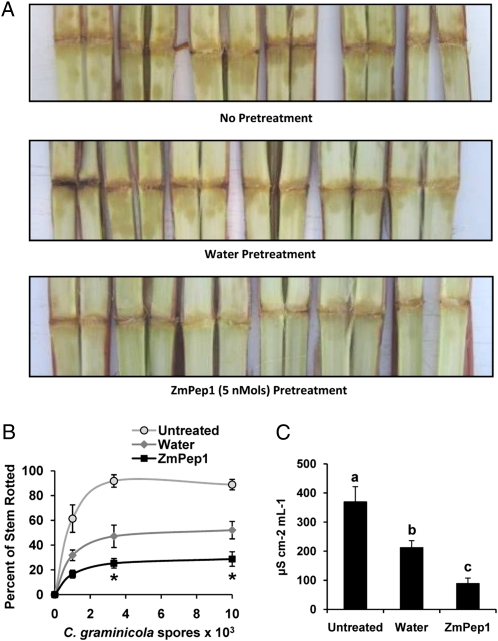
ZmPep1 Enhances Resistance to Anthracnose Stalk Rot
To examine ZmPep1 activity in stems, resistance to anthracnose stalk rot was examined. In plants pretreated with water, the progression of lesion spread from the infected node was less than in control plants, indicating that wounding alone enhanced resistance (Fig. 8A). As compared with either untreated or water-treated plants, the nodes of ZmPep1-treated plants displayed very little necrotic spread.
Figure 8.
ZmPep1-induced resistance to anthracnose stalk rot caused by C. graminicola. A, Necrosis in stems that were pretreated for 3 h with water or 5 nmol of ZmPep1. B, Percentage of stem rotted at 4 d after infection. C, C. graminicola-induced cell death as measured by ion leakage. For all data shown, n = 5 (±se). For the graph in B, asterisks represent significant differences from the water-treated control (P < 0.002). For the graph in C, at the time point of greatest mean change, different letters (a–c) represent significant differences (all ANOVAs, P < 0.001; Tukey test corrections for multiple comparisons, P < 0.05).
After 4 d, greater than 90% of the stalk area was necrotic in untreated control stems that had been inoculated with 3.3 × 103 C. graminicola conidia (Fig. 8B). Stalks of plants that were pretreated with water were 45% to 50% necrotic at high inoculation levels, whereas ZmPep1 pretreatment resulted in only 25% stem rot. Measurements of conductivity to ascertain the extent of cell death as indicated by ion leakage revealed a similar trend. ZmPep1-pretreated stalks had less than a 100 μS cm−2 increase, less than half that of water-pretreated stems and one-quarter that of directly infected controls (Fig. 8C).
ZmPep1 Is an Active Signal in Several Varieties of Maize
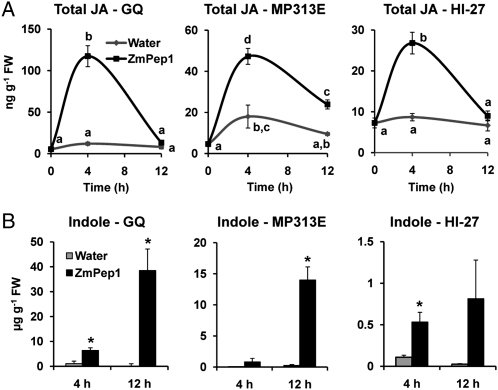
To elucidate whether ZmPep1 modulates responses in other maize varieties, excised leaves of pathogen-resistant lines HI-27 and MP313E were supplied with peptide and levels of JA and indole were quantified. JA accumulated in leaves of all three maize lines after 4 h of ZmPep1 treatment (Fig. 9A). Similarly, indole was observed to increase in ZmPep1-treated leaves of the three lines (Fig. 9B). For both MP313E and HI-27, the magnitude of indole production was less than in GQ, but the peptide-treated leaves were observably induced compared with water-treated leaves. It may be that these varieties are less sensitive to ZmPep1 as a signal or that the selection of indole as a defense marker metabolite is not ideal for all maize lines.
Figure 9.
ZmPep1 promotes the production of JA and defense-related metabolites in multiple maize varieties. A, Time course of induced JA in excised leaves supplied with water or ZmPep1. B, Indole measured in excised leaves. C, Anthranilate levels in excised leaves. ZmPep1 was supplied at 2 nmol g−1 fresh weight. Samples were pools of two leaves, n = 3 (±se). For the graphs in A, different letters (a–c) represent significant differences (all ANOVAs, P < 0.001; Tukey test corrections for multiple comparisons, P < 0.05). For the graphs in B, asterisks represent significant differences from the water-treated control (P < 0.05). FW, Fresh weight.
DISCUSSION
We demonstrate that ZmPep1 acts as a defense-regulating signal and extend the characterization of this family of peptides beyond Arabidopsis. This work examined the molecular and biochemical defenses induced by ZmPep1 that are collectively associated with resistance against invading microorganisms. The maize ZmPROPEP1 ortholog of AtPROPEP1 is functionally homologous, and the gene is transcribed in response to both JA and pathogen infection. As does AtPep1, the ZmPep1 peptide activates numerous components of the innate immune response. This maize peptide-activated defense response was characterized by the production of defense-related phytohormones, induced expression of pathogen defense genes, accumulation of benzoxazinoid defenses, and enhanced resistance to multiple pathogens.
Like other endogenous peptide regulators of defense, ZmPep1 functions through the activation of oxylipin signaling, inducing both expression of JA biosynthetic genes and JA accumulation (Howe et al., 1996; Huffaker and Ryan, 2007). Furthermore, ET is also a component of ZmPep1 signaling; the peptide activates expression of the gene encoding ACC Ox and promotes ET emission in a dose-dependent manner. Coordinated activity of JA and ET signaling regulates pathogen defense responses in many plants (Rojo et al., 2003; Glazebrook, 2005; Bari and Jones, 2009). While the molecular mechanisms regulating pathogen defense responses in maize are not as well characterized as in plants such as Arabidopsis, evidence is accumulating that cooperative JA/ET signaling is a conserved motif of defense initiation. Both JA and ET are produced by maize in response to biotic stress and insect elicitor treatment (Schmelz et al., 2003, 2009). Additionally, elicitor-modulated JA/ET signaling by Trichoderma virens is proposed as the mechanism by which this beneficial fungus activates induced systemic resistance in maize (Djonovic et al., 2007). Our results demonstrating that ZmPep1 regulates JA, ET, and pathogen resistance support the cooperative role of these hormones as signals for maize pathogen defense.
In addition to mediating the production of JA and ET, ZmPep1 also promoted increased transcript abundance for genes encoding antimicrobial and defense signaling proteins. Consistent with the proposed role of ZmPep1 as an endogenous elicitor, many defense-related transcripts were also induced by infection with C. heterostrophus, the exogenous fungal elicitor pectinase. The PR-4 and ECA genes regulated by ZmPep1 are activated by pathogen attack and encode chitinase proteins likely to have direct antifungal activity through degradation of fungal cell walls. In germinating maize embryos, PR-4 gene expression is stimulated by inoculation with fungi and by fungal elicitor extracts; it is inducible in leaves by JA, abscisic acid, and wounding (Bravo et al., 2003). Both PR-4 and ECA transcripts also accumulate in Ustilago maydis-infected ears (Bravo et al., 2003; Doehlemann et al., 2008).
In addition to genes encoding antimicrobial PR proteins, ZmPep1 induced expression of the PRms gene, a homolog of the tobacco (Nicotiana tabacum) PR-1 family that is induced by fungal infection (Casacuberta et al., 1992). Rather than having direct antimicrobial activity, PRms acts as a defense regulator. In both rice (Oryza sativa) and tobacco, constitutive PRms gene expression was found to increase basal levels of defense gene transcripts and to confer enhanced resistance to infection by several pathogens (Murillo et al., 2003; Gómez-Ariza et al., 2007). This up-regulation of defense by PRms is proposed to occur through the modulation of Suc-mediated signaling, raising the intriguing possibility that in addition to activating defense through JA/ET hormone signaling, ZmPep1 may promote disease resistance through PRms-mediated sugar signaling events as well (Gómez-Ariza et al., 2007).
ZmPep1-induced PEX may detoxify reactive oxygen species generated through cellular damage or signaling or may cross-link lignin, cellulose, and extensin to strengthen cell walls against attacking organisms (Lagrimini et al., 1987; Hiraga et al., 2001). SerPIN may act in direct defense, since it is a Ser proteinase inhibitor that could inhibit digestive proteases from both insect and microbial invaders (Ryan, 1989). However, serpin family proteins are also regulators of proteolytic signaling cascades required for innate immune responses in mammals and insects (Law et al., 2006). Furthermore, a serpin in Drosophila melanogaster, termed Necrotic, modulates signaling by spätzle, an endogenous peptide signal mediating Drosophila innate immune responses (Levashina et al., 1999). It remains to be determined whether ZmPep1-induced SerPIN acts in direct defense or as a signaling modulator.
While the antimicrobial and signaling-related genes up-regulated by ZmPep1 are likely factors contributing to induced disease resistance, small molecule defenses are also likely to contribute. Benzoxazinoids are indole-derived hydroxamic acid defenses in poaceous plants that are associated with herbivore and pathogen resistance (Niemeyer, 2009). Cellular damage caused by attacking organisms releases reactive benzoxazinoids from their glycosylated precursors (Frey et al., 2009). Maize seedlings and young tissues have relatively high concentrations of DIMBOA and the glucoside DIMBOA-Glc, which are believed to help protect these essential tissues; however, the role of benzoxazinoids in older plants is not as well defined (Niemeyer, 2009).
Neither DIMBOA-Glc nor free DIMBOA was found to accumulate in response to ZmPep1, but HDMBOA-Glc was induced in ZmPep1-treated leaves. The second methoxyl group on HDMBOA renders the molecule less stable and more reactive than DIMBOA (Maresh et al., 2006). With respect to invading organisms, HDMBOA seems to have multiple functions, capable of acting as both a toxin and a negative effector of pathogenicity. HDMBOA-Glc is a component of maize defense against southwestern corn borer, Diatraea grandiosella, in resistant varieties (Hedin et al., 1993). Southwestern corn borer-resistant maize lines are enriched in HDMBOA content compared with susceptible lines, and HDMBOA was shown to be directly toxic to larvae. HDMBOA is also a predominant constituent of maize root exudates and is postulated to generate a continuously maintained defensive zone in the soil surrounding the roots (Zhang et al., 2000). In root studies, HDMBOA did not act to prevent colonization of roots by Agrobacterium tumefaciens, but it was found to decompose into an o-imidoquinone intermediate that inhibited A. tumefaciens virulence gene expression (Maresh et al., 2006).
Specific accumulation of HDMBOA-Glc is inducible in both wheat (Triticum aestivum) and maize by treatment with JA, pathogen infection, and herbivory (Bücker and Grambow, 1990; Oikawa et al., 2001, 2002, 2004). In these studies, accumulation of HDMBOA-Glc seemed to occur in direct correlation to reduced levels of DIMBOA-Glc, implying that HDMBOA-Glc was generated through methoxylation of existing DIMBOA-Glc pools rather than through de novo hydroxamic acid biosynthesis (Oikawa et al., 2001). For ZmPep1-induced HDMBOA-Glc accumulation, the proportion of HDMBOA-Glc relative to DIMBOA-Glc was increased; however, the increase in HDMBOA-Glc did not come at the expense of DIMBOA-Glc. Rather, we observed that total hydroxamic acid content increased in the ZmPep1-treated leaves of older plants, indicating that the peptide activated de novo synthesis of benzoxazinoids that was channeled into HDMBOA-Glc production. ZmPep1-induced expression of the BX1 gene, encoding an indole glycerol lyase that catalyzes the first committed step in benzoxazinoid production, also supports enhanced metabolic flux into the pathway (Melanson et al., 1997; Frey et al., 2009).
Manipulation of innate immune responses by ZmPep1 caused enhanced disease resistance. ZmPep1-treated plants displayed decreases in both lesion size and cell death in leaves challenged with the necrotroph C. heterostrophus and in stems challenged with the hemibiotroph C. graminicola. Mechanisms of maize resistance to both of these pathogens are still poorly understood. C. heterostrophus is divided into two subgroups based upon toxin production: race T produces toxin, and race O does not. Race O is an endemic pathogen in hot and humid climates and continues to cause disease resulting in lost yield, particularly along the south Atlantic coast (Byrnes et al., 1989). Although a single recessive locus, rhm1, exists that can confer resistance to southern leaf blight disease through an unknown mechanism, most maize lines currently grown rely upon additive quantitative traits that confer partial resistance (Simmons et al., 2001; Balint-Kurti and Carson, 2006). Similar to C. heterostrophus, resistance to C. graminicola is also primarily quantitative and polygenic (Venard and Vaillancourt, 2007). C. graminicola is common in maize fields across the United States, and while the fungus is able to infect most maize tissue, it primarily causes yield losses due to anthracnose stalk rot (Bergstrom and Nicholson, 1999; Venard and Vaillancourt, 2007).
Colletotrichum species are known to actively evade and suppress plant defense, but the fungus was unable to overcome the defense responses preactivated by ZmPep1 treatment (Münch et al., 2008). Transgenic Arabidopsis plants constitutively expressing AtPROPEP1 exhibited increased basal levels of the same genes that were induced in wild-type plants by treatment with AtPep1 (Huffaker et al., 2006). This constitutive induction of basal immunity resulted in increased pathogen resistance (Huffaker et al., 2006). Transgenic maize plants constitutively expressing the ZmPROPEP1 gene may also display higher basal levels of the genes and metabolites that were observed in plants ectopically treated with ZmPep1 peptide. Several molecular studies of maize resistance to attacking organisms have indicated that resistance is associated with increased basal levels of defense gene expression and defense metabolite accumulation similar to those induced by ZmPep1 (Hedin et al., 1993; Niemeyer, 2009; Alessandra et al., 2010). The ability of ZmPep1 to elicit defense signaling and metabolite accumulation in multiple maize lines indicates that constitutive expression through transgenic means could yield results across varieties.
Because endogenous peptide regulators such as ZmPep1 activate multiple defense pathways rather than one gene or metabolite, they may provide a potentially useful strategy to contribute to quantitative resistance through manipulation of a single gene. In crop plants, quantitative disease resistance relies on the additive effects of multiple defenses to provide broad-spectrum partial resistance to many different pathogens (Wisser et al., 2006; Poland et al., 2009). Although quantitative resistance is highly desirable, direct incorporation of this trait into crop development is difficult because of its combinatorial nature; the additive effects that make this resistance robust and versatile also make it difficult to manipulate. Transgenic modulation of peptide signaling has already shown promise as a mechanism for manipulating quantitative resistance. For example, the gene encoding EFR, a Brassicaceae-specific pattern recognition receptor that binds a bacterial peptide MAMP to elicit broad innate immune responses, was ectopically expressed in Nicotiana benthamiana and tomato (Solanum lycopersicum; Lacombe et al., 2010). Transgenic expression of this receptor enhanced resistance to diverse bacterial species by facilitating the recognition of attack and activation of a broad spectrum of defense responses. Constitutive expression of the ZmPROPEP1 gene could similarly confer quantitative resistance effects through simultaneous up-regulation of basal levels of defense responses in maize plants. Furthermore, because orthologs of the precursors to AtPep1 and ZmPep1 have been identified across the plant kingdom, this strategy of endogenous peptide manipulation of defense responses could potentially be used to enhance disease resistance in many diverse plant species.
MATERIALS AND METHODS
Plant and Fungal Materials
Maize (Zea mays) varieties used were B73, HI-27, MP313E, and GQ. All were potted in professional grower’s soil mix (Piedmont Pacific) blended with 14-14-14 Osmocote (Scotts). All varieties were cultivated in a greenhouse under the following conditions: 12-h photoperiod with a minimum of 300 μmol m−2 s−1 photosynthetically active radiation supplied by supplemental lighting. Relative humidity was maintained at 70%, and temperature cycled between 24°C at night and 28°C during the day.
Cochliobolus heterostrophus was isolated from leaf material of an infected maize plant growing in Gainesville, Florida. The specimen was streaked on half-strength potato dextrose agar and subcultured until pure isolates were obtained. The fungus was identified by the Florida Extension Plant Disease Clinic at the University of Florida through macroscopic colony appearance, examination of morphology under both dissecting and light microscopy, and PCR analysis of fungal DNA with species-specific primers. Spore suspensions of C. heterostrophus were prepared in 30% glycerol/0.1% Tween and stored at −80°C. For each bioassay, an aliquot of glycerol stock was used to generate a fresh working culture on half-strength potato dextrose agar (Sigma-Aldrich) that was incubated for 2 weeks at 26°C. Colletotrichum graminicola strain M1.001 was acquired from Dr. Jeffrey Rollins (Department of Plant Pathology, University of Florida), and conidial spore stocks were prepared in 30% glycerol/0.1% Tween and stored at −80°C. For each assay, a fresh working culture was prepared by spotting glycerol stock onto V8-agar plates and incubated for 1.5 to 2 weeks at 26°C.
Peptide and Precursor Gene Identification
The previously identified AtPROPEP1 sequence (Huffaker et al., 2006) was used to query GenBank-registered nucleotide sequences from maize through the National Center for Biotechnology Information TBLASTN version 2.2.7 algorithm (Altschul et al., 1997). Alignments with the AtPROPEP1 sequence revealed GenBank accession DY240150, a full-length cDNA that encodes the ZmPROPEP1 precursor in the −1 frame. To determine possible localization of the protein in the cell, the pSORT prediction program was used (Nakai and Kanehisa, 1991).
Peptide Synthesis
A 23-amino acid peptide corresponding to the predicted ZmPep1 active peptide sequence, VRRRPTTPGRPREGSGGNGGNHH, was synthesized by solid-phase peptide synthesis at the Protein Core Chemistry Facility at the University of Florida using N-(9-fluorenylmethoxycarbonyl)-protected amino acids on a 432A Peptide Synthesizer (Applied Biosystems). The peptide was cleaved from the resin with modified reagent K and HPLC purified on an RP-C18 column using a water-acetonitrile gradient in 0.1% trifluoroacetic acid. The peptide was confirmed to be of the expected Mr (2,452.63) by MS.
Nucleic Acid Purification and Isolation
DNA was isolated from maize leaves using the genomic DNA isolation reagent DNAzol (Invitrogen) as per the instructions provided with the reagent. For RNA isolation, tissues that had been harvested and frozen in liquid nitrogen were ground to a fine powder, and approximately 100 mg of frozen powdered plant material was extracted in 1 mL of Trizol reagent (Invitrogen). RNA isolation was performed as per the Trizol instructions, supplemented by an acid-phenol-chloroform partitioning step to minimize contaminating DNA.
Cloning of the ZmPROPEP1 Gene and cDNA
RNA isolated from young maize leaves was reverse transcribed using the RETROscript kit (Applied Biosystems) as per kit instructions with random decamer primers. The ZmPROPEP1 open reading frame was amplified from the cDNA with the forward primer 5′-GACCTCAGGAAAGGGGAGACCTGGA-3′ and the reverse primer 5′-AAGGAAGCGAACAAGCTAGGGTCACCGTA-3′ using Phusion Hot Start II DNA Polymerase (New England Biolabs). The amplified cDNA was cloned into the pCR BLUNT II TOPO vector using a Zero Blunt PCR cloning kit (Invitrogen) as per kit instructions and transformed by heat shock into TOP10F′ chemically competent Escherichia coli (Invitrogen). Colonies were screened by PCR using the ZmPROPEP1 primers, and plasmids from positive colonies were sequenced using ABI Prism BigDye terminator (Applied Biosystems). All sequencing reactions were run at the DNA Sequencing Core Facility at the University of Florida.
Leaf Bioassays for Analysis of Transcript and Metabolite Abundance
For excision assays, leaf 5 of 3-week-old maize plants was cut and placed in 4-mL glass vials containing either water or a ZmPep1 solution in water. For each treatment and time point, six leaves of leaf stage 5 were assayed. At the time points indicated, the entire leaf was harvested in liquid nitrogen for RNA and metabolite analysis. Zero-hour control leaves were harvested directly from the plant into liquid nitrogen. For intact leaf assays, wax was gently scraped from leaves at two sites on either side of the midrib on leaf 5 of 3-week-old plants. Five microliters of water or of solutions in water of 25 pmol of ZmPep1, 500 μg of pectinase elicitor, or 5 × 103 fungal spores was applied to each site. After the time indicated, a 7.5-cm segment of leaf surrounding the wound sites was harvested in liquid nitrogen for RNA and metabolite analysis.
Semiquantitative Reverse Transcription-PCR
RNA was reverse transcribed using RETROscript reagents (Applied Biosystems) with reactions assembled and incubated as per kit instructions. Semiquantitative PCR was performed as follows. Template cDNA was used at 120 ng per reaction. Each 25-μL reaction had 0.5 units of Platinum Taq polymerase diluted into Platinum 10× PCR buffer (Invitrogen) with 1.5 mm Mg2+, 200 μm each deoxyribonucleotide triphosphate, and 0.4 μm each primer. All primers were designed to be used at an annealing temperature of 56°C, to amplify regions 150 to 350 bp in length, and to span introns when possible; primer sequences are listed in Supplemental Table S1. The Actin1 gene transcript (GenBank accession no. J01238) was used to permit comparisons of relative transcript abundance from sample to sample (Kirchberger et al., 2007; Erb et al., 2009). PCR was performed as follows: 3 min at 94°C, 30 s at 94°C, 30 s at 56°C, 1 min at 72°C, and a final 10 min at 72°C. Cycling time for each transcript was optimized and ranged between 25 and 38 cycles. The number of amplification cycles used for each is listed in Supplemental Table S1.
A 20-μL aliquot of each reaction product was diluted with 2 μL of DNA blue/orange loading dye (Promega Biosciences) and analyzed electrophoretically on a 1% agarose/Tris-acetate-EDTA gel impregnated with ethidium bromide (Promega Biosciences). The gel was visualized on a Gel Doc XR Imaging System (Bio-Rad) using Quantity One version 4.6.2 software (Bio-Rad). A high-resolution image of the gel was captured, and band intensity was measured using the Quantity One program (Bio-Rad). Band intensity of each transcript was normalized by dividing the measured value by the value obtained from measurement of actin band intensity for the same sample. Values obtained for estimation of relative transcript abundance were then defined as fold change in normalized band intensity for each treatment versus normalized band intensity of an untreated control sample.
Measurement of Hormones and Metabolites
Levels of JA, indole, and anthranilic acid were measured using the previously described vapor phase extraction method with GC-MS analysis (Schmelz et al., 2004). Quantification of indole levels in each sample was performed by comparison with an external standard curve. ET emitted by leaves was measured by GC as described previously (Schmelz et al., 2009).
Analysis of Benzoxazinoid Phytoalexins
Benzoxazinoids were extracted and analyzed by HPLC as described by Erb et al. (2009). After 24 h, leaf tissue surrounding the treatment sites was harvested in liquid nitrogen, freeze dried, and extracted in 49:1 methanol:acetic acid prior to analysis. Quantities were estimated using 6-hydroxy-2(3H)-benzoxazolone as an internal standard. HDMBOA-Glc is the predominant hydroxamic acid in 20-d-old maize roots (Cambier et al., 2000); thus, root tissue was used to confirm the HPLC retention time of this ZmPep1-induced metabolite in leaves. HDMBOA is known to be highly unstable (Maresh et al., 2006). Unlike DIMBOA-Glc, even low levels of water in the extracted tissue caused the complete loss of analyzable HDMBOA-Glc.
Leaf Blight Resistance Assays
Intact 2.5-week-old maize plants were infected with C. heterostrophus as follows. On leaves 5 and 6, the wax was gently scraped from each leaf twice on both sides of the midrib, and 5 μL of water or 25 pmol of ZmPep1 was applied to each wound site and allowed to air dry. After 12 to 24 h, 5 μL of C. heterostrophus spores in a 0.1% Tween 20 solution was applied to each wound site and allowed to dry. Each plant was then incubated in open-bottomed glass chambers under greenhouse lights for 3 d with 100% humidified air passed over each plant at 4 L min−1. After 3 d, leaves were photographed and the lesion area measured using ASSESS 2.0 image-analysis software for plant disease quantification by Lakhdar Lamari (American Phytopathological Society). The extent of cell death was estimated through the measurement of ion leakage as described by Torres et al. (2002). Briefly, four leaf disc samples, each with an area of 1 cm2, were collected from infected or uninfected tissues, immersed in 4 mL of water, and vacuum infiltrated for 1 min. After shaking gently for 1 h, the conductivity of the samples was measured in μSiemens at 25°C using a YSI 3100 conductivity meter (YSI, Inc.). To measure total potential conductivity of each sample, the leaf discs in water were microwaved for 1 min, and after cooling to 25°C, the conductivity was remeasured. A comparison of initial conductivity to total potential conductivity of the same leaf discs resulted in a number expressed as a percentage of total conductivity for each sample. C. heterostrophus-induced ion leakage was then defined as the difference between percentage of total conductivity measured for C. heterostrophus-infected samples and that of samples from uninfected leaf tissue in the same assay.
Stalk Rot Resistance Assays
A 1.0-mm-diameter cork borer was used to bore a hole through the second aboveground node in the stalk of 3.5- to 4-week-old plants. The hole was filled with 10 μL of either water or 50 pmol of ZmPep1 to eliminate any air bubbles. A plastic pipette tip filled with 1 mL of either water or 5 nmol of ZmPep1 was then gently inserted into the hole until it was secure. The plant was allowed to take up the full 1 mL of pretreatment solution, which typically occurred within 2 h. After a 3-h pretreatment, the pipette tips were removed and the hole was inoculated with 10 μL of a C. graminicola spore suspension in sterile water. For untreated control plants, a hole was bored through the second node at the time of inoculation. After 4 d, stems were split open and photographed and lesion area was determined using ASSESS 2.0 image-analysis software. A 12.5-cm segment centered around the inoculated node was cut, immersed in 15 mL of water, and vacuum infiltrated for 1 min, and after 1 h of incubation, ion leakage was measured as described to estimate the extent of cell death (Torres et al., 2002). Values obtained for C. graminicola-induced ion leakage were defined as the ratio of conductivity values measured in infected stem samples compared with conductivity measured in wounded control stem samples.
Statistical Analysis
ANOVAs were performed on the quantified levels of metabolites, transcripts, pathogen lesion size, and ion leakage estimates. Treatment effects were investigated when the main effects of the ANOVAs were significant (P < 0.05). Where appropriate, Tukey tests were used to correct for multiple comparisons between control and treatment groups. t tests were also used in limited specific cases to examine significant differences in treatment groups compared with selected controls. With the exception of percentage data, prior to statistical analysis, all data were subjected to square root transformation to compensate for elevated variation associated with larger mean values. The analysis was accomplished with JMP 4.0 statistical discovery software (SAS Institute).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY359573, J01238, NM_001159139, AY488136, AY488135, NM_001111749, EU963425, AY107804, BT039519, EU968115, NM_001154840, and DY240150.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cloned ZmPROPEP1 genes from maize varieties B73 and GQ.
Supplemental Table S1. Primers used for semiquantitative reverse transcription-PCR analysis.
Supplementary Material
Acknowledgments
We thank the following people: G. Pearce (Institute of Biological Chemistry, Washington State University), Y. Yamaguchi (Laboratory of Crop Physiology, Hokkaido University), J. Narváez-Vásquez (Department of Botany and Plant Sciences, University of California, Riverside), and M. Vaughan (U.S. Department of Agriculture, Agricultural Research Service Center for Medical, Agricultural, and Veterinary Entomology) for critical reading of the manuscript; A. Chung (Interdisciplinary Center for Biotechnological Research, University of Florida) for peptide synthesis; B. Forguson (U.S. Department of Agriculture, Agricultural Research Service Center for Medical, Agricultural, and Veterinary Entomology) for care and maintenance of plant materials; C. Harmon (Department of Plant Pathology, University of Florida) for identification of C. heterostrophus; J. Jones (Department of Plant Pathology, University of Kentucky) and L. Vaillancourt (Department of Plant Pathology, University of Florida) for discussions of techniques; J. Rollins (Department of Plant Pathology, University of Florida) for providing C. graminicola; and H. Tang, R. Beck, K. Friman, A. Kuipers, and E. Mok (U.S. Department of Agriculture, Agricultural Research Service Center for Medical, Agricultural, and Veterinary Entomology) for technical support.
References
- Alessandra L, Luca P, Adriano M. (2010) Differential gene expression in kernels and silks of maize lines with contrasting levels of ear rot resistance after Fusarium verticillioides infection. J Plant Physiol 167: 1398–1406 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint-Kurti PJ, Carson ML. (2006) Analysis of quantitative trait loci for resistance to southern leaf blight in juvenile maize. Phytopathology 96: 221–225 [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bergey DR, Howe GA, Ryan CA. (1996) Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc Natl Acad Sci USA 93: 12053–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom GC, Nicholson RL. (1999) The biology of corn anthracnose: knowledge to exploit for improved management. Plant Dis 83: 596–608 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bravo JM, Campo S, Murillo I, Coca M, San Segundo B. (2003) Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol Biol 52: 745–759 [DOI] [PubMed] [Google Scholar]
- Bücker C, Grambow HJ. (1990) Alterations in 1,4-benzoxazinone levels following inoculation with stem rust in wheat leaves carrying various alleles for resistance and their possible role as phytoalexins in moderately resistant leaves. Z Naturforsch C 45: 1151–1155 [Google Scholar]
- Byrnes KJ, Pataky JK, White DG. (1989) Relationships between yield of three maize hybrids and severity of southern leaf blight caused by race O of Bipolaris maydis. Plant Dis 73: 834–840 [Google Scholar]
- Cambier V, Hance T, de Hoffmann E. (2000) Variation of DIMBOA and related compounds content in relation to the age and plant organ in maize. Phytochemistry 53: 223–229 [DOI] [PubMed] [Google Scholar]
- Casacuberta JM, Raventós D, Puigdoménech P, San Segundo B. (1992) Expression of the gene encoding the PR-like protein PRms in germinating maize embryos. Mol Gen Genet 234: 97–104 [DOI] [PubMed] [Google Scholar]
- Chen YC, Siems WF, Pearce G, Ryan CA. (2008) Six peptide wound signals derived from a single precursor protein in Ipomoea batatas leaves activate the expression of the defense gene sporamin. J Biol Chem 283: 11469–11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt DC, Refi-Hind S, Stratmann JW, Lincoln DE. (2010) Systemin and jasmonic acid regulate constitutive and herbivore-induced systemic volatile emissions in tomato, Solanum lycopersicum. Phytochemistry 71: 2024–2037 [DOI] [PubMed] [Google Scholar]
- Djonovic S, Vargas WA, Kolomiets MV, Horndeski M, Wiest A, Kenerley CM. (2007) A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol 145: 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, Poree F, Stitt M, Pons-Kühnemann J, Sonnewald U, Kahmann R, et al. (2008) Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J 56: 181–195 [DOI] [PubMed] [Google Scholar]
- Erb M, Flors V, Karlen D, de Lange E, Planchamp C, D’Alessandro M, Turlings TC, Ton J. (2009) Signal signature of aboveground-induced resistance upon belowground herbivory in maize. Plant J 59: 292–302 [DOI] [PubMed] [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J, Campo S, Rufat M, Estopà M, Messeguer J, San Segundo B, Coca M. (2007) Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Mol Plant Microbe Interact 20: 832–842 [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D, Logemann E, Nürnberger T, Parniske M, Reinold S, Sacks WR, Schmelzer E. (1995) Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc Natl Acad Sci USA 92: 4150–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin PA, Davis FM, Williams WP. (1993) 2-Hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one, a possible toxic factor in corn to the southwestern corn borer. J Chem Ecol 19: 531–542 [DOI] [PubMed] [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H. (2001) A large family of class III plant peroxidases. Plant Cell Physiol 42: 462–468 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. (1996) An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Ryan CA. (2007) Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 104: 10732–10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S, Leroch M, Huynen MA, Wahl M, Neuhaus HE, Tjaden J. (2007) Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J Biol Chem 282: 22481–22491 [DOI] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BP, Staskawicz B, et al. (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. (1987) Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA 84: 7542–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, et al. (2006) An overview of the serpin superfamily. Genome Biol 7: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM. (1999) Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science 285: 1917–1919 [DOI] [PubMed] [Google Scholar]
- Ma W, Smigel A, Verma R, Berkowitz GA. (2009) Cyclic nucleotide gated channels and related signaling components in plant innate immunity. Plant Signal Behav 4: 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresh J, Zhang J, Lynn DG. (2006) The innate immunity of maize and the dynamic chemical strategies regulating two-component signal transduction in Agrobacterium tumefaciens. ACS Chem Biol 1: 165–175 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. (2006) Peptide hormones in plants. Annu Rev Plant Biol 57: 649–674 [DOI] [PubMed] [Google Scholar]
- McGurl B, Pearce G, Orozco-Cardenas M, Ryan CA. (1992) Structure, expression, and antisense inhibition of the systemin precursor gene. Science 255: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Melanson D, Chilton MD, Masters-Moore D, Chilton WS. (1997) A deletion in an indole synthase gene is responsible for the DIMBOA-deficient phenotype of bxbx maize. Proc Natl Acad Sci USA 94: 13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Boland W. (2008) Recognition of herbivory-associated molecular patterns. Plant Physiol 146: 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch S, Lingner U, Floss DS, Ludwig N, Sauer N, Deising HB. (2008) The hemibiotrophic lifestyle of Colletotrichum species. J Plant Physiol 165: 41–51 [DOI] [PubMed] [Google Scholar]
- Murillo I, Roca R, Bortolotti C, Segundo BS. (2003) Engineering photoassimilate partitioning in tobacco plants improves growth and productivity and provides pathogen resistance. Plant J 36: 330–341 [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. (1991) Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11: 95–110 [DOI] [PubMed] [Google Scholar]
- Narváez-Vásquez J, Orozco-Cárdenas ML, Ryan CA. (2007) Systemic wound signaling in tomato leaves is cooperatively regulated by systemin and hydroxyproline-rich glycopeptide signals. Plant Mol Biol 65: 711–718 [DOI] [PubMed] [Google Scholar]
- Niemeyer HM. (2009) Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: key defense chemicals of cereals. J Agric Food Chem 57: 1677–1696 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Ishihara A, Hasegawa M, Kodama O, Iwamura H. (2001) Induced accumulation of 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc) in maize leaves. Phytochemistry 56: 669–675 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Ishihara A, Iwamura H. (2002) Induction of HDMBOA-Glc accumulation and DIMBOA-Glc 4-O-methyltransferase by jasmonic acid in poaceous plants. Phytochemistry 61: 331–337 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Ishihara A, Tanaka C, Mori N, Tsuda M, Iwamura H. (2004) Accumulation of HDMBOA-Glc is induced by biotic stresses prior to the release of MBOA in maize leaves. Phytochemistry 65: 2995–3001 [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. (1991) A polypeptide from tomato leaves activates the expression of proteinase inhibitor genes. Science 253: 895–897 [DOI] [PubMed] [Google Scholar]
- Pearce G, Yamaguchi Y, Barona G, Ryan CA. (2010) A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc Natl Acad Sci USA 107: 14921–14925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Yamaguchi Y, Munske G, Ryan CA. (2008) Structure-activity studies of AtPep1, a plant peptide signal involved in the innate immune response. Peptides 29: 2083–2089 [DOI] [PubMed] [Google Scholar]
- Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14: 21–29 [DOI] [PubMed] [Google Scholar]
- Postel S, Küfner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A, Halter T, Kemmerling B, Nürnberger T. (2010) The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol 89: 169–174 [DOI] [PubMed] [Google Scholar]
- Realini C, Rogers SW, Rechsteiner M. (1994) KEKE motifs: proposed roles in protein-protein association and presentation of peptides by MHC class I receptors. FEBS Lett 348: 109–113 [DOI] [PubMed] [Google Scholar]
- Ren F, Lu YT. (2006) Overexpression of tobacco hydroxyproline-rich glycopeptide systemin precursor A gene in transgenic tobacco enhances resistance against Helicoverpa armigera larvae. Plant Sci 171: 286–292 [Google Scholar]
- Rhoads AR, Friedberg F. (1997) Sequence motifs for calmodulin recognition. FASEB J 11: 331–340 [DOI] [PubMed] [Google Scholar]
- Rojo E, Solano R, Sánchez-Serrano JJ. (2003) Interactions between signaling compounds involved in plant defense. J Plant Growth Regul 22: 82–98 [Google Scholar]
- Romero RM, Roberts MF, Phillipson JD. (1995) Anthranilate synthase in microorganisms and plants. Phytochemistry 39: 263–276 [DOI] [PubMed] [Google Scholar]
- Ryan CA. (1989) Proteinase inhibitor gene families: strategies for transformation to improve plant defenses against herbivores. Bioessays 10: 20–24 [DOI] [PubMed] [Google Scholar]
- Ryan CA, Huffaker A, Yamaguchi Y. (2007) New insights into innate immunity in Arabidopsis. Cell Microbiol 9: 1902–1908 [DOI] [PubMed] [Google Scholar]
- Ryan CA, Pearce G. (2003) Systemins: a functionally defined family of peptide signals that regulate defensive genes in Solanaceae species. Proc Natl Acad Sci USA (Suppl 2) 100: 14577–14580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Alborn HT, Banchio E, Tumlinson JH. (2003) Quantitative relationships between induced jasmonic acid levels and volatile emission in Zea mays during Spodoptera exigua herbivory. Planta 216: 665–673 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Carroll MJ, LeClere S, Phipps SM, Meredith J, Chourey PS, Alborn HT, Teal PE. (2006) Fragments of ATP synthase mediate plant perception of insect attack. Proc Natl Acad Sci USA 103: 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH, III, Teal PEA. (2009) Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc Natl Acad Sci USA 106: 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. (2004) The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J 39: 790–808 [DOI] [PubMed] [Google Scholar]
- Simmons CR, Grant S, Altier DJ, Dowd PF, Crasta O, Folkerts O, Yalpani N. (2001) Maize rhm1 resistance to Bipolaris maydis is associated with few differences in pathogenesis-related proteins and global mRNA profiles. Mol Plant Microbe Interact 14: 947–954 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venard C, Vaillancourt L. (2007) Penetration and colonization of unwounded maize tissues by the maize anthracnose pathogen Colletotrichum graminicola and the related nonpathogen C. sublineolum. Mycologia 99: 368–377 [DOI] [PubMed] [Google Scholar]
- Wisser RJ, Balint-Kurti PJ, Nelson RJ. (2006) The genetic architecture of disease resistance in maize: a synthesis of published studies. Phytopathology 96: 120–129 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Boone L, Kocz R, Zhang C, Binns AN, Lynn DG. (2000) At the maize/Agrobacterium interface: natural factors limiting host transformation. Chem Biol 7: 611–621 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.