Abstract
Background
Prostaglandin E2 (PGE2) levels are frequently elevated in colorectal carcinomas. PGE2 is perceived via four transmembrane G protein coupled receptors (EP1-4), among which the EP4 receptor is most relevant. PGE2/EP4-receptor interaction activates CREB via the ERK/MEK pathway. However, the downstream target genes activated by this pathway remained to be investigated.
Methodology/Principal Findings
Here, we have identified S100P (an EF-hand calcium binding protein) as a novel downstream target. We show by realtime RT-PCR that S100P mRNA levels are elevated in 14/17 (82%) colon tumor tissues as compared to paired adjacent normal colonic tissues. S100P expression is stimulated in the presence of PGE2 in a time dependent manner at mRNA and protein levels in colon, breast and pancreatic cancer cells. Pharmacological and RNAi-mediated inhibition of the EP4 receptor attenuates PGE2-dependent S100P mRNA induction. RNAi-mediated knockdown of CREB inhibits endogenous S100P expression. Furthermore, using luciferase reporter analysis and EMSA we show that mutation and/or deletion of the CRE sequence within the S100P promoter abolished PGE2-mediated transcriptional induction. Finally, we demonstrate that RNAi-mediated knockdown of S100P compromised invadopodia formation, colony growth and motility of colon cancer cells. Interestingly, endogenous knock down of S100P decreases ERK expression levels, suggesting a role for ERK in regulating S100P mediated cell growth and motility.
Conclusions/Significance
Together, our findings show for the first time that S100P expression is regulated by PGE2/EP4-receptor signaling and may participate in a feedback signaling that perpetuates tumor cell growth and migration. Therefore, our data suggest that dysregulated S100P expression resulting from aberrant PGE2/EP4 receptor signaling may have important consequences relevant to colon cancer pathogenesis.
Key words: Colon cancer, CREB, PGE2, EP4 receptor, ERK, S100P, RNAi
PGE2 is an important prostaglandin that has been implicated in colorectal carcinogenesis (CRC).1 PGE2 is elevated in familial adenomatous polyposis (FAP) patients as well as in APC(MIN) mice in a polyp-size dependent manner.2 In addition, exogenously administered PGE2 enhances the growth of intestinal adenomas and worsens CRC.3 PGE2 also protects APC(MIN) mice against NSAID-induced intestinal polyp reduction.4 In conjunction with COX-2, PGE2 plays a multitude of roles in CRC development by deregulating various hallmarks of cancer.5 The effects of PGE2 on cellular responses are mediated by its overall second messenger response. Intracellular signal transduction of PGE2 occurs via four G protein coupled receptors (GPCRs) namely EP1, EP2, EP3 and EP4, among which the EP4 receptor appears to play a significant role in colon cancers. Ligand binding assays show that the EP4 receptor has the highest affinity towards pro-tumorigenic PGE2 ligand.6 There has been a growing appreciation forthe EP4 receptor in recent years, as an important transducer of PGE2 signals leading to cell invasion and motility during tumorigenesis. The EP4 receptor is overexpressed in several different cancers including CRC.7 In addition, the EP4 receptor was determined to be a genetic risk factor in both ulcerative colitis as well as Crohn's disease as determined by genome-wide associations.8 In vitro studies by our group and others indicate that PGE2/EP4-receptor signaling via ERK activation promotes tumorigenic behavior in colon cancer cells.9 Constitutive expression of EP4 receptor promotes proliferation and anchorage-independent growth, demonstrating that the EP4 receptor may also be a key regulator of tumor progression.10 PGE2 stimulates cell proliferation and motility in LS174T colon adenocarcinoma cells through the EP4 receptor-dependent activation of PI3K/AKT signaling.11 It also inhibits apoptosis in human Caco-2 colon cancer cells in an EP4 dependent pathway.12 In addition, a growing quest for the identification of drugs against colorectal and other cancers has encouraged pharmaceutical establishments to consider selective EP4 antagonists as novel therapeutic targets. However, exactly how PGE2/EP4-receptor activation contributes to colorectal cancer development in vivo remains to be determined. The downstream effector genes that are regulated via this pathway are an active area of investigation.
S100P is a member of the S100 family of calcium binding proteins, which share consensus EF-hand motifs. There is considerable evidence implicating S100P in cancer. However, exactly how S100P contributes to colon cancer remains to be clarified. The expression of S100P is upregulated in a number of cancers such as pancreatic,13 breast,14 prostate15 and lung.16 Interestingly, microarray profiling on frozen colon cancer tissue specimens showed elevated S100P levels.17,18 In addition, S100P expression was shown to be enhanced at least 4 fold in a DNA microarray study performed on inflamed colonic tissue from ulcerative colitis and Crohn's disease patients compared to normal volunteers, indicating the relevance of this protein in inflammation-induced colorectal carcinogenesis.19 S100P is a secreted protein and its presence in fine-needle biopsy specimens and pancreatic juice has been positively correlated with extent of cancer spread. Thus elevated levels of S100P has been proposed to be used as an early prognostic indicator for pancreatic cancers.20 Despite this fact, there is limited information available on the upstream signaling components that regulate S100P expression. For instance, in prostate cancers, IL-6 was shown to transcriptionally induce the expression of S100P in an androgen-refractory manner.21 S100P mRNA and protein levels were stimulated in the presence of BMP4 (a member of the TGFβ superfamily), in pancreatic cancers.22 In addition, high concentrations of the selective COX-2 inhibitor, celecoxib, induce S100P expression in human gastric cancer cell lines.23 These studies indicate that S100P can be regulated by transcriptional mechanisms in cancer cells. However, no studies have as yet investigated the mechanism by which S100P expression is regulated in colorectal cancers.
In the present study we show that the expression of S100P in human colorectal tumors is significantly elevated as compared to normal colonic tissues from the same individuals. We further demonstrate that PGE2/EP4-receptor signaling can induce the expression of S100P in a time dependent fashion in LS174T colon cancer cells and in human embryonic kidney (HEK293) cells that overexpress the EP4 receptor. We also provide evidence that the EP4 receptor is essential for S100P expression. Colon cancer cells, in which the EP4 receptor expression is ablated, fail to show S100P induction. In our previous studies we showed that PGE2/EP4-receptor signaling activates CREB via the ERK/MEK pathway.9 Here we show that knockdown of CREB by RNAi drastically reduces endogenous S100P levels. We also show that binding of CREB to the promoter region of s100p gene is required for PGE2-mediated S100P transcription. Finally, we demonstrate that knockdown of S100P decreases endogenous ERK protein levels possibly associated with a decrease in the ability to sustain invadopodia, colony growth and motility in colon cancer cells. Together our findings reveal a novel target of the PGE2/EP4-receptor signaling pathway which may provide an alternative avenue for the management of colorectal cancers.
Results
Intracellular PGE2/EP4-receptor signaling.
Previous studies in our laboratory have shown that PGE2/EP4-receptor signaling activates the transcription factor CREB via the ERK/MEK pathway.9 However, the downstream target genes activated by the PGE2/EP4 → CREB mechanism have not been fully identified. We wanted to investigate what downstream genes may be induced by the PGE2/EP4/ERK/CREB pathway in colon cancer cells. In order to understand the function of CREB protein in colon cancer cell lines, we asked the question whether the transcription factor could affect the growth of colon cancer cells. Two colon cancer cell lines (LS174T and HCA7) were stably transfected with vector control or a dominant negative construct against CREB. The dominant negative protein, termed ACREB, dimerizes with wild-type CREB protein and prevents it from binding DNA sequences. Colon cancer cells stably expressing either pCMV500 vector alone (control) or ACREB construct were plated under G418 selection and allowed to colonize for 3 weeks. Supplemental Figure 1A shows a significant decrease in the number of methylene blue stained colonies in cells transfected with non-functional CREB protein as compared to those transfected with vector alone. We then wanted to investigate the mechanism of growth inhibition. We used LS174T cells transfected with pCMV500 vector alone (control) or ACREB construct and performed Caspase-Glo 3/7 assay. Supplemental Figure 1B shows that apoptotic luminescence, resulting from the specific cleavage of caspases 3 and 7, increases significantly at 48 and 72 hrs in ACREB containing cells compared to those containing vector alone (control). This suggests that knockdown of the transcription factor CREB can significantly diminish colony growth by inducing apoptosis.
Next, in order to identify target genes which are activated by the transcription factor CREB in response to PGE2/EP4-receptor signaling, we performed a microarray. The microarray experiment was designed to identify genes that were differentially regulated in the presence of 1 µM PGE2 on HEK293 cells that have stable ectopic expression of EP4 receptor, a model system used to elucidate signaling events by prostanoid receptor signaling (Sup. Fig. 1C).24–26 A total of 39 genes were significantly upregulated at least 3 fold in PGE2-treated cells (p < 0.001). The data were a culmination of three independent experiments. Among these genes, s100p, a gene encoding for a calcium binding protein, was chosen for further validation because (1) S100P plays an important role in carcinogenesis, (2) S100P shows aberrant expression in gastrointestinal cancers including gastric and pancreatic cancers and (3) the promoter region of the s100p gene contains a CREB Recognition Element (CRE) sequence. These data show, for the first time, that the knockdown of transcription factor CREB can significantly diminish colony growth in colon cancer lines and that S100P could be a potential downstream target of the PGE2/EP4-receptor signaling pathway.
Expression of S100P is elevated in colon tumors.
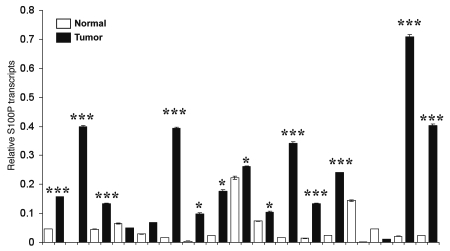
Studies involving microarray profiling of frozen colon cancer specimens as well as tissues from patients with inflammatory bowel disease, have revealed elevated S100P levels.17,19 In order to investigate the expression of S100P in CRC patients, we obtained frozen tumor tissues from surgically resected clinical specimens. We performed quantitative realtime analysis on levels of S100P mRNA from colon tumors and compared it to adjacent normal colonic tissues from 17 patients. Overall, 14 out of 17 (82%) cases displayed an increase in S100P mRNA levels relative to paired normal tissue. Nine of the seventeen cases showed a marked increase of more than 5-fold (cases: 270, 308, 309, 314, 251, 259, 260, 297 and 298). Five cases showed a moderate increase of less than 5-fold (cases: 207, 271, 290, 323 and 234). Finally, three cases (cases: 284, 280 and 285) showed a decrease in S100P mRNA levels compared to adjacent normal tissue from the same individual (Fig. 1). Studies have also shown that S100P protein levels are elevated in epithelial cells during colon cancer.27 We then extracted protein from a subset of patients shown in Figure 1 and confirmed by western blot and ELISA that levels of S100P protein were also elevated in colon tumors as compared to normal colonic tissue (data not shown). Our data suggest that increased mRNA levels of S100P may account for the increased protein levels of S100P seen in colon cancer patients.
Figure 1.
S100P expression is elevated in colon tumors. Human colon tissue from 17 colon cancer patients were obtained from the Arizona Cancer G.I. SPORE tissue resource. The matched normal (open bars) and tumor (filled bars) tissue specimens were processed for RNA and subjected to qRT-PCR analysis for S100P and β-actin. Values represent mean ± SD (*p < 0.05, ***p < 0.001).
Exposure to PGE2 induces the expression of S100P.
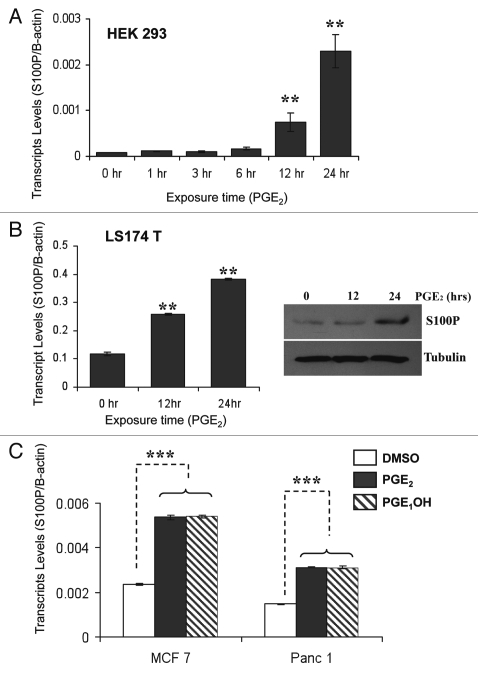
In order to validate our microarray findings, human embryonic kidney HEK293 cells stably expressing high levels of EP4 receptor were treated with either DMSO (vehicle control) or with 1 µM PGE2 for 1, 3, 6, 12 and 24 hours. Drug treatments were performed after at least 20 hours of serum starvation. Realtime analysis of S100P transcripts revealed that S100P message is significantly elevated in the presence of 1 µM PGE2 at 12 and 24 hrs (Fig. 2A). Second, in order to investigate PGE2 dependent response in colon cancer cells, LS174T cells were treated with 1 µM PGE2 for 12 and 24 hours. Figure 2B shows that exposure of LS174T cells to 1 µM PGE2 could induce the expression of the S100P gene by 2.5-fold after 12 hours and up to 4-fold after 24 hours. A similar magnitude of mRNA induction has been observed for the Id-1 and CXCL1 genes in response to PGE2 levels in breast and colon cancer cells respectively.28,29 In addition, S100P protein levels were also induced to high levels at 24 hours (Fig. 2B). Third, to ensure that the induction is universal and not cell-line specific, we demonstrate that S100P is induced in breast and pancreatic cancer cell lines in the presence of PGE2 as well as PGE1OH (EP4 agonist) (Fig. 2C).
Figure 2.
PGE2 stimulates S100P expression. (A) HEK293 cells stably expressing EP4 receptor were treated with 1 µM PGE2 for indicated time points. RNA was extracted from cells and subjected to qRT-PCR analysis for S100P and β-actin. (B) LS174T colon cancer cells treated with 1 µM PGE2 for indicated time. RNA was extracted and subjected to qRT-PCR analysis for S100P and β-actin (left). Total cell protein was extracted from LS174T cells treated for indicated time and analyzed by SDS-PAGE. Western blot was performed using polyclonal S100P antibody (1:1,000). Equal loading was confirmed by probing against α-tubulin (right). (C) MCF7 breast cancer and Panc1 pancreatic cancer cells were also treated as indicated and qRT-PCR was performed. Changes in transcript levels after qRT-PCR are shown as mean ± SD. (**p < 0.01, ***p < 0.001).
Inhibition of the EP4 receptor suppresses PGE2-dependent S100P induction.
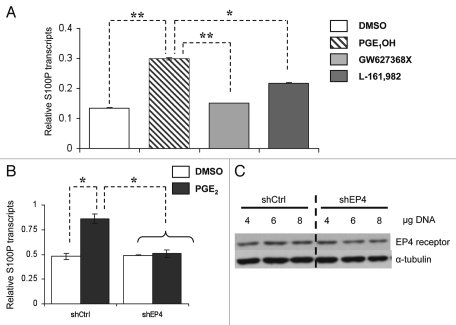
Next, we wanted to establish whether the EP4 receptor plays a role in PGE2-mediated induction of S100P. For this purpose, LS174T cells were exposed to PGE1OH, an EP4-receptor agonist. In addition, cells were exposed to two selective EP4 receptor antagonists: GW627368X and L-161,982. Exposure to PGE1OH alone showed a 2-fold increase in the S100P relative transcript levels, similar to PGE2 mediated induction, whereas treatment with EP4 receptor antagonists repressed the induction of S100P to levels comparable to vehicle control (Fig. 3A). Moreover, exposure to PGE2 resulted in diminished S100P response in shRNA-treated LS174 T cells (shEP4) (Fig. 3B). These levels were comparable to shEP4 cells treated with DMSO (vehicle). By contrast, in shCrtl cells (scrambled control) S100P levels were induced to 2-fold when treated with PGE2 compared to treatment with DMSO. Figure 3C confirms the knockdown of EP4 receptor protein in shEP4 expressing LS174 T cells compared to shCtrl cells. Taken together these results suggest that PGE2-dependent induction of S100P is mediated via the EP4 receptor.
Figure 3.
Inhibition of the EP4 receptor suppresses PGE2-dependent S100P induction. (A) LS174 T cells were treated with EP4 agonist (1 µM PGE1OH) or EP4 antagonist (1 µM GW627368X or 2 µM L-161,982) for 24 hrs. RNA was subjected to qRT-PCR analysis for S100P and α-actin. (B) shRNA-expressing (shEP4) and scrambled control-expressing (shCtrl) LS174 T cells were serum starved and treated with 1µM PGE2 for 24 hrs and qRT-PCR was performed. Differences in relative transcripts are shown as mean ± SD (**p < 0.01, ***p < 0.001). (C) Concentration dependent silencing of the EP4 receptor in shEP4 and shCtrl-expressing LS174T cells is shown by western blotting using EP4-specific primary antibody (1:1,000). Equal loading was confirmed by probing against α-tubulin.
Loss of CREB function depletes S100P.
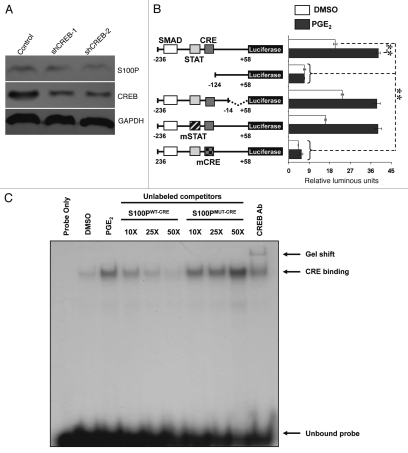
First, in order to establish whether CREB is important and/or required for S100P expression, we knocked down CREB using RNAi. We then determined the levels of S100P protein in LS174T cells transfected with empty vector (control) or two independent constructs expressing shRNA against CREB (shCREB-1 and shCREB-2) by western blot. Figure 4A shows that knockdown of CREB protein results in a decrease in endogenous S100P protein levels, suggesting that CREB directly regulates S100P.
Figure 4.
Loss of CREB function depletes S100P. (A) LS174T cells expressing control, shCREB-1 or shCREB-2 were subjected to western blot analysis using polyclonal S100P-specific antibody (1:1,000). Effective knockdown of CREB and equal loading of total proteins was confirmed by probing against CREB and GAPDH respectively. (B) LS174T cells were transfected with wild-type (solid boxes), deletion or mutant (hashed boxes) S100P promoter constructs. Cells were treated with 1 µM PGE2 (filled bars) or vehicle (DMSO; open bars) after serum starvation. Relative luminous units represent the ratio of luminescence generated by Firefly Luciferase over Renilla Luciferase. Values represent mean ± SD (**p < 0.01). (C) EMSA was performed on nuclear lysates of LS174T cells that were treated with 1µM PGE2 for 30 min. Competitive binding with wild-type (S100PWT-CRE) and mutant (S100PMUT-CRE) probes are shown at increasing concentrations. Gel shift is shown in the presence of 3 µg polyclonal anti-CREB antibody (Ab).
Next, in order to identify specific sequence elements within the s100p promoter that are responsible for the induction of the gene, we conducted promoter reporter assays. Three cis-acting elements (SMAD, STAT and CRE) have been reported to be important for the transcription of the s100p gene. The sequence of the core s100p promoter is depicted in Supplemental Figure 2. Functional analysis of the promoter was performed by transient transfection of three promoter deletion constructs that were cloned into a pGL3-based reporter vector upstream of the firefly luciferase gene (Fig. 4B): (−236/+58) that contains SMAD, STAT and CRE sequences, (−124/+58) lacking all three sequences and (−236/−14) that contains SMAD, STAT and CRE sites, but lacks the proximal region of the s100p promoter. In order to investigate whether SMAD, STAT or CRE sequences are involved in response to PGE2 levels, the three constructs were exposed to vehicle control (DMSO) or 1 µM PGE2. Figure 4B shows that constructs (−236/+58) and (−236/−14) could significantly induce luciferase gene expression in the presence of PGE2 (∼2-fold). However, construct (−124/+58) could not induce luciferase activity. This indicates that SMAD, STAT or CRE sequences could be important for gene expression. Previous studies in HeLa cells show that S100P promoter activation is primarily mediated by the STAT/CRE binding sites.30 In addition, in Panc1 pancreatic cancer cell lines, induction of S100P expression was shown to be independent of SMAD4 binding.22 As the PGE2/EP4-receptor mediated signaling has been shown to activate CREB, we hypothesized that the disruption of the CRE-binding site would abolish luciferase activity. As STAT and CRE sequences are juxtaposed in the S100P promoter region, we wanted to clarify which sequence is responsible for driving S100P transcription. We created mutants of STAT and CRE binding sites by site directed mutagenesis (mSTAT: TGC CAC TG and mCRE: GCC CA). These mutated sequences (underlined) have been shown to effectively abolish binding of STAT and CREB in mammalian, Drosophila and yeast systems.31–33 Figure 4B shows that while mSTAT did not affect luciferase activity, mCRE decreased both basal as well as PGE2-induced S100P expression. It is also important to note that although SMAD-binding site was not mutated in any of the constructs, mCRE alone could completely abolish luciferase activity.
We wanted to further determine whether PGE2 influences CREB binding on the S100P promoter. We generated a radio-labeled 16-mer probe sequence of the S100P promoter, containing the CRE binding site. We then treated LS174T cells with 1 µM PGE2 for various time points, extracted nuclear proteins and performed electrophoretic mobility shift assay (EMSA). We performed a time course experiment and determined that exposure to PGE2 for 30 min resulted in significant nuclear factor binding to S100P promoter (Sup. Fig. 3). We then performed competitive binding assays with non radio-labeled probes containing wild-type (S100PWT-CRE) and mutant (S100PMUT-CRE) CRE-binding sequences in order to confirm CREB binding. Figure 4C shows a dose-dependent loss of nuclear factor binding in the presence of increasing concentrations of S100PWT-CRE. By contrast, S100PMUT-CRE did not show any loss in binding. Finally, the presence of CREB antibody (Ab) resulted in a supershift of the specific band, indicating the oligo occupancy of CREB. Taken together, these data indicate that CREB protein directly regulates S100P levels and PGE2 enhances the promoter activity of S100P via the binding of CREB.
Knockdown of S100P protein levels compromises invadopodia formation, colony growth and tumor cell motility and is associated with reduction in endogenous ERK protein levels.
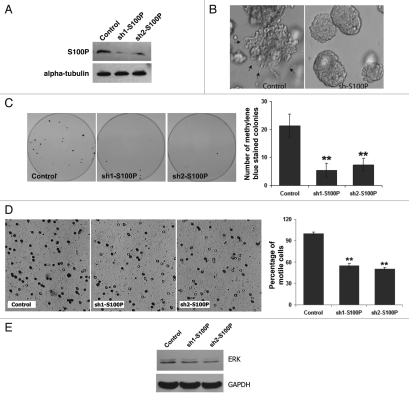
In order to establish the physiological relevance of S100P in colon cancer cells we knocked down the expression of S100P protein by RNAi technology. We transfected LS174T cells with empty pLKO1 vector (control) or two independent shRNA constructs against S100P (sh1-S100P and sh2-S100P). We confirmed the reduction of S100P protein by western blot analysis (Fig. 5A). Next, we seeded LS174T cells stably expressing control or sh1-S100P or sh-2 S100P plasmids on collagen gels and looked for changes in morphology and growth. We observed a dramatic cellular effect in 3D collagen cultures including: smaller colony size, a more rounded phenotype and loss of invadopodia formation in LS174T cells lacking S100P protein compared to controls (Fig. 5B). To confirm that S100P regulates colony growth, we performed methylene blue colony formation assay and observed a drastic decrease in the number of colonies in sh1- and sh2-expressing LS174T cells (sh1-S100P: 5 ± 2 and sh2-S100P: 7 ± 2) compared to control (21 ± 4), p < 0.01 (Fig. 5C). Finally, we analyzed tumor cell motility in a transwell motility assay. After 2 days of incubation, the number of cells penetrating through the membrane (stained purple with crystal violet) were counted and reported as percentage values. The data shows a marked decrease in the percentage of motile tumor cells expressing sh1-S100P (55% ± 3) and sh2-S100P (50% ± 2) compared to control (100% ± 2) (Fig. 5D). Furthermore, in order to determine whether loss of S100P modifies signaling changes within cells, we performed a western blot analysis to look for changes in ERK levels. Figure 5E shows distinct loss of both the subunits (p42 and p44) of endogenous ERK MAP kinase. Together with cell morphology, colony formation and motility assays, these data indicate that loss of S100P modulates downstream signaling that are associated with changes in morphology, colony growth and motility. Thus, we propose that regulation of S100P by the PGE2/EP4 receptor signaling pathway may constitute a feedback regulatory mechanism by which tumor cells perpetuate growth and migration during colon carcinogenesis (Fig. 6).
Figure 5.
Silencing of S100P diminishes invadopodia formation, colony growth and cell motility and is associated with decrease in endogenous ERK levels. LS174T cells were stably transfected with empty pLKO1 vector (control) or two independent shRNA constructs against S100P (sh1-S100P and sh2-S100P). (A) Silencing of S100P protein is confirmed by western blotting using S100P-specific antibody (1:1,000). Equal loading was confirmed by probing against α-tubulin. (B) LS174T cells expressing control and sh-S100P constructs were allowed to grow on collagen gels for 8 days and viewed under light microscope (20× magnification). (C) Methylene blue colony formation assay was performed on control or sh-S100P-expressing LS174T cells after 3 weeks of growth. Methylene blue stained colonies were photographed (left) and counted manually (right). Values represent mean ± SD, (**p < 0.01). (D) Transwell motility assay was performed on cells and the filters were photographed after 2 days of incubation (left). Motile cells (crystal violet positive) were counted after 2 days and are shown as a percentage (right). Values represent mean percentage ± SD (**p < 0.01). (E) Endogenous levels of downstream signaling molecule ERK is shown after western blot analysis of LS174T cells transfected with control, sh1- or sh2-S100P constructs. GAPDH levels indicate equal loading.
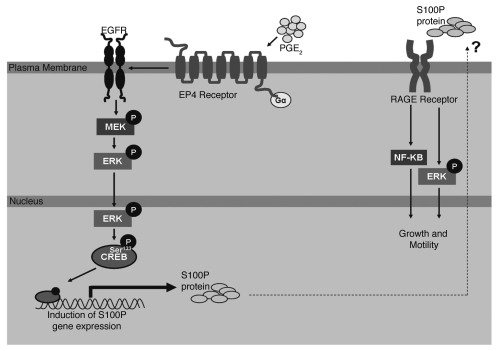
Figure 6.
Model showing PGE2/EP4/CREB/S100P signaling in a feedback regulatory loop.
Discussion
The PGE2/EP4-receptor signaling is implicated in colon tumor progression and metastasis. Thus it is essential to understand how PGE2/EP4-receptor signaling mediates cellular functions in normal physiology and pathologic states such as cancer. Previous studies from our laboratory suggest that the PGE2/EP4-receptor, via ERK, leads to the activation of the transcription factor CREB. Studies employing genetic as well as pharmacological inhibition of CREB have shown that it can suppress growth of cancer cell lines, including endometrial and colon cancer.34,35 However, whether or not CREB activation contributes to colon cancer cell growth has not been previously investigated. We provide new evidence that suppression of CREB activity, by a well-characterized dominant negative mutant construct, suppresses colon cancer cell growth by inducing apoptosis. In the present study, we also focused on identifying the downstream target genes of the PGE2/EP4-receptor in colon cancer cells. We used microarray analysis to find a novel PGE2/EP4-receptor target gene. The gene for calcium binding protein, S100P, was identified as being significantly upregulated by PGE2. This finding is the first of its kind and sheds light on the regulatory mechanisms of this pro-tumorigenic pathway. Altered expression of S100P has been observed in a wide variety of human cancers. Consistent with this literature, we also observed an increase in S100P expression levels in human colon cancers compared to normal specimen.27 Despite this evidence, the upstream regulator of S100P expression, in colon cancer, had not been clarified until now. Our present results reveal that the PGE2/EP4/CREB pathway can upregulate S100P expression in colon as well as other cancer cells (i.e., breast and pancreatic cancer). We further show that PGE2-dependent induction of S100P occurs in the presence of a functional CRE-binding sequence in the promoter region of S100P. Interestingly, we noted an incomplete clearing of the S100P probe in the presence of CREB antibody in our EMSA. This suggests that other transcription factors may also bind the CRE sequence within the S100P promoter. The C/EBP family of transcription factors has been shown to bind the CRE region within the CCR7 promoter in human monocytes.36 Thus, the ability of C/EBP transcription factor to regulate S100P expression in response to PGE2 needs to be further investigated.
Exactly how S100P contributes to the patho-physiology of colorectal cancers remains to be clarified. However, we observed a decrease in colony growth, cell motility and loss of invadopodia formation in colon cancer cells lacking S100P. We also show for the first time a decrease in endogenous levels of ERK MAP kinase with loss of S100P by RNAi. Activation of ERK1/2 is well known to contribute to tumor cell proliferation, invasion and motility. Also, several published reports on pancreatic cancers have shown increased ERK activation (by phosphorylation) by S100P.42–44 Our data suggest that S100P may regulate the expression of ERK and further studies are required to address this issue. Collectively, these studies suggest that elevated S100P levels may participate in tumor cell invasion and migration.
With respect to molecular function, S100P interacts with RAGE and influences intracellular signaling via mitogenic pathways (Fig. 6). The administration of exogenous S100P ligand was shown to increase proliferation (via ERK MAP kinase pathway) as well as cell survival (by constitutive NFκB activation).37,38 At the intracellular level, interaction of S100P with ezrin is necessary for cellular movement via the trans-endothelial passages in lung cancer cells.39 Ezrin is a component of the ERM family of proteins, which crosslink plasma membrane to filamentous actin (F-actin) and needs to be activated in the presence of Ca2+ ions.16 Thus, it is possible that activation of dormant ezrin molecules, by Ca2+ binding S100P protein, assists tumor cells to migrate through otherwise difficult cellular contexts such as the endothelium.
In summary, we have identified S100P as a downstream target gene within the PGE2/EP4-receptor signaling pathway. Transcriptional upregulation of S100P appears to account for increased S100P protein and is mediated by CREB. Combinatorial therapeutic strategies involving the inhibition of the EP4 receptor and S100P activity warrant further investigation.
Materials and Methods
Cell culture and maintenance of cell lines.
LS174T, HCA7, HEK293, MCF7 and Panc1 cell lines (ATCC) were maintained in 1X DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 5 mg/mL penicillin-streptomycin. HEK293 cells stably overexpressing EP4 receptor were maintained in 1X DMEM under 200 µg/mL G418 selection. Cells were plated and allowed to grow for 24 hrs and then serum starved in OptiMEM® I Reduced Serum Medium (Invitrogen) for 20 hrs before drug treatments. All transient transfections were performed using LipofectAMINE 2000 (Invitrogen). PGE2, PGE1OH, GW627368X, L-161,982 (Cayman Chemicals) were prepared in DMSO and maintained at concentrations lower than 0.1%.
Cell survival and colony formation assays.
Cell survival was monitored by seeding 8 × 104 in 6-well plates in triplicate and measured by SRB assay as previously described.40 Apoptosis was measured using Caspase Glo 3/7 Assay (Promega) as per manufacturer's guildelines. Colony formation assay was performed by seeding 500 cells in 10 cm plates and incubating at 37°C for 3 weeks to allow for colonies to form. Media was then removed and colonies were stained in methylene blue dye (0.5% methylene blue and 50% methanol) at room temperature for 10 min. Plates were gently rinsed in water and visible colonies were counted.
DNA constructs and plasmids.
Short hairpin RNA (shRNA) plasmids against a scrambled sequence (shCtrl) or the EP4 receptor (shEP4) were purchased from SuperArray Bioscience Corporation. Dominant negative ACREB plasmids (along with empty vector) and two shRNA plasmids against CREB (with empty vector) were gifts from Dr. Charles Vinson, National Institutes of Health (Bethesda, MD) and Dr. Kathleen Sakamoto, Department of Pathology and Laboratory Medicine, UCLA (Los Angeles, CA). Plasmids (pLKO1) containing shRNA against S100P (with empty vector) were generously donated by Dr. Thiruvengadam Arumugam, Department of Cancer Biology, University of Texas, MD Anderson Cancer Center (Houston, TX).
RNA isolation and realtime RT PCR analysis.
A block of ∼0.5 cm from surgically resected colorectal adenomas or carcinomas was homogenized using Trizol reagent (Invitrogen). RNA was extracted using Qiagen kit reagents. Reverse transcription was performed using 1 µg of RNA from either frozen tissues or cell lines using the iScript cDNA synthesis kit (BioRad Pharmaceuticals). For realtime analysis, 1 µL (50 ng) of the cDNA mix was added to a 25 µL reaction mix containing 1X Roche premix, 0.5 µM forward and reverse primers (S100P-F: 5′-GAT ATT CGG GCA GCG AGG G-3′; S100P-R: 5′-GCA TCC TTG TCT TTT CCA CTC TGC-3′; βactin-F: 5′-GCA AAG ACC TGT ACG CCA A-3′; βactin-R: 5′-GGA GGA GCA ATG ATC TTG ATC TTC-3′). Realtime RT-PCR was performed using Roche SyBr-Green reagents in the LC480 Light Cycler (Roche Diagnostics). Relative quantification was performed using the 2−ΔΔCt method as described.41
Protein extraction and western blot analysis.
Total protein was isolated from cells or tissue sections using protein cell lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate and 1% Protease Inhibitor Cocktail: 1 mM phenyl methane sulphonyl fluoride, 1 mM sodium ortho vanadate and sodium fluoride). Equal amount of proteins were resolved by electrophoresis in a 15% SDS-polyacrylamide gel. Western blotting was performed as previously described.9 S100P-(R & D Systems) or EP4 receptor-(Cayman Chemicals) specific antibodies were used at 1:1,000 followed by horseradish-peroxidase-linked secondary antibody (Santa Cruz Biotechnology) at 1:10,000. CREB-(Santa Cruz) and ERK-(Cell Signaling) specific antibody was used at 1:500 dilution.
Site-directed mutagenesis of S100P promoter.
To assess the promoter activity of the s100p gene, pGL3 based deletion constructs were used.30 The −236/+58 construct containing STAT and CRE binding sites were mutated using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene). Mutated oligos (Sigma-Genosys) were designed based on previous studies (MutSTAT-F: 5′-GGG GAA AGG TGC CAC AAA CGT CAT CAC AAC-3′, MutSTAT-R: 5′-GTT GTG ATG ACG TTT GTG GCA CCT TTC CCC-3′, MutCRE-F: 5′-GGG GAA AGG TTC CAG AAA GCC CAT CAC AAC-3′, MutCRE-R: 5′ GTT GTG ATG GGC TTT CTG GAA CCT TTC CCC-3′).31–33
Dual luciferase promoter reporter assay.
Transient transfections were performed in LS174T cells. After 24 hrs of transfection, cells were serum starved in OptiMEM medium for 20 hrs and treated with 1 µM PGE2 for an additional 24 hrs. Luciferase activity was assayed using the Dual Luciferase Reporter™ Assay (Promega) according to manufacturer's directions and luminescence was measured using the Sirius Luminometer (Berthold Detections Systems).
Electrophoretic mobility shift assay (EMSA).
Nuclear protein extracts were prepared from LS174T cells treated with 1 µM PGE2 for indicated time points as described previously.42 For competition experiments, unlabeled oligonucleotides (Sigma-Genosys), containing wild-type and mutant CRE binding sequences (S100PWT-CRE: 5′-CCA GAA ACG TCA TCA; S100PMUT-CRE: 5′-CCA GAA AGC CCA TCA-3′) were added to the binding reaction 1 hr prior to addition of radio-labeled probe. For antibody supershift assay 3 µg of CREB-specific antibody (Millipore) were incubated with nuclear protein 1 hr prior to probe binding.
Tumor cell morphology.
Colon cancer cells (LS174T) stably expressing empty vector (control) or shRNA plasmids against S100P (sh-S100P) were grown in complete medium for 24 hrs. Subsequently, collagen gels were prepared as previously described using rat tail collagen.43 4 × 104 cells were seeded on collagen gels in 16 mm multiwell dishes (Nunc). Cells were allowed to grow and populate within the collagen layer for 8 days in OptiMEM supplemented with antibiotics. After 8 days, cells were observed under light microscope for changes in morphology.
Transwell motility assay.
4 × 104 LS174T cells stably expressing empty vector (control) or two independent shRNA plasmids against S100P (sh1-S100P and sh2-S100P) were suspended in 150 µL of OptiMEM (with antibiotics). Cells were seeded in the upper chamber of an 8 µm pore Falcon transwell insert (24 well format). The bottom chamber was filled with 600 µL of OptiMEM (with antibiotics). Transwell plates containing cells were incubated for 5 days at 37°C, 5% CO2. Experiment was repeated two times. At the end of the incubation, the medium of the upper and bottom chambers were aspirated and the inserts were placed upside down. The top surface of the inserts was stained with 50 µL of crystal violet stain (0.5% crystal violet in 20% methanol) for 1 min at room temperature. Excess crystal violet solution was removed by plunging the inserts in distilled water several times and then rinsing a second time in distilled water. Non-motile cells inside the insert were removed with a wet cotton swap and the inserts were allowed to dry overnight. Pictures of five fields of each insert were captured under a light microscope and motile cells (cells penetrating through the membrane) were counted as described.44 The numbers of motile cells were expressed as percentage of motile cells over non motile cells. The experiment was repeated three times independently.
All the data is MIAME compliant and the raw data from our microarray studies has deposited in a MIAME compliant database—Gene Expression Omnibus (GEO).
Acknowledgements
This work was supported by the Arizona Cancer Center SPORE Grant CA95060 (MN), Allergan Inc. (JR), and the Lockton Endowment for Pancreatic Cancer Research (CL). Microarray data was generated by the Arizona Cancer Center Genomics Shared Services, supported by NCI grant CA023074-26 and NIEHS grant ES06694. We thank Dr. George Watts for assistance with microarray studies. We are also grateful to Dr. G. Tim Bowden and his laboratory personnel for assistance on the EMSA assays.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/13373
Supplementary Material
References
- 1.Hull MA, Ko SC, Hawcroft G. Prostaglandin EP receptors: targets for treatment and prevention of colorectal cancer? Mol Cancer Ther. 2004;3:1031–1039. [PubMed] [Google Scholar]
- 2.Kettunen HL, Kettunen AS, Rautonen NE. Intestinal immune responses in wild-type and Apcmin/+ mouse, a model for colon cancer. Cancer Res. 2003;63:5136–5142. [PubMed] [Google Scholar]
- 3.Wang D, Buchanan FG, Wang H, Dey SK, DuBois RN. Prostaglandin E2 enhances intestinal adenoma growth via activation of the Ras-mitogen-activated protein kinase cascade. Cancer Res. 2005;65:1822–1829. doi: 10.1158/0008-5472.CAN-04-3671. [DOI] [PubMed] [Google Scholar]
- 4.Hansen-Petrik MB, McEntee MF, Jull B, Shi H, Zemel MB, Whelan J. Prostaglandin E protects intestinal tumors from nonsteroidal anti-inflammatory drug-induced regression in Apc(Min/+) mice. Cancer Res. 2002;62:403–408. [PubMed] [Google Scholar]
- 5.Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, et al. The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–386. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 6.Regan JW. EP2 and EP4 prostanoid receptor signaling. Life Sci. 2003;74:143–153. doi: 10.1016/j.lfs.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 7.Cha YI, DuBois RN. NSAIDs and cancer prevention: targets downstream of COX-2. Annu Rev Med. 2007;58:239–252. doi: 10.1146/annurev.med.57.121304.131253. [DOI] [PubMed] [Google Scholar]
- 8.Budarf ML, Labbe C, David G, Rioux JD. GWA studies: rewriting the story of IBD. Trends Genet. 2009;25:137–146. doi: 10.1016/j.tig.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Cherukuri DP, Chen XB, Goulet AC, Young RN, Han Y, Heimark RL, et al. The EP4 receptor antagonist, L-161,982, blocks prostaglandin E2-induced signal transduction and cell proliferation in HCA-7 colon cancer cells. Exp Cell Res. 2007;313:2969–2979. doi: 10.1016/j.yexcr.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chell SD, Witherden IR, Dobson RR, Moorghen M, Herman AA, Qualtrough D, et al. Increased EP4 receptor expression in colorectal cancer progression promotes cell growth and anchorage independence. Cancer Res. 2006;66:3106–3113. doi: 10.1158/0008-5472.CAN-05-3702. [DOI] [PubMed] [Google Scholar]
- 11.Sheng H, Shao J, Washington MK, DuBois RN. Prostaglandin E2 increases growth and motility of colorectal carcinoma cells. J Biol Chem. 2001;276:18075–18081. doi: 10.1074/jbc.M009689200. [DOI] [PubMed] [Google Scholar]
- 12.Leone V, di Palma A, Ricchi P, Acquaviva F, Giannouli M, Di Prisco AM, et al. PGE2 inhibits apoptosis in human adenocarcinoma Caco-2 cell line through Ras-PI3K association and cAMP-dependent kinase A activation. Am J Physiol Gastrointest Liver Physiol. 2007;293:673–681. doi: 10.1152/ajpgi.00584.2006. [DOI] [PubMed] [Google Scholar]
- 13.Arumugam T, Ramachandran V, Logsdon CD. Effect of cromolyn on S100P interactions with RAGE and pancreatic cancer growth and invasion in mouse models. J Natl Cancer Inst. 2006;98:1806–1818. doi: 10.1093/jnci/djj498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G, Platt-Higgins A, Carroll J, de Silva Rudland S, Winstanley J, Barraclough R, et al. Induction of metastasis by S100P in a rat mammary model and its association with poor survival of breast cancer patients. Cancer Res. 2006;66:1199–1207. doi: 10.1158/0008-5472.CAN-05-2605. [DOI] [PubMed] [Google Scholar]
- 15.Mousses S, Bubendorf L, Wagner U, Hostetter G, Kononen J, Cornelison R, et al. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res. 2002;62:1256–1260. [PubMed] [Google Scholar]
- 16.Bulk E, Hascher A, Liersch R, Mesters RM, Diederichs S, Sargin B, et al. Adjuvant therapy with small hairpin RNA interference prevents non-small cell lung cancer metastasis development in mice. Cancer Res. 2008;68:1896–1904. doi: 10.1158/0008-5472.CAN-07-2390. [DOI] [PubMed] [Google Scholar]
- 17.Kita H, Hikichi Y, Hikami K, Tsuneyama K, Cui ZG, Osawa H, et al. Differential gene expression between flat adenoma and normal mucosa in the colon in a microarray analysis. J Gastroenterol. 2006;41:1053–1063. doi: 10.1007/s00535-006-1894-y. [DOI] [PubMed] [Google Scholar]
- 18.Birkenkamp-Demtroder K, Olesen SH, Sorensen FB, Laurberg S, Laiho P, Aaltonen LA, et al. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut. 2005;54:374–384. doi: 10.1136/gut.2003.036848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445–456. doi: 10.1093/hmg/10.5.445. [DOI] [PubMed] [Google Scholar]
- 20.Ohuchida K, Mizumoto K, Egami T, Yamaguchi H, Fujii K, Konomi H, et al. S100P is an early developmental marker of pancreatic carcinogenesis. Clin Cancer Res. 2006;12:5411–5416. doi: 10.1158/1078-0432.CCR-06-0298. [DOI] [PubMed] [Google Scholar]
- 21.Hammacher A, Thompson EW, Williams ED. Interleukin-6 is a potent inducer of S100P, which is upregulated in androgen-refractory and metastatic prostate cancer. Int J Biochem Cell Biol. 2005;37:442–450. doi: 10.1016/j.biocel.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Hamada S, Satoh K, Hirota M, Fujibuchi W, Kanno A, Umino J, et al. Expression of the calcium-binding protein S100P is regulated by bone morphogenetic protein in pancreatic duct epithelial cell lines. Cancer Sci. 2009;100:103–110. doi: 10.1111/j.1349-7006.2008.00993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Namba T, Homan T, Nishimura T, Mima S, Hoshino T, Mizushima T. Upregulation of S100P expression by non-steroidal anti-inflammatory drugs and its role in anti-tumorigenic effects. J Biol Chem. 2009;284:4158–4167. doi: 10.1074/jbc.M806051200. [DOI] [PubMed] [Google Scholar]
- 24.Fujino H, Regan JW. EP prostanoid receptor coupling to a pertussis toxin-sensitive inhibitory G protein. Mol Pharmacol. 2006;69:5–10. doi: 10.1124/mol.105.017749. [DOI] [PubMed] [Google Scholar]
- 25.Fujino H, West KA, Regan JW. Phosphorylation of glycogen synthase kinase-3 and stimulation of T-cell factor signaling following activation of EP2 and EP4 prostanoid receptors by prostaglandin E2. J Biol Chem. 2002;277:2614–2619. doi: 10.1074/jbc.M109440200. [DOI] [PubMed] [Google Scholar]
- 26.Fujino H, Xu W, Regan JW. Prostaglandin E2 induced functional expression of early growth response factor-1 by EP4, but not EP2, prostanoid receptors via the phosphatidylinositol 3-kinase and extracellular signal-regulated kinases. J Biol Chem. 2003;278:12151–12156. doi: 10.1074/jbc.M212665200. [DOI] [PubMed] [Google Scholar]
- 27.Parkkila S, Pan PW, Ward A, Gibadulinova A, Oveckova I, Pastorekova S, et al. The calcium-binding protein S100P in normal and malignant human tissues. BMC Clin Pathol. 2008;8:2. doi: 10.1186/1472-6890-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subbaramaiah K, Benezra R, Hudis C, Dannenberg AJ. Cyclooxygenase-2-derived prostaglandin E2 stimulates Id-1 transcription. J Biol Chem. 2008;283:33955–33968. doi: 10.1074/jbc.M805490200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wang D, Wang H, Brown J, Daikoku T, Ning W, Shi Q, et al. CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. J Exp Med. 2006;203:941–951. doi: 10.1084/jem.20052124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibadulinova A, Oveckova I, Parkkila S, Pastorekova S, Pastorek J. Key promoter elements involved in transcriptional activation of the cancer-related gene coding for S100P calcium-binding protein. Oncol Rep. 2008;20:391–396. [PubMed] [Google Scholar]
- 31.Eresh S, Riese J, Jackson DB, Bohmann D, Bienz M. A CREB-binding site as a target for decapentaplegic signalling during Drosophila endoderm induction. EMBO J. 1997;16:2014–2022. doi: 10.1093/emboj/16.8.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu CR, Ortaldo JR, Curiel RE, Young HA, Anderson SK, Gosselin P. Role of a STAT binding site in the regulation of the human perforin promoter. J Immunol. 1999;162:2785–2790. [PubMed] [Google Scholar]
- 33.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catalano S, Giordano C, Rizza P, Gu G, Barone I, Bonofiglio D, et al. Evidence that leptin through STAT and CREB signaling enhances cyclin D1 expression and promotes human endometrial cancer proliferation. J Cell Physiol. 2009;218:490–500. doi: 10.1002/jcp.21622. [DOI] [PubMed] [Google Scholar]
- 35.Corona G, Deiana M, Incani A, Vauzour D, Dessi MA, Spencer JP. Inhibition of p38/CREB phosphorylation and COX-2 expression by olive oil polyphenols underlies their anti-proliferative effects. Biochem Biophys Res Commun. 2007;362:606–611. doi: 10.1016/j.bbrc.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 36.Cote SC, Pasvanis S, Bounou S, Dumais N. CCR7-specific migration to CCL19 and CCL21 is induced by PGE stimulation in human monocytes: Involvement of EP/EP receptors activation. Mol Immunol. 2009;46:2682–2693. doi: 10.1016/j.molimm.2008.08.269. [DOI] [PubMed] [Google Scholar]
- 37.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 38.Fuentes MK, Nigavekar SS, Arumugam T, et al. RAGE activation by S100P in colon cancer stimulates growth, migration and cell signaling pathways. Dis Colon Rectum. 2007;50:1230–1240. doi: 10.1007/s10350-006-0850-5. [DOI] [PubMed] [Google Scholar]
- 39.Koltzscher M, Neumann C, Konig S, Gerke V. Ca2+-dependent binding and activation of dormant ezrin by dimeric S100P. Mol Biol Cell. 2003;14:2372–2384. doi: 10.1091/mbc.E02-09-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Gonzales M, Bowden GT. Ultraviolet B (UVB) induction of the c-fos promoter is mediated by phospho-cAMP response element binding protein (CREB) binding to CRE and c-fos activator protein 1 site (FAP1) cis elements. Gene. 2002;293:169–179. doi: 10.1016/s0378-1119(02)00723-0. [DOI] [PubMed] [Google Scholar]
- 43.Mercado-Pimentel ME, Hubbard AD, Runyan RB. Endoglin and Alk5 regulate epithelial-mesenchymal transformation during cardiac valve formation. Dev Biol. 2007;304:420–432. doi: 10.1016/j.ydbio.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez-Diaz S, Valle N, Garcia JM, Peña C, Freije JM, Quesada V, et al. Cystatin D is a candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J Clin Invest. 2009;119:2343–2358. doi: 10.1172/JCI37205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.