Abstract
A large body of emerging evidence indicates a functional interaction between the kallikrein-related peptidases (KLKs) and proteases of the thrombostasis axis. These interactions appear relevant for both normal health as well as pathologies associated with inflammation, tissue injury, and remodeling. Regulatory interactions between the KLKs and thrombostasis proteases could impact several serious human diseases, including neurodegeneration and cancer. The emerging network of specific interactions between these two protease families appears to be complex, and much work remains to elucidate it. Complete understanding how this functional network resolves over time, given specific initial conditions, and how it might be controllably manipulated, will probably contribute to the emergence of novel diagnostics and therapeutic agents for major diseases.
Keywords: activation cascade, kallikrein, kallikrein-related peptidase (KLK), plasmin, thrombogenesis, thrombolysis, thrombostasis
Introduction
The study of the human kallikrein-related peptidases (KLKs; for a recent review see Sotiropoulou et al., 2009) has witnessed remarkable progress over the past decade. Initial studies of the properties of KLK proteins from tissue extracts were obfuscated by the lack of both an understanding regarding the total number of uniquely different KLK proteins and an associated commonly accepted nomenclature. The deciphering of the genomic organization of the human KLK gene locus (Gan et al., 2000; Harvey et al., 2000; Yousef et al., 2000), as well as the adoption of a commonly accepted nomenclature (Lundwall et al., 2006), resolved these two fundamental issues. The vast body of work has associated several cancer pathologies with differential regulation or expression of individual members of the KLK family, and has served to elevate the importance of the KLKs in serious human disease and their diagnosis (Diamandis et al., 2000; Diamandis and Yousef, 2001; Yousef and Diamandis, 2001, 2003; Borgoño et al., 2004). Studies of the functional properties of KLK proteins have progressed from individual members (Lilja, 1985; Watt et al., 1986; Lundstrom and Egelrud, 1991; Kishi et al., 1997; Little et al., 1997; Brattsand and Egelrud, 1999; Nelson et al., 1999) to the interaction among sets of KLKs active in specific tissues (Lovgren et al., 1997; Brattsand et al., 2005; Michael et al., 2006). Similarly, initial studies of KLK substrate specificities has progressed from analysis of individual substrates under a single buffer condition to specificity profiling using peptide or phage libraries (Janssen et al., 2004; Debela et al., 2006; Li et al., 2008), as well as studies of the effects of a broad range of cosolvents upon enzymatic activity (Angelo et al., 2006). Furthermore, the early association of KLKs with important pathologies has progressed to detailed studies of signaling through specific receptor molecules (e.g., the PARs), providing a molecular description of the role of specific KLKs in hormone-like signal transduction pathways (Oikonomopoulou et al., 2006b; Hollenberg et al., 2008; Ramsay et al., 2008a; Vandell et al., 2008), as well as widespread acceptance of disease-specific association of particular KLKs (particularly KLK3 with prostate cancer) and with significant diagnostic utility (Stamey et al., 1987; Catalona et al., 1991; Lilja et al., 2008).
Because the KLKs are initially secreted as inactive pro-forms that must be proteolytically cleaved to achieve functional activity, there has been substantial interest in the potential for activation cascades or networks among sets of coexpressed KLKs. This set of activation interactions among the KLKs, defining a network of regulatory interactions among the KLKs, is termed the KLK ‘activome’. As with other forms of characterization, studies of the KLK activome have progressed from individual pairwise studies (Lovgren et al., 1997; Denmeade et al., 2001) to high-throughput efforts to provide a comprehensive description of the entire KLK activome potential (Yoon et al., 2007, 2009). These and other studies have demonstrated a vast potential for self-activation and reciprocal cross-activation among the KLKs, resulting in complex networks that contrast with more classical linear-type cascades (e.g., the blood-clotting cascade). Interest is expanding to understand how the KLK family might functionally interact with members of other major protease families, such as the matrix metalloproteases and thrombostasis enzymes. The purpose of this review is to summarize available data related to the functional interactions between the KLKs and proteases of the thrombostasis system (i.e., both thrombogenic and thrombolytic proteases). A thorough review of such interactions was provided by Borgoño and Diamandis in 2004 (Borgoño and Diamandis, 2004); however, significant additional new data has been reported. These data provide compelling evidence that a substantial functional intersection exists between these two major protease families.
Proteolytic activation of pro-KLKs by thrombostasis proteases
Plasmin and urokinase-type plasminogen activator (uPA) have both been shown to activate pro-KLK6, and plasmin is currently the most efficient known activator of pro-KLK6 (Blaber et al., 2007; Yoon et al., 2008). Pro-KLK11 can be activated by both thrombolytic and thrombogenic proteases, including plasmin, uPA, factor Xa, and plasma kallikrein; of these proteases, plasma kallikrein is the most efficient activator (Yoon et al., 2008). Pro-KLK12 can similarly be activated by a set of both thrombolytic and thrombogenic proteases, including plasmin, uPA, thrombin, and plasma kallikrein; however, in this case thrombin is the most efficient activator. Indeed, the activation profile of thrombin against the KLK pro-sequences as substrates indicates that thrombin has a significant and unique specificity for pro-KLK12 (Yoon et al., 2008). Pro-KLK14, like pro-KLK6, can be activated by the thrombolytic proteases plasmin and uPA, with plasmin being the more efficient activator of the two (Yoon et al., 2008).
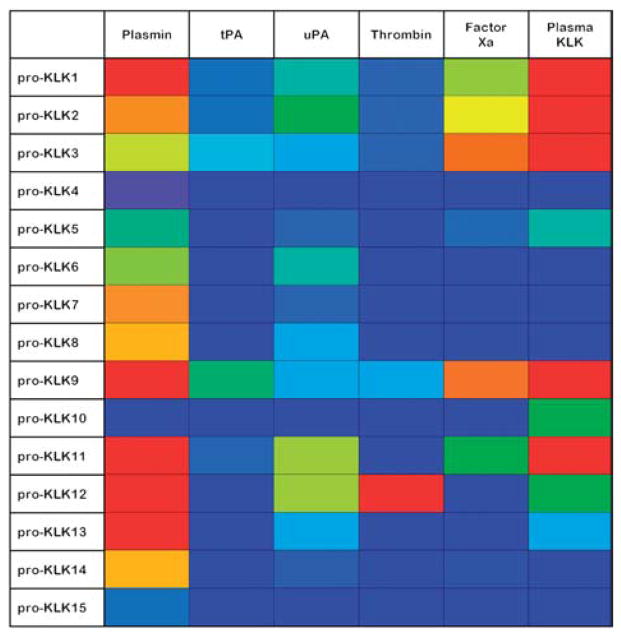
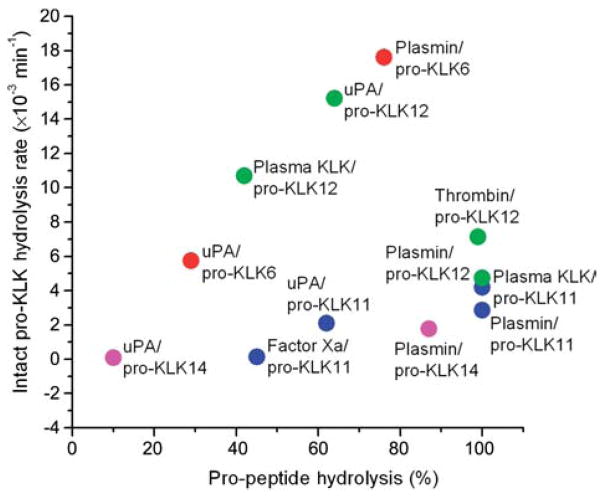
Plasmin, and to a lesser extent uPA, stand out as ‘general activators’ of the KLK pro-peptide sequences; in this regard, both Arg- and Lys-P1 pro-peptides appear efficiently hydrolyzed. Factor Xa and plasma kallikrein exhibit a similar activation profile against the KLK pro-peptide sequences, with a pronounced Arg selectivity, and with plasma kallikrein generally the more active of the two proteases (Yoon et al., 2008). Figure 1 shows a ‘heat map’ indicating the percent hydrolysis of KLK pro-peptides by different thrombostasis proteases. In this representation the extent of hydrolysis is indicated by the gradation of hue, where red=100%, and blue=0%, and the standard spectrum of colors (i.e., red, orange, yellow, green, blue) define the intermediate percent hydrolysis values. Available data comparing the hydrolysis rates by thrombostasis proteases of KLK pro-peptide sequences versus the intact native pro-KLK protein (Figure 2) suggest that some interactions can utilize exosites separate from the pro-peptide region to enhance catalysis. In particular, Figure 2 indicates that the uPA/pro-KLK6, plasma KLK/pro-KLK12, uPA/pro-KLK12, and plasmin/pro-KLK6 hydrolytic rates are enhanced with the intact pro-KLK protein, thereby identifying the presence of potential exosite interactions.
Figure 1. A ‘heat map’ indicating the percent hydrolysis of KLK pro-peptides by different thrombostasis proteases.
In this representation the extent of hydrolysis is indicated by the gradation of hue, where red=100%, and dark blue=0%, and the standard spectrum of colors (i.e., red, orange, yellow, green, blue) define the intermediate percent hydrolysis values. The indicated hydrolysis is for pH 7.4 conditions over 24 h (Yoon et al., 2008).
Figure 2. A comparison of the percent hydrolysis of KLK pro-peptides (abscissa) with the hydrolysis rate for intact pro-KLK proteins (ordinate) (Yoon et al., 2008).
Data points are colored according to the individual pro-KLK protein being hydrolyzed. The plot identifies hydrolyses that exhibit enhanced rates for the intact pro-KLK relative to the pro-KLK peptide (e.g., uPA/pro-KLK6, plasma KLK/pro-KLK12, uPA/pro-KLK12, and plasmin/pro-KLK6), thereby identifying potential exosite interactions present within the intact pro-KLK protein.
Proteolytic activation of thrombostasis proteases by mature KLKs
(MOUSE)Klk1 (mouse mGK-6) can activate single-chain uPA (independent of plasmin) in plasminogen-deficient mice (List et al., 2000). The urine from plasminogen−/− mice contained active two-chain uPA as well as a protease capable of activating exogenously added pro-uPA. Mass spectrometry and peptide mapping identified this protease as mGK-6 the mouse ortholog of (HUMAN)KLK1 (true tissue kallikrein). These results demonstrate that KLK1 is an activator of pro-uPA in the urinary tract; furthermore, as KLK1 occurs in other tissues, in addition to plasma, it is also a likely physiological activator of pro-uPA in other locations and tissue microenvironments.
(MOUSE)Klk1 can convert single-chain tissue plasminogen activator (tPA) to two-chain tPA (Rajapakse et al., 2007). Single-chain tPA and two-chain tPA are different in plasminogen activating ability and affinity to fibrins. It has been shown that two-chain tPA has ten times the plasminogen activating ability as compared with single-chain tPA (Japanese Patent Laid-Open No. 118717/1984). Single-chain tPA has very low plasmin-generating activity in the absence of cofactor fibrin, whereas two-chain tPA shows full activity without fibrin (Stubbs et al., 1998). Thus, (MOUSE)Klk1 could trigger activation of the tPA/plasmin system in the absence of fibrin (Rajapakse et al., 2007).
As regards the ability of other members of the KLK family to proteolytically activate thrombostasis proteases, KLK2 has been shown to activate single-chain uPA, leading to the generation of plasmin (Frenette et al., 1997), KLK4 can activate single-chain uPA, leading to the generation of plasmin (Takayama et al., 2001), and KLK8 has also been shown to have a single-chain tPA converting activity (Rajapakse et al., 2005).
Proteolytic inactivation/degradation of thrombostasis proteases/proteins
KLK3 from human seminal plasma can degrade fibrinogen (Watt et al., 1986), producing fibrin/fibrinogen degradation products that can profoundly impair the hemostatic process. KLK4 can digest the α-chain of fibrinogen; as KLK4 expression is upregulated in prostate cancer its potential involvement in cancer pathology could be via the degradation of collagen and fibrinogen in the extracellular matrix, thereby facilitating cancer cell invasion (Obiezu et al., 2006). KLK5, and to a lesser extent KLK8, have been shown to degrade both the α- and β-chains of fibrinogen (Brattsand et al., 2009). Cryptic tPA and plasminogen binding sites are located within the fibrinogen molecule, and a portion of the α-chain contains both plasminogen and tPA binding sites (Tsurupa and Medved, 2001). Thus, KLK4, 5, and 8 cleavage of fibrinogen α-chain can regulate functional interactions between components of the thrombolytic system. Internal cleavage of uPA by KLK5, potentially resulting in functional inactivation of uPA, is predicted based upon positional scanning synthetic combinatorial peptide libraries (Borgoño et al., 2007a).
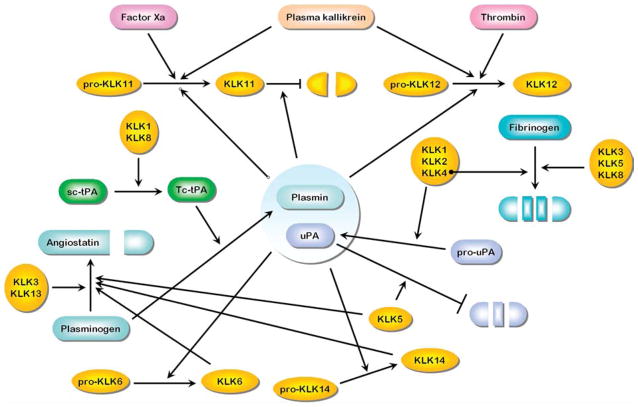
The interactions described above, as well as the prior description of pro-KLK activations by thrombostasis proteases, and the proteolytic activation of thrombostasis proteases by mature KLKs, are summarized in Figure 3. In this Figure the arrowheads indicate specific proteolytic activities, typically resulting in activation of inactive protease pro-forms. In some cases, the specific proteolytic activity results in inactivation of the target protease (indicated by a blunted line).
Figure 3. Interactions between the thrombostasis and KLK axes involving activation and proteolytic processing (see text for individual references).
Arrowheads indicate specific proteolytic activities, typically resulting in activation of inactive pro-forms. In some cases, the specific proteolytic activity results in inactivation of the target protease (indicated by a blunted line).
Generation of angiostatin-like fragments from plasminogen by KLKs
Angiostatin is a fragment of plasminogen (containing kringle domains 1–4) that is a biologically active inhibitor of angiogenesis (O’Reilly et al., 1994). Angiostatin is produced by the proteolytic cleavage of plasminogen by metalloelastase (matrix metalloprotease 12) (Dong et al., 1997). KLK3 (prostate-specific antigen) has been shown to be able to convert Lys-plasminogen to biologically active angiostatin-like fragments by cleavage between Glu439 and Ala440 located between kringle domains 4 and 5 (Heidtmann et al., 1999). A study of the enzymatic properties of KLK5 suggested that it can potentially release angiostatin from plasminogen, as well as ‘cystatin-like domain 3’ from low-molecular weight kininogen, and fibrinopeptide B and peptide β15–42 from the Bβ chain of fibrinogen (Michael et al., 2005). Related processing of plasminogen to release angiostatin-like fragments has also been reported for KLK6 (Bayes et al., 2004), KLK13 (Sotiropoulou et al., 2003), and KLK14 (Borgoño et al., 2007c).
Protease-activated receptor (PAR) signaling
Protease-activated receptors (PARs) are G-protein-coupled receptors that can be activated by proteolytic cleavage and unmasking of a tethered receptor-triggering ligand (for review see Traynelis and Trejo, 2007). Cleavage downstream of such positions can ‘disarm’ PARS – rendering them incapable of subsequent proteolytic activation (Hansen et al., 2008). Activated PARs trigger responses ranging from vaso-dilation to intestinal inflammation, increased cytokine production, and increased nociception (Hansen et al., 2008; Hollenberg et al., 2008; Ramsay et al., 2008b). PARs play a key role in the body’s innate immune defense system as a primary trigger of the inflammatory response and pain sensation owing to tissue injury or remodeling caused by pathogenic processes (Hollenberg et al., 2008). The coagulation cascade and PARs together provide a mechanism that links tissue injury to cellular responses, and PARs account for the majority of the cellular effects of thrombin (Coughlin, 2005).
In vivo, the enzymes of the coagulation cascade are physiological regulators of PAR activity. Thrombin can activate PARs 1, 3, and 4 in vivo (Coughlin, 2005; Ludeman et al., 2005). Factor-VIIa/Xa complex can activate both PAR1 and PAR2 (Ruf et al., 2003; Ruf and Mueller, 2006; Versteeg and Ruf, 2006). Plasmin can both activate and disarm PAR1 (Kimura et al., 1996; Kuliopulos et al., 1999) and can activate PAR4 (Quinton et al., 2004). In certain circumstances factor Xa can activate PAR1 (Blanc-Brude et al., 2005; Bhattacharjee et al., 2008), and PAR2 can be activated by factor Xa (Camerer et al., 2000). Fibroblasts appear to be the only cell type in which the effects of factor Xa are mediated mainly via PAR1 and not PAR2 (Blanc-Brude et al., 2005). PAR4 can be activated by several different proteases, including thrombin (Kahn et al., 1998; Xu et al., 1998). Activation of PAR4 can play a key role in generating two hallmarks of the inflammatory response: edema and granulocyte infiltration.
(MOUSE)Klk1 activates PAR4 in a rodent paw edema model (Houle et al., 2005); thus, the kallikreinkinin system is an important contributor to the inflammatory response. KLK4 activates both PAR1 and PAR2 but not PAR4 (Ramsay et al., 2008a). KLK14 can both activate and disarm PAR1, thereby preventing its activation by thrombin (Oikonomopoulou et al., 2006a). Immunohistochemical analysis demonstrates the coexpression of KLK4 and PAR2 in primary prostate cancer and bone metastases, indicating that KLK4 signaling via PAR2 could be important in prostate cancer. PAR2 is activated by KLK5, 6, and 14 (Oikonomopoulou et al., 2006a,b). KLK14 is as potent as thrombin for the activation of PAR4 (Oikonomopoulou et al., 2006a). KLK6 can activate PAR1 on NSC34 neurons and both PAR1 and PAR2 on Neu7 astrocytes (Vandell et al., 2008).
The above data highlight the potential for functional overlap between the kallikrein-related peptidases and thrombostasis system in PAR signaling and regulation (Figure 4A). Like thrombin, the KLKs are now considered as important ‘hormonal’ regulators of tissue function (Oikonomopoulou et al., 2006b).
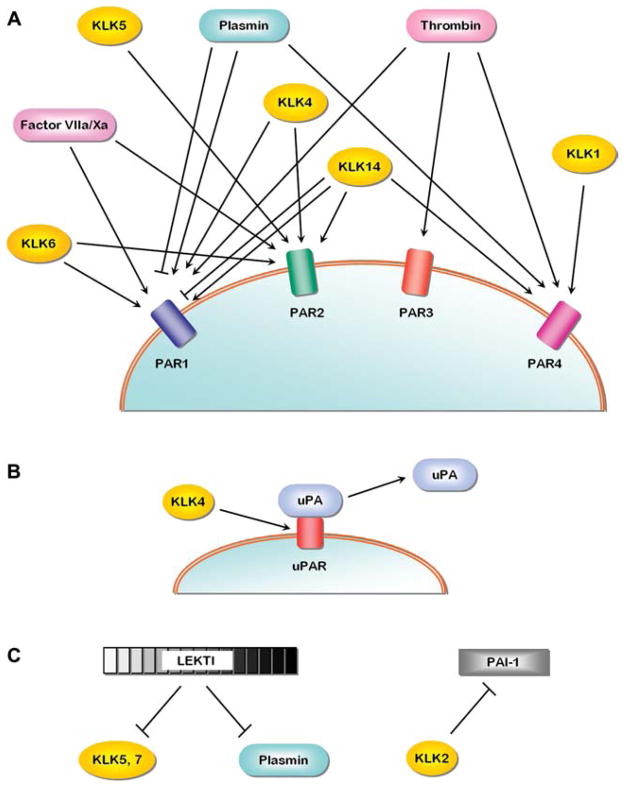
Figure 4. Additional regulatory interactions involving the KLKs.
(A) Intersection between the thrombostasis and KLK axes and PAR signaling; (B) control of uPAR binding of uPA by KLK4; (C) intersection of thrombostasis and KLK axes related to specific inhibitors (see text for individual references). The meaning of the arrowheads and blunted lines follows the convention defined in the legend to Figure 3.
uPAR cleavage
uPAR is a surface receptor for uPA that can result in the localization of plasmin-generating activity to the surface of cells, and is therefore a key element in processes affecting cell migration and tissue remodeling (Blasi and Carmeliet, 2002). Furthermore, by interacting with other molecules (e.g., vitronectin, integrin adhesion proteins, caveolin, and a G-protein-coupled receptor) uPAR can facilitate the initiation of several intracellular signal transduction pathways that involve cytosolic and transmembrane kinases, cytoskeletal components, and others (see the review by Blasi and Carmeliet, 2002).
The function of uPAR is regulated in part through proteolytic cleavage that can lead to the shedding of uPA-binding domain. KLK4 regulates the function of uPAR; KLK4 cleaves soluble recombinant uPAR both in its D1–D2 linker sequence and at the carboxy terminus of D3 (Beaufort et al., 2006). As the D1 amino-terminal domain of uPAR is required for high-affinity interactions between uPA and uPAR (Ploug, 2003), this action of KLK4 upon uPAR would effectively reduce the localization of plasmin-generating activity at the cell surface. The uPAR interactions involving members of the KLK family and the thrombostasis proteases are illustrated in Figure 4B.
Inhibitor function
Lymphoepithelial Kazal-type-related inhibitor (LEKTI, product of the SPINK5 gene; Chavanas et al., 2000) contains 15 different serine protease inhibitory domains (Mägert et al., 1999). The inhibitory functions of the LEKTI domains are diverse and can inhibit plasmin (Mitsudo et al., 2003; Egelrud et al., 2005) as well as specific KLKs (including KLK5 and KLK7) (Egelrud et al., 2005; Schechter et al., 2005). The balance between KLKs and LEKTI is essential for normal skin desquamation and barrier function (Chavanas et al., 2000; Ekholm et al., 2000; Komatsu et al., 2002, 2008; Bitoun et al., 2003; Caubet et al., 2004; Egelrud et al., 2005; Schechter et al., 2005). The presence of KLKs and LEKTI can both be detected in serum (Mägert et al., 1999; Yousef and Diamandis, 2001), and thus potentially interact functionally with plasmin and other thrombostasis proteases.
Growth hormone (hGH) is proteolytically processed by plasmin and thrombin in both the pituitary and periphery (Baumann, 1991; Garcia-Barros et al., 2000). KLK5–8 and KLK10–14, as well as LEKTI are also expressed in the pituitary, localized to hGH-producing cells; thus, thrombostasis proteases, KLKs, and LETKI have been postulated to be involved in the regulation of hGH processing (Komatsu et al., 2007). Therefore, stratum corneum, blood, and pituitary provide emerging evidence of important in vivo regulatory interactions between specific KLKs, thrombostasis proteases, and their inhibitors.
KLK5 is inhibited by α-2-antiplasmin (Michael et al., 2005) and is also predicted based on positional scanning synthetic combinatorial peptide libraries (Borgoño et al., 2007b). Newly identified SPINK9 inhibitor (product of the SPINK9 gene) from human skin has been shown to inhibit KLK5 (Brattsand et al., 2009; Meyer-Hoffert et al., 2009). Although SPINK9 does not appear to inhibit other proteases (including thrombin and plasmin), its effective inhibition of KLK5 eliminates the degradation of fibrinogen by KLK5.
Other hydrolysis reactions involving KLKs
A putative proteolytic inactivator of KLK11 is plasmin (however, this cleavage can only partially inactivate KLK11; Luo et al., 2006) and both proteases are colocalized in semen. Mature KLK2 was shown to inactivate plasminogen activator inhibitor-1 (PAI-1) the primary inhibitor of uPA (Mikolajczyk et al., 1999), thus potentially effectively increasing the relative levels of active uPA. KLK3 has also been shown to bind to and inactivate protein C inhibitor (PAI-3/PCI) (Catalona et al., 1991) which is also an inhibitor of uPA.
Thrombin can activate MMP-2 (Lafleur et al., 2001). MMP20 can activate (PORCINE)pro-Klk4 (Ryu et al., 2002) an activity that is essential for the maturation (deproteination) of enamel (Lu et al., 2008). Thus, there is additional uncharacterized potential for intersection of these three different protease families.
Summary
One area of current interest in KLK research is an integrated understanding of the functional role of the KLKs in health and disease. In this regard, study of the intersection between the KLK and thrombostasis axes suggests an important and extensive interaction. KLKs are expressed in a wide range of tissues at both the mRNA and protein levels, with highest expression levels within select major tissues and lower levels of expression in many others (Clements et al., 2001; Yousef and Diamandis, 2001; Komatsu et al., 2003). Of particular note is that contact between thrombostasis proteins in plasma, and secreted KLKs in the extracellular matrix, is facilitated under conditions of vascular permeability such as occurs during inflammation, edema, and tissue injury; thus, functional interaction between these two protease axes defines pathogenic conditions. Reports of coexpression of thrombostasis proteases and members of the KLK family are emerging in disease states, particularly cancer (Pettus et al., 2009). Research into the functional interaction between the KLKs and thrombostasis proteases is therefore likely to result in significant new advances in the diagnosis and treatment of important diseases associated with inflammation, tissue injury, and remodeling.
Presumably, only a subset of functional interactions between the KLK and thrombostasis axes has been identified to date. The mesh-like network of the activation connectivities within the KLK activome is much more complex in contrast to more simple schemes of linear activation cascades (e.g., the coagulation cascade), and when combined with the additional interactions involving the thrombostasis axis, the complexity is increased. It is difficult to simply look at such networks and comprehend how they would resolve over time for a particular initial condition or stimulus. However, such networks can be considered as a series of simultaneous rate equations, the majority of which can be approximated by Michaelis-Menten kinetics. Simulations of such networks can be constructed, assuming that the relevant kinetic constants and initial concentrations of components are known. Rate data are currently reported mostly for particular KLK/peptide substrate combinations, and there is limited data for intact protein substrates. Efforts to develop methods to quantify in vivo concentrations of particular KLKs have also been reported (Oikonomopoulou et al., 2008). In principle, kinetic rate data for different enzyme/substrate combinations, produced through the effort of independent investigators, can be combined to facilitate construction of the overall network. As such kinetic data are keenly sensitive to buffer conditions of pH and cosolvents, only data collected under identical conditions can effectively be combined. In this regard, phosphate buffered saline (0.15 M NaCl, pH 7.4) might serve as a useful, commonly accepted condition. However, another challenge in understanding KLK regulation is knowing the exact physiological condition of KLK action. Studies have shown that the presence of metal ions, salts, and glycans can substantially alter the kinetic properties of KLKs (Lovgren et al., 1999; Angelo et al., 2006). Additionally, determining solute concentrations within local physiological environments is a challenge. Thus, functional networks determined under ‘standard’ conditions are probably highly plastic under actual physiological conditions.
References
- Angelo PF, Lima AR, Alves FM, Blaber SI, Scarisbrick IA, Blaber M, Juliano L, Juliano MA. Substrate specificity of human kallikrein 6: salt and glycosaminoglycan effects. J Biol Chem. 2006;281:3116–3126. doi: 10.1074/jbc.M510096200. [DOI] [PubMed] [Google Scholar]
- Baumann G. Growth hormone heterogeneity: genes, iso-hormones, variants, and binding proteins. Endocr Rev. 1991;12:424–449. doi: 10.1210/edrv-12-4-424. [DOI] [PubMed] [Google Scholar]
- Bayes A, Tsetsenis T, Ventura S, Vendrell J, Aviles FX, Sotiropoulou G. Human kallikrein 6 activity is regulated via an autoproteolytic mechanism of activation/inactivation. Biol Chem. 2004;385:517–524. doi: 10.1515/BC.2004.061. [DOI] [PubMed] [Google Scholar]
- Beaufort N, Debela M, Creutzburg S, Kellermann J, Bode W, Schmitt M, Pidard D, Magdolen V. Interplay of human tissue kallikrein 4 (hK4) with the plasminogen activation system: hK4 regulates the structure and functions of the urokinase-type plasminogen activator receptor (uPAR) Biol Chem. 2006;387:217–222. doi: 10.1515/BC.2006.029. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee G, Ahamed J, Pawlinski R, Liu C, Mackman N, Ruf W, Edgington TS. Factor Xa binding to annexin 2 mediates signal transduction via protease-activated receptor 1. Circ Res. 2008;102:457–464. doi: 10.1161/CIRCRESAHA.107.167759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Micheloni A, Lamant L, Bonnart C, Tartaglia-Polcini A, Cobbold C, Al Saati T, Mariotti F, Mazereeuw-Hautier J, Boralevi F, et al. LEKTI proteolytic processing in human primary keratinocytes, tissue distribution and defective expression in Netherton syndrome. Hum Mol Genet. 2003;12:2417–2430. doi: 10.1093/hmg/ddg247. [DOI] [PubMed] [Google Scholar]
- Blaber SI, Yoon H, Scarisbrick IA, Juliano MA, Blaber M. The autolytic regulation of human kallikrein-related peptidase 6. Biochemistry. 2007;46:5209–5217. doi: 10.1021/bi6025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Brude OP, Archer F, Leoni P, Derian C, Bolsover S, Laurent GJ, Chambers RC. Factor Xa stimulates fibroblast procollagen production, proliferation, and calcium signaling via PAR1 activation. Exp Cell Res. 2005;304:16–27. doi: 10.1016/j.yexcr.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- Borgoño CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4:876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- Borgoño CA, MIacovos MP, Diamandis EP. Human tissue kallikreins: physiologic roles and applications in cancer. Mol Cancer Res. 2004;2:257–280. [PubMed] [Google Scholar]
- Borgoño CA, Gavigan JA, Alves J, Bowles B, Harris JL, Sotiropoulou G, Diamandis EP. Defining the extended substrate specificity of kallikrein 1-related peptidases. Biol Chem. 2007a;388:1215–1225. doi: 10.1515/BC.2007.124. [DOI] [PubMed] [Google Scholar]
- Borgoño CA, Michael IP, Komatsu N, Jayakumar A, Kapadia R, Clayman GL, Sotiropoulou G, Diamandis EP. A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J Biol Chem. 2007b;282:3640–3652. doi: 10.1074/jbc.M607567200. [DOI] [PubMed] [Google Scholar]
- Borgoño CA, Michael IP, Shaw JLV, Luo LY, Ghosh MC, Soosaipilla A, Grass L, Katsaros D, Diamandis EP. Expression and functional characterization of the cancer-related serine protease, human tissue kallikrein 14. J Biol Chem. 2007c;282:2405–2422. doi: 10.1074/jbc.M608348200. [DOI] [PubMed] [Google Scholar]
- Brattsand M, Egelrud T. Purification, molecular cloning, and expression of a human stratum corneum trypsin-like serine protease with possible function in desquamation. J Biol Chem. 1999;274:30033–30040. doi: 10.1074/jbc.274.42.30033. [DOI] [PubMed] [Google Scholar]
- Brattsand M, Stefansson K, Lundh C, Haasum Y, Egelrud T. A proteolytic cascade of kallikreins in the stratum corneum. J Invest Dermatol. 2005;124:198–203. doi: 10.1111/j.0022-202X.2004.23547.x. [DOI] [PubMed] [Google Scholar]
- Brattsand M, Stefansson K, Hubiche T, Nilsson SK, Egelrud T. SPINK9: a selective, skin-specific Kazal-type serine protease inhibitor. J Invest Dermatol. 2009;129:1656–1665. doi: 10.1038/jid.2008.448. [DOI] [PubMed] [Google Scholar]
- Camerer E, Huang W, Coughlin SR. Tissue factor-and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc Natl Acad Sci USA. 2000;97:5255–5260. doi: 10.1073/pnas.97.10.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalona WJ, Smith DS, Ratliff TL, Dodds KM, Coplen DE, Yuan JJ, Petros JA, Andriole GL. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991;324:1156–1161. doi: 10.1056/NEJM199104253241702. [DOI] [PubMed] [Google Scholar]
- Caubet C, Jonca N, Brattsand M, Guerrin M, Bernard D, Schmidt R, Egelrud T, Simon M, Serre G. Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J Invest Dermatol. 2004;122:1235–1244. doi: 10.1111/j.0022-202X.2004.22512.x. [DOI] [PubMed] [Google Scholar]
- Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, Bonafé JL, Wilkinson J, Taïeb A, Barrandon Y, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25:141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- Clements J, Hooper J, Dong Y, Harvey T. The expanded human kallikrein (KLK) gene family: genomic organisation, tissue-specific expression and potential functions. Biol Chem. 2001;382:5–14. doi: 10.1515/BC.2001.002. [DOI] [PubMed] [Google Scholar]
- Coughlin SR. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost. 2005;3:1800–1814. doi: 10.1111/j.1538-7836.2005.01377.x. [DOI] [PubMed] [Google Scholar]
- Debela M, Magdolen V, Schechter N, Valachova M, Lottspeich F, Craik CS, Choe Y, Bode W, Goettig P. Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J Biol Chem. 2006;281:25678–25688. doi: 10.1074/jbc.M602372200. [DOI] [PubMed] [Google Scholar]
- Denmeade SR, Lovgren J, Khan SR, Lilja H, Isaacs JT. Activation of latent protease function of pro-hK2, but not pro-PSA, involves autoprocessing. Prostate. 2001;48:122–126. doi: 10.1002/pros.1088. [DOI] [PubMed] [Google Scholar]
- Diamandis EP, Yousef GM. Human tissue kallikrein gene family: a rich source of novel disease biomarkers. Expert Rev Mol Diagn. 2001;1:182–190. doi: 10.1586/14737159.1.2.182. [DOI] [PubMed] [Google Scholar]
- Diamandis EP, Yousef GM, Luo LY, Magklara A, Obiezu CV. The new human kallikrein gene family: implications in carcinogenesis. Trends Endocrinol Metab. 2000;11:54–60. doi: 10.1016/s1043-2760(99)00225-8. [DOI] [PubMed] [Google Scholar]
- Dong Z, Kumar R, Yang X, Fidler JJ. Macrophage-derived mealloelastase is responsible for the generation of angiostatin in Lewis lung carcinoma. Cell. 1997;88:801–810. doi: 10.1016/s0092-8674(00)81926-1. [DOI] [PubMed] [Google Scholar]
- Egelrud T, Brattsand M, Kreutzmann P, Walden M, Vitzithum K, Marx UC, Forssmann WG, Mägert HJ. hK5 and hK7, two serine proteinases abundant in human skin, are inhibited by LEKTI domain 6. Br J Dermatol. 2005;153:1200–1203. doi: 10.1111/j.1365-2133.2005.06834.x. [DOI] [PubMed] [Google Scholar]
- Ekholm IE, Brattsand M, Egelrud T. Stratum corneum tryptic enzyme in normal epidermis: a missing link in the desquamation process? J Invest Dermatol. 2000;114:56–63. doi: 10.1046/j.1523-1747.2000.00820.x. [DOI] [PubMed] [Google Scholar]
- Frenette G, Tremblay RR, Lazure C, Dube JY. Prostatic kallikrein hK2, but not prostate-specific antigen (hK3), activates single-chain urokinase-type plasminogen activator. Int J Cancer. 1997;71:897–899. doi: 10.1002/(sici)1097-0215(19970529)71:5<897::aid-ijc31>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Gan L, Lee I, Smith R, Argonza-Barrett R, Lei H, McCuaig J, Moss P, Paeper B, Wang K. Sequencing and expression analysis of the serine protease gene cluster located in chromosome 19q13 region. Gene. 2000;257:119–130. doi: 10.1016/s0378-1119(00)00382-6. [DOI] [PubMed] [Google Scholar]
- Garcia-Barros M, Devesa J, Arce VM. Proteolytic processing of human growth hormone (GH) by rat tissues in vitro: influence of sex and age. J Endocrinol Invest. 2000;23:748–754. doi: 10.1007/BF03345065. [DOI] [PubMed] [Google Scholar]
- Hansen KK, Oikonomopoulou K, Baruch A, Ramachandran R, Beck P, Diamandis EP, Hollenberg MD. Proteinases as hormones: targets and mechanisms for proteolytic signaling. Biol Chem. 2008;389:971–982. doi: 10.1515/BC.2008.120. [DOI] [PubMed] [Google Scholar]
- Harvey TJ, Hooper JD, Myers SA, Stephenson SA, Ash-worth LK, Clements JA. Tissue-specific expression patterns and fine mapping of the human kallikrein (KLK) locus on proximal 19q13.4. J Biol Chem. 2000;275:37397–37406. doi: 10.1074/jbc.M004525200. [DOI] [PubMed] [Google Scholar]
- Heidtmann HH, Nettelbeck DM, Mingels A, Jager R, Welker HG, Kontermann RE. Generation of angiostatin-like fragments from plasminogen by prostate-specific antigen. Br J Cancer. 1999;81:1269–1273. doi: 10.1038/sj.bjc.6692167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg MD, Oikonomopoulou K, Hansen KK, Saifeddine M, Ramachandran R, Diamandis EP. Kallikreins and proteinase-mediated signaling: proteinase-activated receptors (PARs) and the pathophysiology of inflammatory diseases and cancer. Biol Chem. 2008;389:643–651. doi: 10.1515/BC.2008.077. [DOI] [PubMed] [Google Scholar]
- Houle S, Papez MD, Ferazzini M, Hollenberg MD, Vergnolle N. Neutrophils and the kallikreinkinin system in proteinase-activated receptor 4-mediated inflammation in rodents. Br J Pharmacol. 2005;146:670–678. doi: 10.1038/sj.bjp.0706371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen S, Jakobsen CM, Rosen DM, Ricklis RM, Reineke U, Christensen SB, Lilja H, Denmeade SR. Screening a combinatorial peptide library to develop a human glandular kallikrein 2-activated prodrug as targeted therapy for prostate cancer. Mol Cancer Ther. 2004;3:1439–1450. [PubMed] [Google Scholar]
- Kahn ML, Zheng YW, Huang W, Bigornia V, Zeng D, Moff S, Farese RVJ, Tam C, Coughlin SR. A dual thrombin receptor system for platelet activation. Nature. 1998;394:690–694. doi: 10.1038/29325. [DOI] [PubMed] [Google Scholar]
- Kimura M, Andersen TT, Fenton JW, II, Bahou WF, Aviv A. Plasmin-platelet interaction involves cleavage of functional thrombin receptor. Am J Physiol. 1996;271:C54–C60. doi: 10.1152/ajpcell.1996.271.1.C54. [DOI] [PubMed] [Google Scholar]
- Kishi T, Kato M, Shimizu T, Kato K, Matsumoto K, Yoshida S, Shiosaka S, Hakoshima T. Crystallization and preliminary X-ray analysis of neuropsin, a serine protease expressed in the limbic system of mouse brain. J Struct Biol. 1997;118:248–251. doi: 10.1006/jsbi.1997.3862. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Takata M, Otsuki N, Ohka R, Amano O, Takehara K, Saijoh K. Elevated stratum corneum hydrolytic activity in Netherton syndrome suggests an inhibitory regulation of desquamation by SPINK5-derived peptides. J Invest Dermatol. 2002;118:436–443. doi: 10.1046/j.0022-202x.2001.01663.x. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Takata M, Otsuki N, Toyama T, Ohka R, Takehara K, Saijoh K. Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J Invest Dermatol. 2003;121:542–549. doi: 10.1046/j.1523-1747.2003.12363.x. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Saijoh K, Otsuki N, Kishi T, Michael IP, Obiezu CV, Borgono CA, Takehara K, Jayakumar A, Wu HK, et al. Proteolytic processing of human growth hormone by multiple tissue kallikreins and regulation by the serine protease inhibitor Kazal-Type5 (SPINK5) protein. Clin Chim Acta. 2007;377:228–236. doi: 10.1016/j.cca.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Saijoh K, Jayakumar A, Clayman GL, Tohyama M, Suga Y, Mizuno Y, Tsukamoto K, Taniuchi K, Takehara K, et al. Correlation between SPINK5 gene mutations and clinical manifestations in Netherton syndrome patients. J Invest Dermatol. 2008;128:1148–1159. doi: 10.1038/sj.jid.5701153. [DOI] [PubMed] [Google Scholar]
- Kuliopulos A, Covic L, Seeley SK, Sheridan PJ, Helin J, Costello CE. Plasmin desensitization of the PAR1 thrombin receptor: kinetics, sites of truncation, and implications for thrombolytic therapy. Biochemistry. 1999;38:4572–4585. doi: 10.1021/bi9824792. [DOI] [PubMed] [Google Scholar]
- Lafleur MA, Hollenberg MD, Atkinson SJ, Knäuper V, Murphy G, Edwards DR. Activation of pro-(matrix metalloproteinase-2) (pro-MMP-2) by thrombin is membrane-type-MMP-dependent in human umbilical vein endothelial cells and generates a distinct 63 kDa active species. Biochem J. 2001;357:107–115. doi: 10.1042/0264-6021:3570107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HX, Hwang BY, Laxmikanthan G, Blaber SI, Blaber M, Golubkov PA, Ren P, Iverson BL, Georgiou G. The substrate specificity of human kallikrein 1 and 6 determined by phage display. Protein Sci. 2008;17:664–672. doi: 10.1110/ps.073333208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J Clin Invest. 1985;76:1899–1903. doi: 10.1172/JCI112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- List K, Jensen ON, Bugge TH, Lund LR, Ploug M, Danø K, Behrendt N. Plasminogen-independent initiation of the pro-urokinase activation cascade in vivo. Activation of pro-urokinase by glandular kallikrein (mGK-6) in plasminogen-deficient mice. Biochemistry. 2000;39:508–515. doi: 10.1021/bi991701f. [DOI] [PubMed] [Google Scholar]
- Little SP, Dixon EP, Norris F, Buckley W, Becker GW, Johnson M, Dobbins JR, Wyrick T, Miller JR, MacKellar W, et al. Zyme, a novel and potentially amyloidogenic enzyme cDNA isolated from Alzheimer’s disease brain. J Biol Chem. 1997;272:25135–25142. doi: 10.1074/jbc.272.40.25135. [DOI] [PubMed] [Google Scholar]
- Lovgren J, Rajakoski K, Karp M, Lundwall A, Lilja H. Activation of the zymogen form of prostate-specific antigen by human glandular kallikrein 2. Biochem Biophys Res Commun. 1997;238:549–555. doi: 10.1006/bbrc.1997.7333. [DOI] [PubMed] [Google Scholar]
- Lovgren J, Airas K, Lilja H. Enzymatic action of human glandular kallikrein 2 (hK2). Substrate specificity and regulation by Zn2+ and extracellular protease inhibitors. Eur J Biochem. 1999;262:781–789. doi: 10.1046/j.1432-1327.1999.00433.x. [DOI] [PubMed] [Google Scholar]
- Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludeman MJ, Kataoka H, Srinivasan Y, Esmon NL, Esmon CT, Coughlin SR. PAR1 cleavage and signaling in response to activated protein C and thrombin. J Biol Chem. 2005;280:13122–13128. doi: 10.1074/jbc.M410381200. [DOI] [PubMed] [Google Scholar]
- Lundstrom A, Egelrud T. Stratum corneum chymotryptic enzyme: a proteinase which may be generally present in the stratum corneum and with a possible involvement in desquamation. Acta Derm Venereol. 1991;71:471–474. [PubMed] [Google Scholar]
- Lundwall A, Band V, Blaber M, Clements JA, Courty Y, Diamandis EP, Fritz H, Lilja H, Malm J, Maltais LJ, et al. A comprehensive nomenclature for serine proteases with homology to tissue kallikreins. Biol Chem. 2006;387:637–641. doi: 10.1515/BC.2006.082. [DOI] [PubMed] [Google Scholar]
- Luo LY, Shan SJC, Elliott MB, Soosaipilla A, Diamandis EP. Purification and characterization of human kallikrein 11, a candidate prostate and ovarian cancer biomarker, from seminal plasma. Hum Cancer Biol. 2006;12:742–750. doi: 10.1158/1078-0432.CCR-05-1696. [DOI] [PubMed] [Google Scholar]
- Mägert HJ, Ständker L, Kreutzmann P, Zucht HD, Reinecke M, Sommerhoff CP, Fritz H, Forssmann WG. LEKTI, a novel 15-domain type of human serine proteinase inhibitor. J Biol Chem. 1999;274:21499–21502. doi: 10.1074/jbc.274.31.21499. [DOI] [PubMed] [Google Scholar]
- Meyer-Hoffert U, Wu Z, Schröder JM. Identification of lympho-epithelial Kazal-type inhibitor 2 in human skin as a kallikrein-related peptidase 5-specific protease inhibitor. PLoS One. 2009;4:e4372. doi: 10.1371/journal.pone.0004372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael IP, Sotiropoulou G, Pampalakis G, Magklara A, Ghosh M, Wasney GA, Diamandis EP. Biochemical and enzymatic characterization of human kallikrein 5 (hK5), a novel serine protease potentially involved in cancer progression. J Biol Chem. 2005;280:14628–14635. doi: 10.1074/jbc.M408132200. [DOI] [PubMed] [Google Scholar]
- Michael IP, Pampalakis G, Mikolajczyk SD, Malm J, Sotiropoulou G, Diamandis EP. Human tissue kallikrein 5 (hK5) is a member of a proteolytic cascade pathway involved in seminal clot liquefaction and potentially in prostate cancer progression. J Biol Chem. 2006;281:12743–12750. doi: 10.1074/jbc.M600326200. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk SD, Millar LS, Kumar A, Saedi MS. Prostatic human kallikrein 2 inactivates and complexes with plasminogen activator inhibitor-1. Int J Cancer. 1999;81:438–442. doi: 10.1002/(sici)1097-0215(19990505)81:3<438::aid-ijc18>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Mitsudo K, Jayakumar A, Henderson Y, Frederick MJ, Kang Y, Wang M, El-Naggar AK, Clayman GL. Inhibition of serine proteinases plasmin, trypsin, subtilisin A, cathepsin G, and elastase by LEKTI: a kinetic analysis. Biochemistry. 2003;42:3874–3881. doi: 10.1021/bi027029v. [DOI] [PubMed] [Google Scholar]
- Nelson PS, Gan L, Ferguson C, Moss P, Gelinas R, Hood L, Wang K. Molecular cloning and characterization of prostase, an androgen-regulated serine protease with prostate-restricted expression. Proc Natl Acad Sci USA. 1999;96:3114–3119. doi: 10.1073/pnas.96.6.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly MS, Holmgren L, Shing Y, Chen C, Rosenthal RA, Moses M, Lane WS, Cao Y, Sage EH, Folkman J. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by Lewis lung carcinoma. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- Obiezu CV, Michael IP, Levesque MA, Diamandis EP. Human kallikrein 4: enzymatic activity, inhibition, and degradation of extracellular matrix proteins. Biol Chem. 2006;387:749–759. doi: 10.1515/BC.2006.094. [DOI] [PubMed] [Google Scholar]
- Oikonomopoulou K, Hansen KK, Saifeddine M, Tea I, Blaber M, Blaber SI, Scarisbrick I, Andrade-Gordon P, Cottrell GS, Bunnett NW, et al. Proteinase-activated receptors, targets for kallikrein signaling. J Biol Chem. 2006a;281:32095–32112. doi: 10.1074/jbc.M513138200. [DOI] [PubMed] [Google Scholar]
- Oikonomopoulou K, Hansen KK, Saifeddine M, Vergnolle N, Tea I, Blaber M, Blaber SI, Scarisbrick I, Diamandis EP, Hollenberg MD. Kallikrein-mediated cell signalling: targeting proteinase-activated receptors (PARs) Biol Chem. 2006b;387:817–824. doi: 10.1515/BC.2006.104. [DOI] [PubMed] [Google Scholar]
- Oikonomopoulou K, Hansen KK, Baruch A, Hollenberg MD, Diamandis EP. Immunofluormetric activity-based probe analysis of active KLK6 in biological fluids. Biol Chem. 2008;389:747–756. doi: 10.1515/BC.2008.086. [DOI] [PubMed] [Google Scholar]
- Pettus JR, Johnson JJ, Shi Z, Davis JW, Koblinski J, Ghosh S, Liu Y, Ravosa MJ, Frazier S, Stack MS. Multiple kallikrein (KLK 5, 7, 8, and 10) expression in squamous cell carcinoma of the oral cavity. Histol Histophathol. 2009;24:197–207. doi: 10.14670/hh-24.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploug M. Structure-function relationships in the interaction between the urokinase-type plasminogen activator and its receptor. Curr Pharm Des. 2003;9:1499–1528. doi: 10.2174/1381612033454630. [DOI] [PubMed] [Google Scholar]
- Quinton TM, Kim S, Derian CK, Jin J, Kunapuli SP. Plasmin-mediated activation of platelets occurs by cleavage of protease-activated receptor 4. J Biol Chem. 2004;279:18434–18439. doi: 10.1074/jbc.M401431200. [DOI] [PubMed] [Google Scholar]
- Rajapakse S, Ogiwara K, Takano N, Moriyama A, Takahashi T. Biochemical characterization of human kallikrein 8 and its possible involvement in the degradation of extracellular matrix proteins. FEBS Lett. 2005;579:6879–6884. doi: 10.1016/j.febslet.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Rajapakse S, Yamano N, Ogiwara K, Hirata K, Takahashi S, Takahashi T. Estrogen-dependent expression of the tissue kallikrein gene (Klk1) in the mouse uterus and its implications for endometrial tissue growth. Mol Reprod Dev. 2007;74:1053–1063. doi: 10.1002/mrd.20567. [DOI] [PubMed] [Google Scholar]
- Ramsay AJ, Dong Y, Hunt ML, Linn M, Samaratunga H, Clements JA, Hooper JD. Kallikrein-related peptidase 4 (KLK4) initiates intracellular signaling via protease-activated receptors (PARs). KLK4 and PAR-2 are co-expressed during prostate cancer progression. J Biol Chem. 2008a;283:12293–12304. doi: 10.1074/jbc.M709493200. [DOI] [PubMed] [Google Scholar]
- Ramsay AJ, Reid JC, Adams MN, Samaratunga H, Dong Y, Clements JA, Hooper JD. Prostatic trypsin-like kallikrein-related peptidases (KLKs) and other prostate-expressed tryptic proteinases as regulators of signalling via proteinase-activated receptors (PARs) Biol Chem. 2008b;389:653–668. doi: 10.1515/BC.2008.078. [DOI] [PubMed] [Google Scholar]
- Ruf W, Mueller BM. Thrombin generation and the pathogenesis of cancer. Semin Thromb Hemost. 2006;32(Suppl 1):61–68. doi: 10.1055/s-2006-939555. [DOI] [PubMed] [Google Scholar]
- Ruf W, Dorfleutner A, Riewald M. Specificity of coagulation factor signaling. J Thromb Haemost. 2003;1:1495–1503. doi: 10.1046/j.1538-7836.2003.00300.x. [DOI] [PubMed] [Google Scholar]
- Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, Bartlett JD, Simmer JP. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci. 2002;110:358–365. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- Schechter NM, Choi EJ, Wang ZM, Hanakawa Y, Stanley JR, Kang Y, Clayman GL, Jayakumar A. Inhibition of human kallikreins 5 and 7 by the serine protease inhibitor lympho-epithelial Kazal-type inhibitor (LEKTI) Biol Chem. 2005;386:1173–1184. doi: 10.1515/BC.2005.134. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G, Rogakos V, Tsetsenis T, Pampalakis G, Zafiropoulos N, Simillides G, Yiotakis A, Diamandis EP. Emerging interest in the kallikrein gene family for understanding and diagnosing cancer. Oncol Res. 2003;13:381–391. doi: 10.3727/096504003108748393. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G, Pampalakis G, Diamandis EP. Functional roles of human kallikrein-related peptidases. J Biol Chem. 2009;284:32989–32994. doi: 10.1074/jbc.R109.027946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamey TA, Yang N, Hay AR, McNal JE, Freiha FS, Redwine E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med. 1987;15:909–916. doi: 10.1056/NEJM198710083171501. [DOI] [PubMed] [Google Scholar]
- Stubbs MT, Renatus M, Bode W. An active zymogen: unravelling the mystery of tissue-type plasminogen activator. Biol Chem. 1998;379:95–103. doi: 10.1515/bchm.1998.379.2.95. [DOI] [PubMed] [Google Scholar]
- Takayama TK, McMullen BA, Nelson PS, Matsumura M, Fujikawa K. Characterization of hK4 (prostase), a prostate-specific serine protease: activation of the precursor of prostate specific antigen (pro-PSA) and single-chain urokinase-type plasminogen activator and degradation of prostatic acid phosphatase. Biochemistry. 2001;40:15341–15348. doi: 10.1021/bi015775e. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Trejo J. Protease-activated receptor signaling: new roles and regulatory mechanisms. Curr Opin Hematol. 2007;14:230–235. doi: 10.1097/MOH.0b013e3280dce568. [DOI] [PubMed] [Google Scholar]
- Tsurupa G, Medved L. Identification and characterization of novel tPA- and plasminogen-binding sites within fibrin(ogen) aC-domains. Biochemistry. 2001;40:801–808. doi: 10.1021/bi001789t. [DOI] [PubMed] [Google Scholar]
- Vandell AG, Larson N, Laxmikanthan G, Panos M, Blaber SI, Blaber M, Scarisbrick IA. Protease-activated receptor dependent and independent signaling by kallikreins 1 and 6 in CNS neuron and astroglial cell lines. J Neurochem. 2008;107:855–870. doi: 10.1111/j.1471-4159.2008.05658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg HH, Ruf W. Emerging insights in tissue factor-dependent signaling events. Semin Thromb Hemost. 2006;32:24–32. doi: 10.1055/s-2006-933337. [DOI] [PubMed] [Google Scholar]
- Watt KWK, Lee PJ, M’Timkulu T, Chan WP, Loor R. Human prostate-specific antigen: structural and functional similarity with serine proteases. Proc Natl Acad Sci USA. 1986;83:3166–3170. doi: 10.1073/pnas.83.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WF, Andersen H, Whitmore TE, Presnell SR, Yee DP, Ching A, Gilbert T, Davie EW, Foster DC. Cloning and characterization of human protease-activated receptor 4. Proc Natl Acad Sci USA. 1998;95:6642–6646. doi: 10.1073/pnas.95.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Laxmikanthan G, Lee J, Blaber SI, Rodriguez A, Kogot JM, Scarisbrick IA, Blaber M. Activation profiles and regulatory cascades of the human kallikrein-related proteases. J Biol Chem. 2007;282:31852–31864. doi: 10.1074/jbc.M705190200. [DOI] [PubMed] [Google Scholar]
- Yoon H, Blaber SI, Evans DM, Trim J, Juliano MA, Scarisbrick IA, Blaber M. Activation profiles of human kallikrein-related peptidases by proteases of the thrombostasis axis. Protein Sci. 2008;17:1998–2007. doi: 10.1110/ps.036715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Blaber SI, Debela M, Goettig P, Scarisbrick IA, Blaber M. A completed KLK activome profile: investigation of activation profiles of KLK9, 10 and 15. Biol Chem. 2009;390:373–377. doi: 10.1515/BC.2009.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef GM, Diamandis EP. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr Rev. 2001;22:184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
- Yousef GM, Diamandis EP. An overview of the kallikrein gene families in humans and other species: emerging candidate tumour markers. Clin Biochem. 2003;36:443–452. doi: 10.1016/s0009-9120(03)00055-9. [DOI] [PubMed] [Google Scholar]
- Yousef GM, Chang A, Scorilas A, Diamandis EP. Genomic organization of the human kallikrein gene family on chromosome 19q13.3–q13.4. Biochem Biophys Res Commun. 2000;276:125–133. doi: 10.1006/bbrc.2000.3448. [DOI] [PubMed] [Google Scholar]