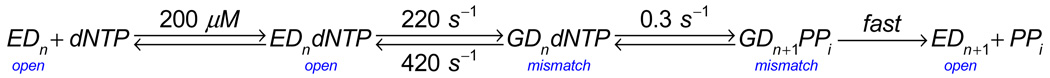Abstract
This review summarizes our current understanding of the structural, kinetic and thermodynamic basis for the extraordinary accuracy of high fidelity DNA polymerases. High fidelity DNA polymerases, such as the enzyme responsible for the replication of bacteriophage T7 DNA, discriminate against similar substrates with an accuracy that approaches one error in a million base pairs while copying DNA at a rate of approximately 300 base pairs per second. When the polymerase does make an error, it stalls, giving time for the slower proofreading exonuclease to remove the mismatch so that the overall error frequency approaches one in a billion. Structural analysis reveals a large change in conformation after nucleotide binding from an open to a closed state. Kinetic analysis has shown that the substrate-induced structural change plays a key role in the discrimination between correct and incorrect base pairs by governing whether a nucleotide will be retained and incorporated or rapidly released.
Keywords: DNA polymerase, mechanism, kinetics, transient, stopped-flow, fluorescence, quench-flow, conformational change, enzyme dynamics
Most enzymes exhibit a large change in structure upon binding substrate, and it is apparent that some of the substrate binding energy is used to organize the active site and orienting the substrate for catalysis. Similarly, the binding of a nucleotide substrate to a high-fidelity DNA polymerase induces a change in structure of the enzyme from an open state in the absence of nucleotide to a closed state after binding nucleotide (Figure 1). Several high resolution structures have been published with enzymes trapped in the closed state with the nucleotide and a dideoxy-terminated DNA primer to prevent chemistry [1–5]. The bound substrate in the closed state is surrounded by residues that promote catalysis as described in more detail below. The structure of the E-DNA complex in the absence of substrate shows an open active site so the conformational change from the open to the closed state is large and complex with movements in the backbone over distances up to 15 Å and changes in the packing of the helices so that the motion is not a simple hinge rotation of a rigid body. We are just beginning to understand the role of this complex movement on DNA polymerase specificity as will be detailed in this review.
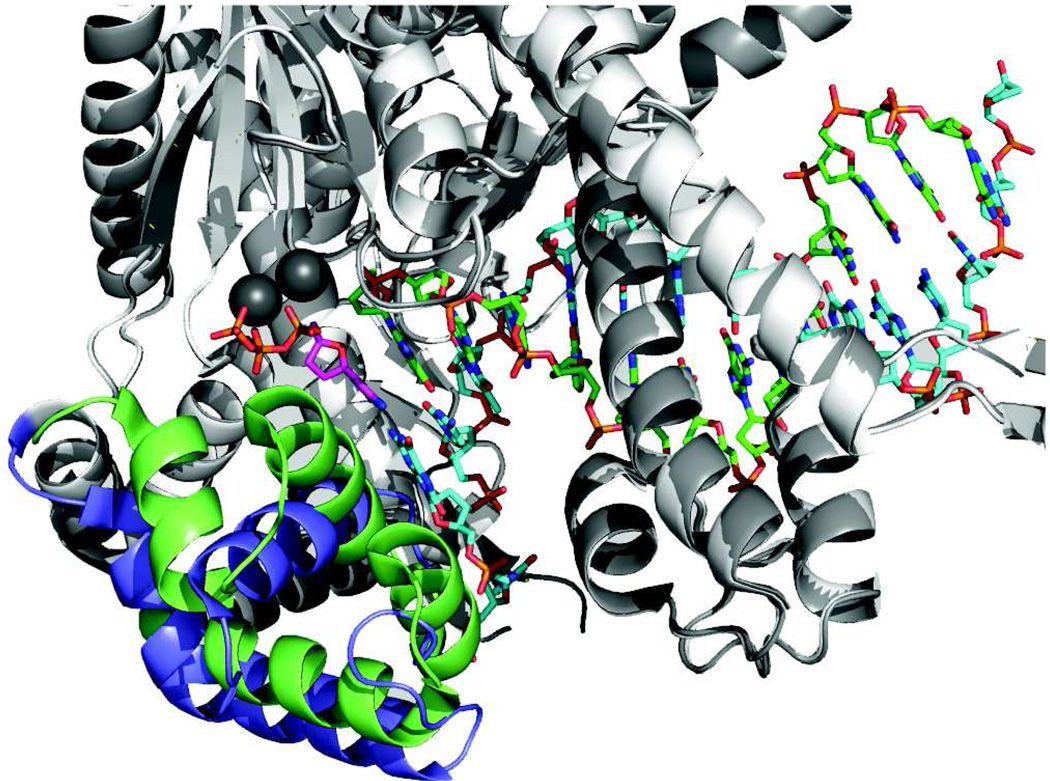
Figure 1. Conformational change upon nucleotide binding.
Structures of T7 DNA polymerase in the presence (green and dark gray) and absence (blue and light gray) of nucleotide are aligned to show the changes in structure of the recognition domain (green or blue). Very little change in structure was seen in the remainder of the protein (light or dark gray), which provided a basis for the alignment. The metal ions are dark gray, the incoming nucleotide is magenta, the primer strand is green and the template strand is cyan. The recognition domain is defined as residues N502 to P560. Drawn using Pymol with PDB files 1tk5 (with nucleotide) and 1tk0 (without nucleotide) [62].
It appears obvious that one role of the substrate-induced conformational change is to allow the rapid binding of substrates (and release of products) in the open state while affording optimal alignment of catalytic residues surrounding the substrate to promote catalysis in the closed state. However, the contributions of different conformational states to specificity and efficiency go beyond this simple reasoning, and as we have learned recently, the rate of the conformational change governs the rate of incorporation. Although many structural studies attempt to define aspects of protein structure that determine enzyme specificity, it is important to note that specificity is a purely kinetic property that is difficult to predict from structure alone.
The role that changes in enzyme structure play in specificity and efficiency has been controversial. Theoretical studies have argued against a role for conformational changes in enzyme specificity. For example, it has been argued that an induced-fit mechanism involving a two-step binding reaction can occur with the same free energy change as a one-step binding mechanism and therefore induced-fit cannot alter the net binding constant and therefore cannot change specificity beyond that which could be achieved in a single step [6]. Moreover, it has been suggested that if an enzyme was pre-organized in an optimal configuration for catalysis, then, substrate binding energy would not be wasted in re-orienting the enzyme to achieve the closed state, and, accordingly, more binding energy would be available to do the work of catalysis. However, these arguments are flawed in that they are based upon the inherent assumption that substrate binding and enzyme isomerization are rapid reactions that come to equilibrium on a time scale much faster than the chemistry step. Certainly, it is true that if the binding and isomerization reactions are at equilibrium then the pathway does not matter. However, when that is not the case, the pathways of binding and enzyme isomerization are critical.
Another theoretical point of contention is the question of whether the transition state structure is the same for a desired substrate as for one that is disfavored [7]. In the case of DNA polymerases, the reaction centers are identical for correct and incorrect base pairs, and specificity is a function of the structure of the base pair formed between the incoming dNTP and the templating base, presumably leading to misalignment. Nonetheless, the question remains whether catalysis to incorporate correct and incorrect base pairs occurs from similar or dissimilar enzyme structures. Moreover, the enzymes must rapidly sample each of the four nucleotides, but then differentiate among them to favor incorporation of the one nucleotide that forms a proper base pair with the template.
Questions addressing the role of conformational changes in polymerase specificity can be approached by direct measurement and quantification of the steps leading to catalysis. Although detailed kinetic studies have been conducted on several polymerases [8], in this review results obtained in studies of the high fidelity T7 DNA polymerase will be used to illustrate concepts.
Kinetics of nucleotide binding and incorporation
For more than a decade, we promoted a model in which the conformational change was thought to be rate-limiting and was followed by a very fast chemistry step and fast pyrophosphate release [9–12]. This afforded a simplified scheme where nucleotide incorporation was governed by a single rate-limiting step, defined by kpol, after equilibrium binding of nucleotide to the open complex with an apparent dissociation constant, Kd,app (Scheme 1).
Scheme 1.
If k2 is slow relative to the chemistry step (k3), then the rate of polymerization (kpol) is defined by k2, the apparent dissociation constant (Kd,app) is 1/K1, and the specificity constant is determined by kcat/Km = kpol/Kd,app. This model afforded a direct method to quantify specificity by measurement of the concentration dependence of nucleotide incorporation in rapid-quench single turnover kinetic studies [13, 14]. By monitoring a single incorporation event on the time scale of one reaction turnover, rapid quench methods overcome the serious errors introduced in attempting to measure kcat/Km for a processive enzyme by steady state methods which usually measure the rate of DNA release from the enzyme. As an aside, it should be pointed out that studies using steady state methods to monitor the kinetics of single nucleotide incorporation on processive enzymes are suspect and should be disregarded. Particularly troublesome are rates reported in percent gel band extension per minute [15], which are difficult to interpret mechanistically. Although one could, in principle, make accurate measurements in the steady state by working at sufficiently low nucleotide concentrations (nM), that is generally not the case. The specificity constants for correct nucleotides are typically underestimated in steady state measurements by a factor of up to 100 and the effects of mutations or altered substrates are masked. This has led to greatly underestimated values for discrimination [16] and seriously flawed conclusions [12, 17, 18].
Rapid quench single turnover kinetic studies provide the best method to measure the kinetic parameters governing sequential nucleotide incorporation during processive synthesis such that kcat/Km = kpol/Kd,app. However, we now know that the simplifications of Scheme 1 that allow the assignment of Kd,app as the nucleotide dissociation constant are incorrect. Rather, the apparent dissociation constant measured in a single turnover experiment should be regarded as a Km value, which can be much lower than the true Kd (1/K1) for the initial collision complex (EDndNTP) [19]. This will be explained more clearly below.
The central question that we and others have attempted to address is to determine whether the conformational change or the chemistry step is rate limiting. Initial studies were based upon measuring the thio-elemental effect, relying upon substitution of sulfur for a non-bridging oxygen on the α-phosphate of the incoming dNTP to slow the rate of the chemical reaction. Because the thio-elemental effect was small, it was reasoned that the chemistry step must be fast following a rate-limiting conformational change step [10, 11]. Other studies, including results showing that dNTP binding appeared to be tighter during incorporation by HIV reverse transcriptase in the presence of a nonnucleoside inhibitor which slowed the rate of chemistry, supported the notion of a conformational change leading to tighter nucleotide binding and preceding chemistry [20].
Ming-Daw Tsai and his students were the first to provide evidence indicating that the conformational change was faster than chemistry, based upon studies using a fluorescence signal arising from 2-aminopurine in the template strand [21, 22]. This method was also used in studies on the Klenow fragment of Pol I [23]. These studies stimulated our studies in which we placed a fluorophore on the fingers domain of T7 DNA polymerase to monitor changes in enzyme structure (Figure 2). We confirmed that the conformational change was faster than chemistry. Moreover, comprehensive analysis of the kinetics made us realize that comparing the rates of chemistry and the conformational change was only part of the story. It is equally important to determine the relative rates of chemistry and the reverse of the conformational change leading to nucleotide release. Specificity is not only determined by the relative magnitudes of k2 and k3, but also by the relative rates of k−2 and k3 as described below.
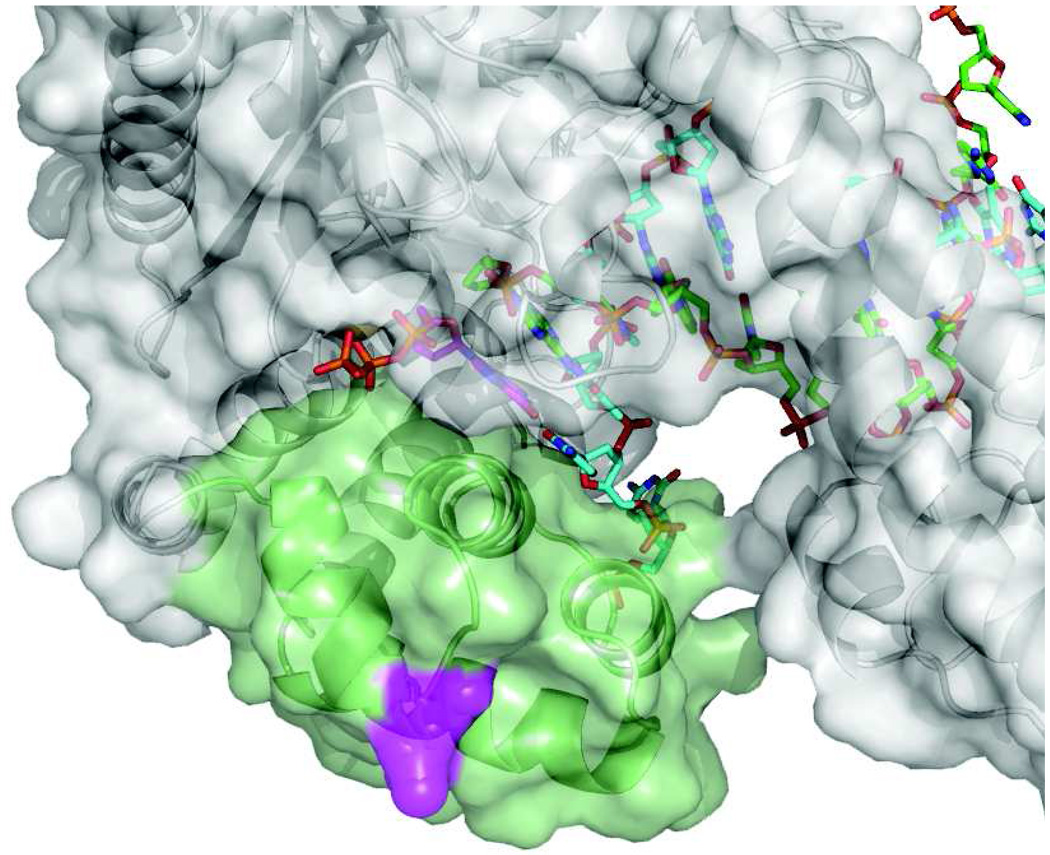
Figure 2. Location of the MDCC label.
The location of the MDCC label on the surface of the recognition domain is shown in magenta. The recognition domain is green. From 1tk5.pdb.
The incorporation of a correct nucleotide by T7 DNA polymerase is governed by the rates shown in Scheme 2 [19]. where 1/K1 = 28 µM, defining the dissociation constant in forming the collision complex. Weak binding is followed by a fast conformational change leading to much tighter binding (K2 = 400), which is then followed by the chemical reaction. All evidence suggests that pyrophosphate release and translocation are fast, as described below, so this simple model accounts for the sequential nucleotide incorporation during processive synthesis and these rate constants can be used to compute the specificity constant, kcat/Km. For this three-step model, kcat/Km can be derived as:
Interestingly, because k−2 is small relative to k3, the rate of the chemical reaction drops out of the expression so that the specificity constant reduces to:
The specificity constant further reduces to kcat/Km = K1k2 = 24 µsM−1s−1 because k−1 >> k2. This term represents the apparent second order rate constant for substrate binding for a two-step binding reaction with a weak rapid equilibrium followed by a fast isomerization. Thus nucleotide selectivity during correct incorporation is based solely upon the rate at which the substrate binds to the enzyme including the isomerization to the form the tight FDndNTP complex. Once this complex forms, the rate of chemistry does not affect specificity because chemistry is faster than the rate at which the enzyme opens to release bound substrate. That is, once tightly bound, the substrate is committed to go forward and, therefore, it is the binding steps alone that dictate specificity.
Scheme 2.
Kinetics of correct nucleotide binding
In contrast the binding and incorporation of a mismatched nucleotide (defined by the identity of the templating base) is governed by very different kinetics as shown in Scheme 3.
Scheme 3.
Kinetics of misincorporation
The initial binding of the mismatch to the collision complex is about tenfold weaker than for a correct base pair. However, the large selectivity against a mismatch occurs with the isomerization to the GDndNTP state from which the rate of chemistry is reduced 1000-fold, while the rate of release of the bound nucleotide is increased 300-fold, relative to a correct nucleotide. Because chemistry is slow relative to nucleotide release, the binding and isomerization come to equilibrium and, therefore, the specificity constant is determined by the product of two equilibrium constants and the rate of chemistry, kcat/Km = K1K2k3 = 0.0008 µM−1s−1.
Nucleotide selectivity
According to this analysis, nucleotide selectivity is governed by the kinetic partitioning of the enzyme-bound nucleotide species. That is, the probability that a bound nucleotide is incorporated is given simply by the ratio of k3/(k−2+k3). For a correct nucleotide, k−2 is slow relative to k3, so once a nucleotide is bound, it is incorporated most (99.6%) of the time. In contrast, for a bound mismatch, k3 is greatly reduced and k−2 is greatly increased, so it the mismatch is released most (99.9%) of the time. Thus, the changes in enzyme structure following nucleotide binding govern the fate of the bound nucleotide, and the conformational change plays an essential role in establishing enzyme selectivity as dictated by the kinetic partitioning of the FDndNTP (or GDndNTP) state.
We would like ascribe a certain degree of nucleotide selectivity to each step in the pathway in an attempt to define the free energy contribution of the initial binding, the conformational change, and chemistry to net selectivity [24]. However, this is not so simple. Discrimination is defined as the ratio of the kcat/Km values derived for correct and incorrect base pairs:
However, for our current model, the discrimination involves the ratio of different kinetic constants for correct and incorrect base pairs:
From this analysis, we can only analyze the effect of changes in K1 for the free energy contribution to discrimination in binding of nucleotide to the open complex.
This level of discrimination is comparable to that observed for a single hydrogen bond. Therefore, these results suggest that the incoming dNTP forms a base pair with the templating base in the open E.DNA complex. Although the locations of the incoming dNTP and the templating base in the open complex are not known, these data rule out models suggesting that the dNTP binds first to the enzyme and is then delivered to the template during the conformational change step. Rather, the base pair must form first.
We can also compute the net free energy contribution for discrimination during catalysis by comparing k3 for correct and incorrect base pairs to derive a ΔΔG value of 4.2 kcal/mol. However, this value does not translate linearly to the net nucleotide discrimination because the value of k3 for correct base pair incorporation cancels from the expression defining specificity. Nonetheless it represents the real changes in transition state stabilization affecting the chemical reaction at the active site in comparing a correct base pair and a mismatch, which are most probably related to misalignment of the reactive groups in the mismatch.
Understanding the role of the conformational change in specificity is complex. Because the conformational change is faster than chemistry, the specificity constant defined by K1k2 is greater than would be realized in a pathway in which the conformational change was omitted and the specificity was determined solely by the product of the contributions due to the initial binding and chemistry (K1k3, derived by using the numbering in Scheme 1 but bypassing step 2). It is reasonable to suppose that there are limits during evolution on the extent to which an enzyme can achieve discrimination in the chemistry step alone in that further changes in structure that might increase the rate of chemistry for a correct base may also increase the rate of incorporation of a mismatch. The conformational change step allows a disconnect between the rate constants governing the incorporation of a correct base and those governing the incorporation of a mismatch.
The conformational coupling between enzyme structure and fidelity affords further discrimination by altering the structure in response to whether a correct base pair binds to the open E.DNA complex. It should also be noted that our data indicate that the mismatch recognition state, GDndNTP is different from the active, closed conformation, FDndNTP, formed after binding a correct base pair. While a correct base induced a decrease in fluorescence of our reporter group, the binding of a mismatch led to an increase, suggesting that there are at least three states: open, closed and mismatch recognition [19]. The fluorescence data, coupled with the observed slower rate of incorporation, led us to postulate a unique mismatch recognition state in which substrate binding energy is used to misalign catalytic residues and slow the rate of catalysis while promoting nucleotide release. This is in contrast to the binding of a correct base in which substrate binding energy is used to organize the active site, hold the nucleotide tightly, and promote catalysis. This new model defines the real power underlying the role of induced fit in enzyme specificity. While the correct substrate induces a structure to facilitate catalysis, the wrong substrate induces a structure to slow catalysis and promote substrate release. This mechanism for achieving increased selectivity may be universal [25].
Relating structure to kinetics
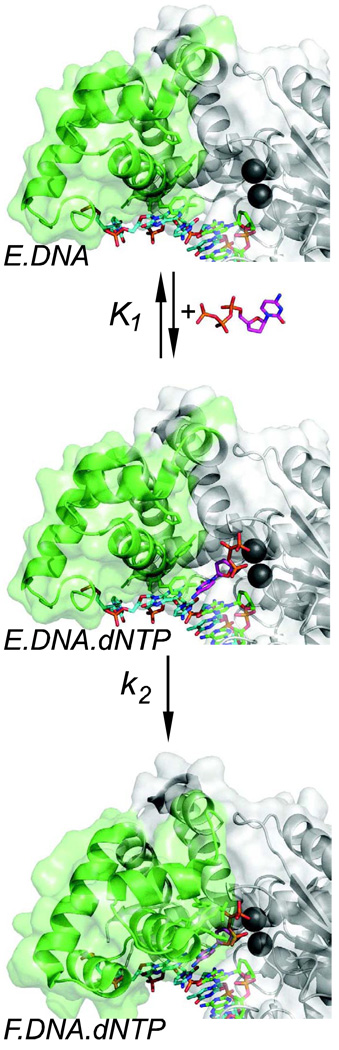
According to our working model, specificity is dictated by the changes in enzyme structure that occur after the nucleotide first binds to the open complex (Figure 3). Little is known about the structure of the initial open EDndNTP complex or where the dNTP binds. However, the observed difference in binding energy between correct and incorrect base pairs in the EDndNTP complex is comparable to what is expected for hydrogen bonding between base pairs (ΔΔG = 1–2 kcal/mole), suggesting that the dNTP interacts with the template base in the open complex via hydrogen bonds. We propose that the shape of the base pair determines the fate of the weakly bound nucleotide during the conformational change step. A correct base pair induces a change in enzyme structure in which the enzyme closes around the base pair to form a tight, catalytic complex. If a mismatched nucleotide is bound, the enzyme does not close, but rather proceeds to a structure which promotes nucleotide release while reducing the rate of catalysis.
Figure 3. Pathway of nucleotide binding.
The two-step sequence for nucleotide binding is shown with structures of the open E.DNA complex (tko.pdb) and the closed F.DNA.dNTP complex (tk5.pdb). The structure of the open E.DNA.dNTP complex is not known and is modeled here by placing dNTP into the empty E.DNA complex.
A graphic analogy can be made based upon the tasting and swallowing of food. If the initial taste is good, the mouth closes, and in most instances this represents that step at which a commitment is made to swallow the food. In contrast, if the initial taste is bad, that induces an altered configuration of the mouth which promotes release and inhibits swallowing.
Although our model provides a satisfying thermodynamic description of the role of conformational changes in enzyme specificity and efficiency, there are many questions that remain unanswered. In particular, structures of the empty E-DNA state show more disorder in the vicinity of the active site suggesting a more flexible structure than seen the closed E-DNAdd-dNTP state. In addition, there is no structure that shows the binding of a mismatch as an incoming nucleotide. Our kinetic data suggest that the mismatch recognition state is not a single state but a mixture of states, and that may preclude crystallization. Moreover, we do not know the location of the templating base or how the nucleotide first binds to the open E-DNA complex and it may not be possible to obtain a crystal structure of the open complex with nucleotide bound. We can to infer that the dNTP forms a base pair with the templating base because specificity is seen in the initial EDndNTP complex. Following the initial formation of the open EDndNTP complex, we known nothing about how the initial weak interactions trigger a conformational change to the closed state for a correct base, but trigger the formation of a mismatch-recognition state for an incorrect base. This remains as an area of active investigation.
Pyrophosphate release and translocation
Following the chemistry step, the enzyme must release pyrophosphate and then translocates to allow the binding of the next nucleotide. Very little is known about these steps because all evidence suggests that both reactions are normally much faster than chemistry. Evidence for fast pyrophosphate release and translocation come from two experiments. First, the incorporation of two nucleotides in rapid succession occurs as a simple two-step reaction (A→B→C) without any evidence for a kinetically significant step between the two incorporation events [11]. Second, direct measurement of the rate of pyrophosphate release in a single turnover experiment showed that the rate of the chemical reaction and the rate of pyrophosphate release were coincident [26, 27]. Because the data indicate that pyrophosphate release is fast following chemistry, there are also little data to assess the reversibility of the chemical reaction at the active site. In single turnover experiments, the rapid release of pyrophosphate drives the reaction to completion. Measurements of the kinetics of the synthesis of a nucleoside triphosphate after adding pyrophosphate from solution are problematic due to the weak binding of pyrophosphate and the low solubility of Mg-pyrophosphate [11].
The incorporation of 8-oxo-dGTP and AZT-triphosphate by the human mitochondrial DNA polymerase provides an exception to the general rule that pyrophosphate release is fast. Analysis of the burst kinetics during incorporation of 8-oxo-dGTP showed that the amplitude of the burst was dependent upon the nucleotide concentration implying that the chemical reaction came to equilibrium at that active site of the enzyme [28]. However, if pyrophosphate release is fast and largely irreversible, then chemistry cannot come to equilibrium in a single turnover, leading to the suggestion that pyrophosphate release must be slow after the incorporation of 8-oxodGTP. Subsequent analysis of the incorporation of the nucleoside analog AZT revealed the same phenomena, and direct measurement showed that pyrophosphate release was extremely slow following the incorporation of AZT [26]. The reversible chemistry and slow release of pyrophosphate decreases the specificity constant for the incorporation of AZT and 8-oxo-dGTP. Although the structural and thermodynamic basis for this effect is unknown, the results can be rationalized in terms of the physiological challenges of replicating DNA in the highly oxidative environment of the mitochondria. Perhaps the mitochondrial DNA polymerase has evolved this unique means of discriminating against 8-oxo-dGTP, a major oxidative product that accumulates to high concentrations in the mitochondria. Interestingly, in the evolution of resistant to AZT by HIV reverse transcriptase, it appears as though translocation is disfavored which increases the rate of removal of AZT from the DNA by pyrophosphorolysis [3, 29, 30].
The most reasonable model for translocation is based upon a fast diffusion of the DNA between the N- and P-sites. When the DNA is bound in the P-site (product site), pyrophosphate can bind to reverse the chemical reaction by the process of pyrophosphorolysis producing dNTP. When DNA is in the N-site (the post-translocation state), it can bind the next nucleotide leading to primer extension. As the DNA rapidly diffuses between the N and P sites, the binding of dNTP captures the DNA at the N-site and the conformational change then locks the nucleotide and DNA in the closed state. Alternatively, pyrophosphate can trap the DNA at the P-site. This model was proposed some time ago [12] and has recently been rediscovered and renamed as a “Brownian ratchet” model [31]. If the equilibrium constant for translocation favors the P-site, then it will appear kinetically and thermodynamically as if dNTP binding drives translocation. Alternatively, if the equilibrium constant favors the N-site, then the nucleotide simply binds to the enzyme after translocation. In either case, translocation is usually fast and not kinetically significant.
Chemistry of DNA polymerization
Two metal ion mechanism
The polymerization reaction proceeds by a simple nucleophilic attack of the 3'OH of the primer on the α-phosphate of the incoming dNTP followed by the elimination of pyrophosphate as illustrated in Figure 4. Depending the degree of bond formation and bond breakage during the transition state, the reaction may proceed through a pentavalent intermediate, in which there are 5 oxygen atoms bound to the α-phosphate, and which decays by the elimination of pyrophosphate. The reaction uses a "two metal ion" mechanism in which metal ion A activates the 3'OH as a metal hydroxide while both metals A and B stabilizes the developing negative charge on the α-phosphate in the transition state. The two metal ion mechanism appears to be universal, accounting for the basic mechanism by which non-homologous RNA and DNA polymerases catalyze polymerization reactions and as the mechanism by which many enzymes and ribozymes catalyze exonuclease reactions [32–34].
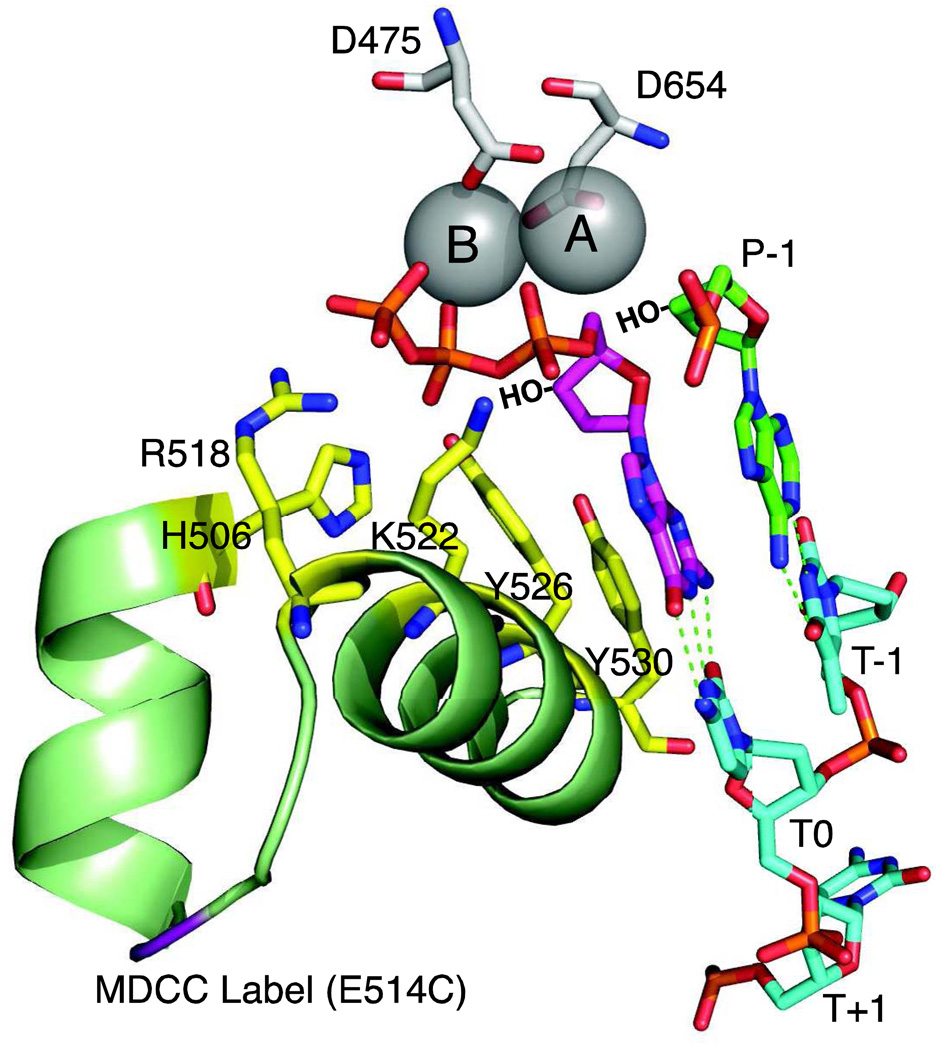
Figure 4. Polymerase active site residues.
The active site of T7 DNA polymerase is shown derived from 1T7P.pdb [1]. Aspartate residues D475 and D654 hold two metal ions (A and B) in place. Important catalytic residues from the fingers domain, (R518, H506, K522, Y526 and Y530, shown in yellow) make contact only in the closed state. The incoming dGTP is shown in magenta, the primer is in green and the template in cyan. Template positions are labeled T−1 through T+1. The 3'OH groups, lacking in the crystal structure, are shown by HO−. The site of the MDCC label is shown in purple.
The nucleotide binds to the enzyme as a Mg-dNTP−2 complex (metal B in Figure 4). Although it has been suggested that the second metal ion (A) binds after the conformational change [35, 36], it is perhaps reasonable to suggest that metal A binds in a rapid-equilibrium reaction such that it equilibrates will all forms of the enzyme, but with higher affinity to the closed (FDndNTP) complex. The addition of EDTA to chelate free metal ions stops the reaction as fast as adding HCl [11], suggesting that metal A dissociates rapidly.
Calcium supports nucleotide binding but not chemistry, so it serves as a useful substitute to probe nucleotide binding [36]. Manganese (Mn+2) supports chemistry, but leads to markedly decreased fidelity by accelerating the rate of incorporation of mismatches [37, 38], so that it is routinely used to generate random mutations during PCR [39]. The effect of Mn+2 on fidelity provides insight into the important role of geometry in fidelity. Mg+2 enforces tetrahedral geometry in the arrangement of its ligands, whereas Mn+2 accommodates square planar, tetrahedral, and octahedral coordination. These data suggest that the lower fidelity observed in the presence of Mn+2 is a result of the increased ability to accelerate the rate of reaction of misaligned substrates [40]. Interestingly, Mn+2 is also used in studies attempting to get some activity from the hepatitis C RNA-dependent RNA polymerase, because attempts to reconstitute physiological activity have so far failed [41, 42].
Catalytic residues
The two metal ions alone are not sufficient for optimal specificity and efficiency of DNA replication. Mutagenesis has shown that residues in the fingers domain that contact the incoming nucleotide and templating base are critical for catalysis and fidelity. Cathy Joyce and her colleagues have defined which amino acids are important for catalysis by mutagenesis of homologous residues in the Klenow fragment of E. coli Pol I [43–48]. In particular, positively charged residues R518, H506, and K522 contact the β-and γ-phosphates of the incoming dNTP, contributing to charge neutralization and alignment of the α-phosphate for reaction (Figure 4). The tyrosine residue Y526 stacks with the incoming dNTP forms a critical H-bond to the ribose. Mutations in Y530 lead to decreased fidelity.
These residues may be distant from the incoming base during the initial binding of the nucleotide to the E-DNA open complex. The conformational change, which is triggered by the binding of a correct base pair, brings these residues into contact with the incoming dNTP and templating base to align the reactants and facilitate catalysis by the two metal ions. Thus, one can think of the two metal ions as forming the core catalytic center, but movements in the recognition domain provide the basis for selective activity by bringing a correct incoming dNTP into proper alignment with the primer 3’OH for reaction and by providing additional catalytic residues. How these same residues and others in the active site recognize a mismatch and disfavor catalysis remains as an important mystery.
Base pair hydrogen bonds
Numerous publications by Kool and his colleagues suggested that hydrogen bonds between base pairs were unimportant for nucleotide selectivity [49–58]. Rather, they argued that nucleotide specificity was determined solely by steric effects in selecting base pairs of the proper size and geometry. However, subsequent analysis showed that these conclusions were a result of faulty kinetic analysis of the rates of incorporation of nucleotide analogues using steady state methods. Careful kinetic analysis has shown that hydrogen bonds contribute at least 2.4 kcal/mol to the specificity constant governing nucleotide incorporation [18, 59]. In addition, hydrogen bonds between base pairs are absolutely essential in the terminal base pair for efficient addition of the next base and for the selectivity of the proofreading exonuclease [18, 60], which is dependent upon the kinetic partitioning between forward polymerization and the transfer of the DNA primer from the polymerase to the exonuclease active site [9]. Thus hydrogen bonds are essential elements contributing to nucleotide selectivity and efficiency of incorporation and proofreading. Moreover, because the strength of a hydrogen bond is strongly dependent upon angles of the two dipoles, it is likely that hydrogen bonds contribute significantly to base pair geometry as well. Certainly, this explains the failure of analogs lacking hydrogen bonds in the terminal base pair of a primer:template DNA to serve as an efficient substrate for the next round of polymerization [17]. Moreover, it is known that a mismatch in the primer:template greatly inhibits extension [9]. Therefore, it is reasonable to suppose that interactions between the protein and the terminal base pair “test” for proper base pair geometry and free energy such that a mismatch is recognized leading to an altered enzyme structure that inhibits the incorporation of the next base.
Thio-phosphate elemental effects
Sulfur substitution of one of the non-bridging oxygen atoms on the α-phosphate of the incoming dNTP alters the electronic structure of the transition state to slow the rate of the chemical reaction. There are two non-bridging oxygen atoms. The Rp oxygen is liganded to the metal ions, so substitution with sulfur greatly inhibits the rate of the chemical reaction (Figure 5). However, substitution of the Sp oxygen with sulfur allows for much smaller effects on catalysis that were thought to be due solely to the change in transition state structure. Early estimates suggested a 100-fold effect of the thio substitution based upon model studies in solution; accordingly an observed 3-fold effect on the rate of the incorporation reaction was interpreted to mean that chemistry was not rate limiting [10, 11]. Moreover, the 60-fold effect during misincorporation indicated that chemistry was rate limiting with an incorrect base pair. However, subsequent analysis of ribozymes indicated a much smaller inherent elemental effect on chemistry [61]. Our recent unpublished studies on T7 polymerase support the conclusion that the majority of the observed elemental effect is due to steric effects of the larger sulfur atom (Z. Jin and K. A. Johnson, manuscript in preparation).
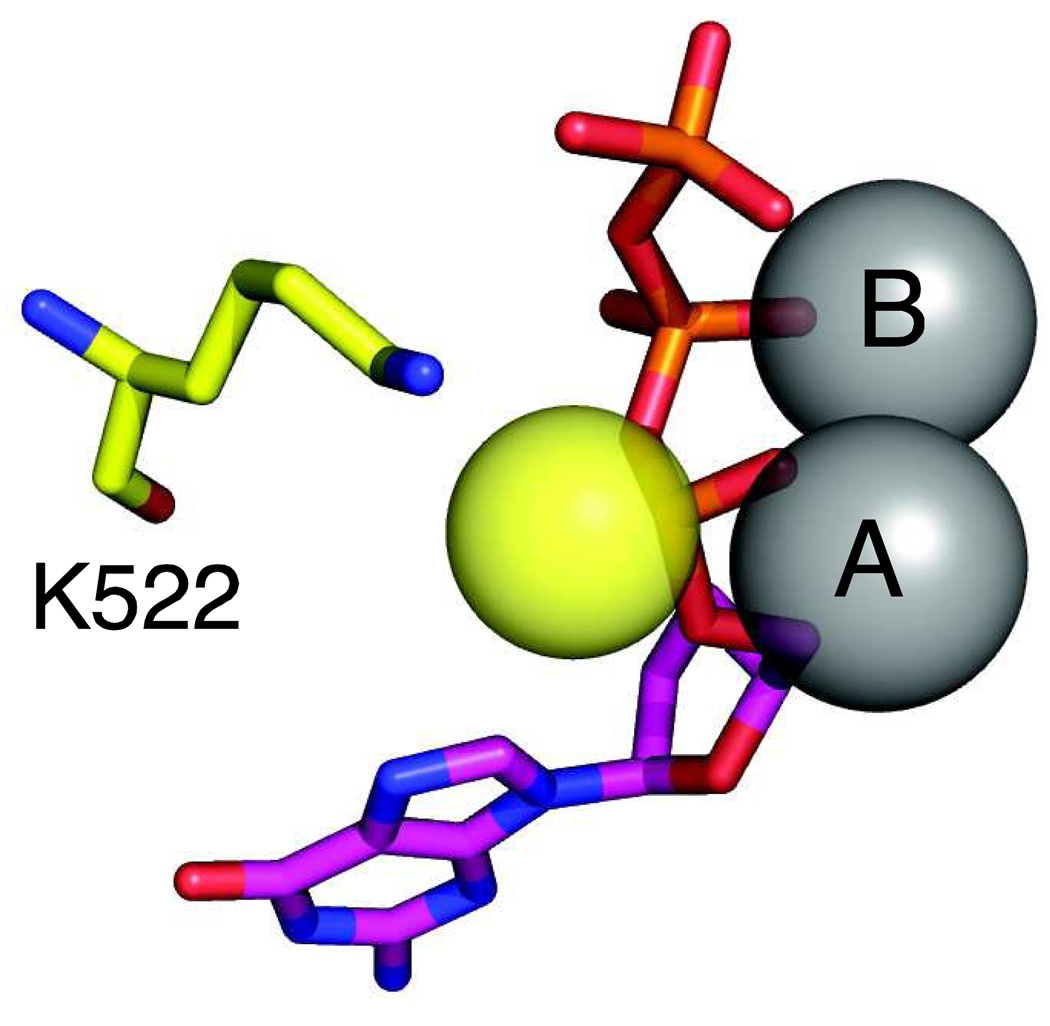
Figure 5. Alpha-thio nucleotide.
The structure of the dNTP-αS(Sp) is shown with the sulfur represented by the yellow sphere. Also shown is the residue K522 which forms a bond to the Sp oxygen on the α-phosphate and may cause steric effects upon sulfur substitution. The nucleotide is shown in magenta and the metal ions A and B are gray. Drawn from 1T7P.pdb.
Outstanding questions
Our current high resolution structural and kinetic data raise as many questions as they provide answers to define the mechanistic basis for nucleotide discrimination. Where does dNTP first bind? Where is the templating base? How does initial binding trigger the conformation change? We do not know where and how the dNTP first binds, other that to know that it appears to form hydrogen bonds with the templating base. We have a good crystal structure of the closed ternary E.DNA.dNTP complex for several enzymes. However, the open E.DNA complex shows varying degrees of disorder which limit our understanding of the structure of the enzyme. The structures of the closed complex help to define the interactions that lead to catalysis, but the pathway from the open to the closed state is unknown. In particular, we need to determine how the initial binding of the dNTP “tickles” the enzyme through the initial weak contacts that then grow in strength as the enzyme proceeds to the closed complex. Perhaps even more of a puzzle is to understand how the initial weak binding of a mismatch is recognized and tickles to the enzyme to proceed to a unique mismatch recognition state which disfavors catalysis while promoting release. Thus the process is much more complex than can be addressed simply by examination of the structure of the closed complex. Nucleotide selectivity is a function of the dynamics of the enzyme structure in ways that cannot be understood by examination of a single static structure.
Acknowledgements
Supported by NIH grants GM044613, GM071404, and GM084741 and a grant from the Welch Foundation (F-1604).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 2.Huang HF, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-I reverse transcriptase: Implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 3.Sarafianos SG, Clark AD, Jr, Tuske S, Squire CJ, Das K, Sheng D, Ilankumaran P, Ramesha AR, Kroth H, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. Trapping HIV-1 reverse transcriptase before and after translocation on DNA. J.Biol.Chem. 2003;278:16280–16288. doi: 10.1074/jbc.M212911200. [DOI] [PubMed] [Google Scholar]
- 4.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of Ternary Complexes of Rat Dna-Polymerase-Beta, A Dna Template-Primer, and Ddctp. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 5.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fersht AR. Enzyme Structure and Mechanism. 3rd ed. New York: Freeman; 1999. [Google Scholar]
- 7.Post CB, Ray WJ., Jr Reexamination of induced fit as a determinant of substrate specificity in enzymatic reactions. Biochemistry. 1995;34:15881–15885. doi: 10.1021/bi00049a001. [DOI] [PubMed] [Google Scholar]
- 8.Berdis AJ. Mechanisms of DNA polymerases. Chem Rev. 2009;109:2862–2879. doi: 10.1021/cr800530b. [DOI] [PubMed] [Google Scholar]
- 9.Donlin MJ, Patel SS, Johnson KA. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- 10.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 11.Patel SS, Wong I, Johnson KA. Pre-Steady-State Kinetic-Analysis of Processive Dna-Replication Including Complete Characterization of An Exonuclease-Deficient Mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 12.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 13.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 14.Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. The.Enzymes. 1992;XX:1–61. [Google Scholar]
- 15.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 16.Lim SE, Copeland WC. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase gamma. J.Biol.Chem. 2001;276:23616–23623. doi: 10.1074/jbc.M101114200. [DOI] [PubMed] [Google Scholar]
- 17.Lee HR, Helquist SA, Kool ET, Johnson KA. Base pair hydrogen bonds are essential for proofreading selectivity by the human mitochondrial DNA polymerase. J Biol Chem. 2008;283:14411–14416. doi: 10.1074/jbc.M705006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J Biol Chem. 2008;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunlap CA, Tsai MD. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase beta. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 22.Showalter AK, Tsai MD. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 23.Purohit V, Grindley ND, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 24.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 25.Johnson KA. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J Biol Chem. 2008;283:26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanes JW, Johnson KA. A novel mechanism of selectivity against AZT by the human mitochondrial DNA polymerase. Nucleic Acids Res. 2007;35:6973–6983. doi: 10.1093/nar/gkm695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanes JW, Johnson KA. Real-time measurement of pyrophosphate release kinetics. Anal Biochem. 2008;372:125–127. doi: 10.1016/j.ab.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanes JW, Thal DM, Johnson KA. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J. Biol. Chem. 2006;281:36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 29.Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci U S A. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotte M. Effects of nucleotides and nucleotide analogue inhibitors of HIV-1 reverse transcriptase in a ratchet model of polymerase translocation. Curr.Pharm.Des. 2006;12:1867–1877. doi: 10.2174/138161206776873626. [DOI] [PubMed] [Google Scholar]
- 32.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J.Biol.Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 34.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 35.Bakhtina M, Roettger MP, Kumar S, Tsai MD. A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry. 2007;46:5463–5472. doi: 10.1021/bi700084w. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Lee HR, Konigsberg W. Effect of A and B metal ion site occupancy on conformational changes in an RB69 DNA polymerase ternary complex. Biochemistry. 2009;48:2075–2086. doi: 10.1021/bi801627h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabor S, Richardson CC , Effect of Manganese Ions on the Incorporation of Dideoxynucleotides by Bacteriophage-T7 Dna-Polymerase and Escherichia-Coli Dna-Polymerase-I. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hays H, Berdis AJ. Manganese substantially alters the dynamics of translesion DNA synthesis. Biochemistry. 2002;41:4771–4778. doi: 10.1021/bi0120648. [DOI] [PubMed] [Google Scholar]
- 39.Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 40.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta. Biochemistry. 1996;35:12762–12777. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 41.Liu C, Chopra R, Swanberg S, Olland S, O'Connell J, Herrmann S. Elongation of synthetic RNA templates by hepatitis C virus NS5B polymerase. J Biol Chem. 2004;279:10738–10746. doi: 10.1074/jbc.M310062200. [DOI] [PubMed] [Google Scholar]
- 42.Cramer J, Jaeger J, Restle T. Biochemical and pre-steady-state kinetic characterization of the hepatitis C virus RNA polymerase (NS5BDelta21, HC-J4) Biochemistry. 2006;45:3610–3619. doi: 10.1021/bi051483s. [DOI] [PubMed] [Google Scholar]
- 43.Astatke M, Grindley ND, Joyce CM. How E. coli DNA polymerase I (Klenow fragment) distinguishes between deoxy- and dideoxynucleotides. J.Mol.Biol. 1998;278:147–165. doi: 10.1006/jmbi.1998.1672. [DOI] [PubMed] [Google Scholar]
- 44.Lam WC, Van der Schans EJ, Joyce CM, Millar DP. Effects of mutations on the partitioning of DNA substrates between the polymerase and 3'-5' exonuclease sites of DNA polymerase I (Klenow fragment) Biochemistry. 1998;37:1513–1522. doi: 10.1021/bi9720181. [DOI] [PubMed] [Google Scholar]
- 45.Astatke M, Ng KM, Grindley NDF, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin TC, Wang CX, Joyce CM, Konigsberg WH. 3'-5' Exonucleolytic activity of DNA polymerases: structural features that allow kinetic discrimination between ribo- and deoxyribonucleotide residues. Biochemistry. 2001;40:8749–8755. doi: 10.1021/bi0105936. [DOI] [PubMed] [Google Scholar]
- 47.Lam WC, Thompson EH, Potapova O, Sun XC, Joyce CM, Millar DP. 3'-5' exonuclease of Klenow fragment: role of amino acid residues within the single-stranded DNA binding region in exonucleolysis and duplex DNA melting. Biochemistry. 2002;41:3943–3951. doi: 10.1021/bi0120603. [DOI] [PubMed] [Google Scholar]
- 48.Thompson EH, Bailey MF, Van der Schans EJ, Joyce CM, Millar DP. Determinants of DNA mismatch recognition within the polymerase domain of the Klenow fragment. Biochemistry. 2002;41:713–722. doi: 10.1021/bi0114271. [DOI] [PubMed] [Google Scholar]
- 49.Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc.Natl.Acad.Sci.U.S.A. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guckian KM, Kool ET. Highly precise shape mimicry by a difluorotoluene deoxynucleoside, a replication-competent substitute for thymidine. Angewandte Chemie-International Edition. 1997;36:2825–2828. doi: 10.1002/anie.199728251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moran S, Ren RXF, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moran S, Ren RXF, Rumney S, Kool ET. Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. Journal of the American Chemical Society. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morales JC, Kool ET. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat.Struct.Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 54.Guckian KM, Morales JC, Kool ET. Structure and base pairing properties of a replicable nonpolar isostere for deoxyadenosine. Journal of Organic Chemistry. 1998;63:9652–9656. doi: 10.1021/jo9805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matray TJ, Kool ET. Selective and stable DNA base pairing without hydrogen bonds. Journal of the American Chemical Society. 1998;120:6191–6192. doi: 10.1021/ja9803310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Neill BM, Ratto JE, Good KL, Tahmassebi DC, Helquist SA, Morales JC, Kool ET. A highly effective nonpolar isostere of deoxyguanosine: synthesis, structure, stacking, and base pairing. J.Org.Chem. 2002;67:5869–5875. doi: 10.1021/jo025884e. [DOI] [PubMed] [Google Scholar]
- 57.Helquist SA, Qu J, Morales JC, Kool ET. Replication of nonpolar shape mimics of guanine and cytosine by two A-family DNA polymerases. Abstracts of Papers of the American Chemical Society. 2004;228:U166–U166. [Google Scholar]
- 58.Kool ET, Sintim HO. The difluorotoluene debate--a decade later. Chem Commun.(Camb.) 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 59.Potapova O, Chan C, DeLucia AM, Helquist SA, Kool ET, Grindley NDF, Joyce CM. DNA polymerase catalysis in the absence of Watson-Crick hydrogen bonds: Analysis by single-turnover kinetics. Biochemistry. 2006;45:890–898. doi: 10.1021/bi051792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morales JC, Kool ET. Importance of terminal base pair hydrogen-bonding in 3'-end proofreading by the Klenow fragment of DNA polymerase I. Biochemistry. 2000;39:2626–2632. doi: 10.1021/bi992173a. [DOI] [PubMed] [Google Scholar]
- 61.Herschlag D, Piccirilli JA, Cech TR. Ribozyme-catalyzed and nonenzymatic reactions of phosphate diesters: rate effects upon substitution of sulfur for a nonbridging phosphoryl oxygen atom. Biochemistry. 1991;30:4844–4854. doi: 10.1021/bi00234a003. [DOI] [PubMed] [Google Scholar]
- 62.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]