Abstract
We describe a method for direct, quantitative, in vivo lipid profiling of oil-producing microalgae using single-cell laser-trapping Raman spectroscopy. This approach is demonstrated in the quantitative determination of the degree of unsaturation and transition temperatures of constituent lipids within microalgae. These properties are important markers for determining engine compatibility and performance metrics of algal biodiesel. We show that these factors can be directly measured from a single living microalgal cell held in place with an optical trap while simultaneously collecting Raman data. Cellular response to different growth conditions is monitored in real time. Our approach circumvents the need for lipid extraction and analysis that is both slow and invasive. Furthermore, this technique yields real-time chemical information in a label-free manner, thus eliminating the limitations of impermeability, toxicity, and specificity of the fluorescent probes common in currently used protocols. Although the single-cell Raman spectroscopy demonstrated here is focused on the study of the microalgal lipids with biofuel applications, the analytical capability and quantitation algorithms demonstrated are applicable to many different organisms and should prove useful for a diverse range of applications in lipidomics.
Keywords: lipid analysis, bioenergy
The global concerns surrounding unabated fossil fuel consumption and the risk of significant environmental impact caused by the associated greenhouse gas emissions, compounded by potential challenges associated with land-based biofuels, have renewed significant interest in microalgae as an alternative feedstock for the production of biodiesel and other biofuels (1). Microalgae hold considerable promise because of their ability to synthesize and store lipids, such as fatty acids and triacylglycerols (TAGs), which can be readily converted into biodiesel (fatty acid methyl or ethyl esters) through relatively simple chemical reactions (2). Small yet efficient, microalgae are attractive for many reasons, including their rapid, cost-effective, and resource-efficient production on nonarable land or photobioreactors (3), with impaired water, and for especially significant lipid production—up to 20–50% of their total dry weight, with examples of up to 80% under certain conditions reported (4). It has been estimated that lipid production of microalgae could be 30 times more efficient in terms of relative production of lipids per acre per year than any other terrestrial plant oil feedstock (2, 5).
Under optimal growth conditions, microalgae synthesize fatty acids in the form of various glycerol-based membrane lipids primarily for structural and functional roles (6). In contrast, adverse environmental or metabolic stress conditions such as nutrient limitation, commonly referred to as “lipid trigger” conditions, result in an increase in carbon partitioning and accumulation of substantial proportions of neutral lipids (20–50% of dry weight), primarily in the form of TAGs. The TAGs are a form of efficient energy storage within the microalgae and are stored as dense lipid bodies. It is also reported that the lipid trigger conditions result in low growth rates for the algae, thereby placing an upper limit on the total lipid production that can be achieved under these conditions (5, 7). Novel strain selection as well as genetic and physiological manipulations may offer solutions that shift this balance between growth rate and lipid biosynthesis, but they require facile methods that establish correlations among lipid trigger conditions, growth rate, and lipid production.
Lipid content within microalgae, as in the general field of lipidomics (8, 9), is commonly analyzed by using solvent extraction–based methodologies, with the lipids subsequently quantified gravimetrically (10–12). Further separation and analysis of lipids can be carried out by using chromatography (GC or HPLC) (13), mass spectrometry (14), or NMR (15). These steps provide compositional identification of the extracted lipids but are relatively time- and effort-intensive. Moreover, the information linking cellular dynamics and functionality is often lost during the homogenization step typically required for these assays, and therefore it becomes less useful for understanding and controlling the fundamental biological processes needed for developing physiological manipulation and metabolic engineering methods. Staining microalgae cells with environmentally sensitive lipid-soluble fluorescent probes (e.g., Nile red) that can discriminate between neutral and polar lipid environments has been shown to allow single-cell lipid analyses, including high-throughput techniques (16, 17). Although useful, these approaches are limited by the availability of photostable fluorescent lipid probes with a well-characterized partitioning preference. For some species of microalgae, these fluorescent probes can be toxic or impermeable, and their fluorescence intensity can be modulated by other molecules such as carotenoids (SI Appendix, Fig. S1) (18, 19). Moreover, these approaches provide little chemical information about the lipids, such as the composition, degree of unsaturation, and chain length. Because these chemical characteristics ultimately determine the quality of the derived biofuel—including viscosity, cloud point, burning efficiency, cetane rating, and stability against oxidation and polymerization (20)—alternative approaches that offer direct and rapid quantification of lipid properties are highly desirable.
Raman spectroscopy offers an attractive alternative for deriving direct, quantitative chemical information in a label-free, nondestructive, and real-time manner at the single-cell level without requiring any exogenous modification of samples (21). Specifically, laser-trapping Raman spectroscopy (LTRS), a combination of a near-IR optical trap and a micro-Raman system (22–25), is particularly well suited to meet these requirements. LTRS affords in vivo spectroscopy from single living cells, producing spectra with significantly higher signal-to-noise ratios, without any preparation step (Movies S1 and S2), than conventional Raman measurements performed on bulky, dried, or immobilized algal samples (26–28) and thus enables a different level of quantitative analysis. As demonstrated in this work, a Raman spectral library of model microalgal lipids can be conveniently generated and applied readily to obtain quantitative information such as the degree of unsaturation (quantified by the average number of C═C bonds per lipid molecule), lipid chain length, and melting temperature (Tm) of the constituent fatty acids in microalgae by using single-cell measurements. We have obtained and interpreted in vivo single-cell Raman spectra of several algal species of interest to bioenergy, including Botryococcus braunii, Neochloris oleoabundans, and Chlamydomonas reinhardtii. Our approach of rapidly interrogating single cells for information related to growth and lipid production should not only pave the way for direct and rapid screening of microalgae but also enable the optimization and selection of species and growth conditions as well as rapidly test the efficacy of physiological and genetic manipulations.
Results
Experimental Setup.
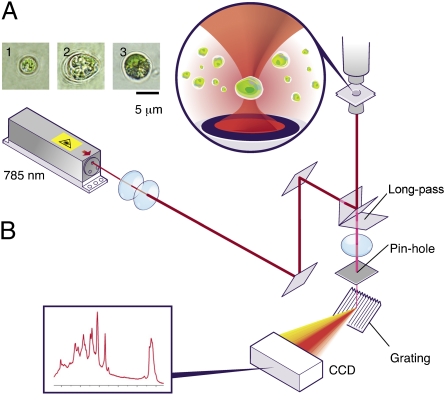
The single-cell LTRS setup combines an optical trap and a micro-Raman system with a confocal pinhole setup in a single custom-built configuration. The micro-Raman system consists of a 785-nm laser (also used for the optical trap), an inverted microscope with a 60× 1.2 N.A. lens fitted with a filter set to clean the laser and block Rayleigh scattering, a spectrograph, and a CCD detector (Fig. 1). For samples of microalgae suspended in their native growth media or other aqueous buffer, the laser focused by the high-N.A. objective forms an optical trap that immobilizes a cell and also allows it to be lifted several micrometers from the bottom to avoid surface-induced perturbations. Signals from regions outside the sample point are rejected by the confocal pinhole, affording high signal-to-noise ratio needed for single-cell spectroscopy. Because a trapped cell is in a large reservoir of aqueous buffer, the temperature within the trap is stable near the bath temperature (29). Because no loss of activity of a biflagellate C. reinhardtii cell has been observed after a 10-min exposure in the laser trap, this system is thought to have a minimal negative impact on the state of health of the cells (Movie S1). Typical acquisition times for well-resolved spectra of pure fatty acids were found to be <1 s and, for a microalgal cell, <10 s (Materials and Methods).
Fig. 1.
Single-cell LTRS setup. (A) Individual microalgal cells immobilized by the optical trap, imaged by bright-field microscopy: (1) N. oleoabundans; (2) B. braunii; and (3) C. reinhardtii. (B) Instrument layout showing the 785-nm laser beam used as both trap beam and Raman excitation beam, excitation path, Raman scattering path, positions of objective lens, pin hole, grating, and CCD.
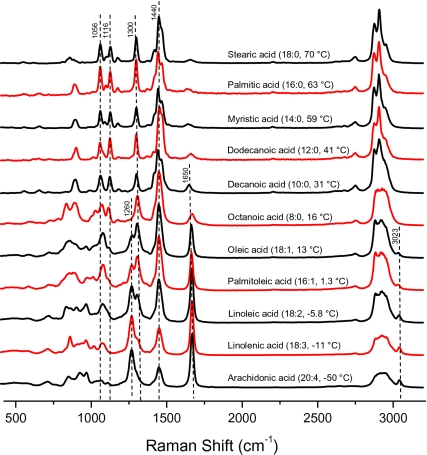
Raman Spectral Library of Algal Model Lipids.
A spectral library was assembled from Raman spectra of several single fatty acids typically found in algal lipid extracts (Fig. 2). Several interrelated diagnostic Raman markers, directly associated with lipid unsaturation, and the respective phase state deduced from the data in Fig. 2 are used to guide the analyses. First, Raman bands corresponding to (i) the ═C—H cis stretch at 1,260 cm−1, (ii) the C═C stretch at 1,650 cm−1, and (iii) the ═C—H stretch at 3,023 cm−1 provide unequivocal evidence for chain unsaturation. Second, the presence and relative intensities of C—C gauche stretch at 1,075 cm−1 and C—C trans stretches at 1,056 cm−1 and 1,116 cm−1 can be used to assess the phase state of the constituent lipids because fatty acid tails are packed in an orderly trans conformation in the solid phase, whereas gauche conformers populate the more disordered, fluid phases (30). Third, peaks assigned to saturated CH2 bonds, such as the CH2 twist at 1,300 cm−1, the CH2 bend at 1,440 cm−1, and the CH2 symmetric and asymmetric stretches at ∼2,800–3,000 cm−1, are typically strong in saturated fatty acids. For instance, between arachidonic acid (20:4, Tm = −50 °C) and stearic acid (18:0, Tm = 70 °C), the unsaturation markers, 1,260 cm−1, 1,650 cm−1, and 3,023 cm−1 consistently diminish, whereas the relative intensities of the bands related to the CH2 bond increase. The gauche conformers are gradually suppressed and intensities caused by trans conformers increase for lipids with higher Tm. These markers provide a semiquantitative classification for the presence and concentration of specific lipid types in unknown samples. Semiquantitative information related to the chain length, unsaturation, and Tm can be deduced from the Raman spectra. This information is typically inaccessible in fluorescence-based data.
Fig. 2.
Raman spectra library of 11 pure fatty acids (model lipids), listed in descending order of their respective Tm.
In Vivo Raman Spectra of Microalgal Cells.
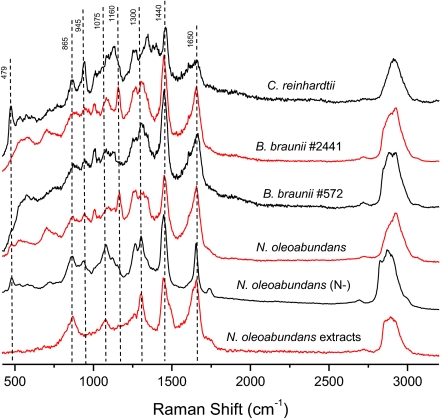
Single-cell LTRS delivers a high-quality Raman spectrum of a single cell that is held steady in a laser trap. Representative LTRS spectra of single cells of several strains, N. oleoabundans cells cultured in nitrate-depleted media, and lipids extracted from N. oleoabundans are shown in Fig. 3. The extracted microalgal lipids show all major lipid Raman bands, which are also represented in the microalgae cells. Raman spectra can also identify other important components such as proteins, carbohydrates, and pigments (see Table 1 for band assignments). Information about proteins can be obtained from the amide I and III bands (1,600–1,700 cm−1 and 1,200–1,350 cm−1, respectively) and from the distinctive phenylalanine symmetric stretching (ring breathing) at 1,004 cm−1. In addition, carotenoids, characterized by bands at 1,008 cm−1, 1,160 cm−1, and 1,537 cm−1, and carbohydrates, seen at 479 cm−1, 945 cm−1, and 1,082 cm−1, can help provide insights into queries related to algal biology.
Fig. 3.
In vivo single-cell LTRS spectra of several species of microalgae, from top to bottom: C. reinhardtii, B. braunii (no. 2441; UTEX), B. braunii (no. 572; UTEX), N. oleoabundans, N. oleoabundans grown in nitrate-depleted media (N−), and the spectrum of algal lipids extracted from normal N. oleoabundans by the chloroform/methanol method.
Table 1.
Assignments for Raman bands
| Wave number (cm−1) | Components | Assignments |
| 479 | Carbohydrate | C— C—C deformation |
| 744 | Carbohydrate, chlorophyll a | H—C—O stretches and chlorophyll a N—C—C deformation |
| 854 | Carbohydrate | Hemiacetal stretches and methylene deformation |
| 865 | Phospholipid | C4N+ and O—C—C—N symmetric stretches |
| 945 | Carbohydrate, protein | α-helix C—C backbone stretches, C—O—C stretches |
| 1,004–1,008 | Protein, carotenoid | Phenylalanine ring breath, carotene C—H bending |
| 1,056, 1,116 | Lipid | Alkyl C—C trans and gauche stretches |
| 1,075 | Lipid | Alkyl C—C gauche stretches |
| 1,082 | Carbohydrate | Carbohydrate C—O—H bending |
| 1,120 | Carbohydrate | C—O—H deformation, C—O and C—C stretches |
| 1,160 | Carotenoid | Carotene C—H stretches |
| 1,200–1,350 | Protein | Amide III |
| 1,260 | Lipid | Alkyl ═C—H cis stretches |
| 1,300 | Lipid | Alkyl C—H2 twist |
| 1,340 | Carbohydrate, chlorophyll a | Carbohydrate C—H2 deformation and C—O—H bending, chlorophyll a C—N stretches |
| 1,440 | Lipid | Alkyl C—H2 bend |
| 1,537 | Carotenoid | Carotene C═C stretches |
| 1,600–1,700 | Protein | Amide I |
| 1,650 | Lipid | Alkyl C═C stretches |
| 1,736 | Lipid | Ester C═O stretches |
| 2,850–2,930 | Lipid, carbohydrate | C—H2, C—H3 asymmetric and symmetric stretches |
| 3,023 | Lipid | Alkyl ═C—H stretches |
A comparison of these in vivo single-cell Raman spectra of B. braunii and N. oleoabundans with the spectra of other types of biomass, such as terrestrial plants (35), bacteria (36), and human cells (24), shows the lipid signatures (e.g., 1,440 and 1,650 cm−1) of the microalgae to be substantially stronger than other components, confirming high lipid content in the microalgae cells. Although the Raman bands (Table 1) mentioned here and in the spectral libraries correspond to compounds found abundantly in algal cells, the LTRS technique is sufficiently sensitive to allow the determination of compounds generated during early-stage lipid accumulation (37).
To determine the ability of Raman spectroscopy to characterize alterations of the lipid profiles within single cells after a change of their environment, the N. oleoabundans cells were stimulated by culturing in nitrate-depleted media and observed by single-cell LTRS. The overall spectral signatures reveal a significant increase in bands attributed to the lipids under nitrate-depleted conditions, confirming a net increase in the lipid content. Although this increase in lipid content, especially TAGs, can also be monitored by fluorescence methods, such as Nile red, for selected strains of microalgae using the cell-culture suspension (SI Appendix, Fig. S1), the single-cell LTRS approach is applicable to all microalgal cells and furthermore provides additional information regarding the chemical characteristics of these compositional changes. The spectra indicate an obvious increase in the overall lipid content (e.g., enhancement of 1,440 cm−1, 1,736 cm−1, and 2,820–3,030 cm−1 bands) that occurs concurrently with a significant decrease in protein content, which is evident from the relative intensities of the protein-associated amide bands (e.g., 1,004 cm−1, amide I and amide III band) (Table 1). This finding is consistent with the notion that, within single cells, nitrate starvation induces an increase in the total lipids present per cell (38).
One major challenge in this approach is the utilization of LTRS measurements of the increase in the total lipid present in a single cell to accurately represent the bulk characteristics of the same response in an algal culture. To address this challenge, statistically significant average behavior reflecting bulk characteristics can be deduced by collecting and analyzing data over multiple cells in single populations (SI Appendix, Fig. S2). This data set indicates that LTRS is able to provide sufficient information to accurately represent the response in a bulk cell population. Additionally, there could be a microfluidics interface with the LTRS unit that would be capable of providing information on tens of hundreds of microalgal cells within minutes.
Quantitative Analysis of Lipid Unsaturation.
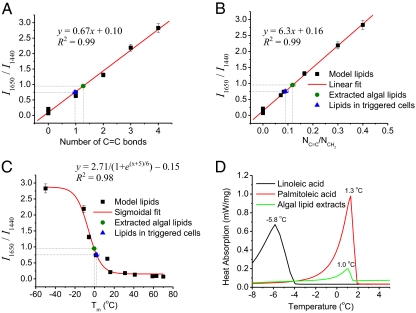
In addition to the qualitative information determined above from the single-cell Raman spectra, quantitation is also possible. Beer's law indicates that the spontaneous Raman scattering depends linearly on the analyte concentration (39, 40). This relationship indicates that, by measuring relevant experimentally acquired Raman bands assigned to specific lipid types (or proteins), concentrations of the corresponding molecular species can be estimated. In actual practice, however, the inherent irreproducibility in condensed-phase Raman spectroscopy measurements limits the direct applicability of Beer's law for absolute determination of concentrations from single peaks. The use of ratiometric analyses involving a comparison of Raman intensities of two independent, but molecularly related, peaks in the same spectrum circumvent this limitation. They also provide important diagnostic markers. For instance, the ratio of Raman intensity of the C═C stretch and the intensity of CH2 bend, I1,650/I1,440, displays a linear dependence (R2 = 0.99) with the ratio of the number of C═C bonds and the number of CH2 bonds for the model lipids (Fig. 4A). The relationship is also linear (R2 = 0.99) when the ratio I1,650/I1,440 is plotted simply as a function of the number of C═C bonds per lipid molecule (Fig. 4B). This approach also eliminates spectral artifacts caused by source-intensity fluctuations, sample-to-sample variation, and other uncontrolled experimental parameters, thus yielding reliable quantitative information.
Fig. 4.
Quantitative analysis of degree of unsaturation, chain length, and Tm of microalgal lipid extracts and lipids in single cells by LTRS spectroscopy and validation by DSC method. (A) The ratio of the intensity of the 1,650 cm−1 band (C═C stretch) to the 1,440 cm−1 band (CH2 twist). I1,650/I1,440 of pure fatty acid standards (model lipids) are plotted versus the ratio of the number of C═C bonds to CH2 bonds and fitted linearly (R2 = 0.99, n = 11). Then I1,650/I1,440 is measured on the lipids extracted from normal N. oleoabundans cells (extracted algal lipids), and the lipids in the individual N. oleoabundans cells grown in nitrate-depleted media (triggered cells) is fitted on the curve to predict the lipids’ molecular structure. (B) I1,650/I1,440 of the model lipids are plotted verses the number of C═C bonds per molecule and fitted linearly (R2 = 0.99, n = 11) to predict the degree of unsaturation of the extracted algal lipids and the lipids in the triggered cells. (C) I1,650/I1,440 of the model lipids are plotted versus their published Tm and fitted with a sigmoidal Boltzmann function (R2 = 0.98, n = 11) to predict the Tm of the extracted algal lipids and the lipids in the triggered cells. (D) DSC measurement of the Tm of the extracted algal lipids confirms the prediction based on its Raman spectra.
This ratiometric method provides a quantitative means with which to estimate the average number of C═C bonds when the ratio of I1,650/I1,440 can be measured. For example, I1,650/I1,440 obtained from the lipids extracted from N. oleoabundans in normal growth condition and from the triggered individual cells are plotted on the graphs shown in Fig. 4 A and B (see SI Appendix, Note S1 for fitting parameters). These results suggest that the extracted algal lipids have, on average, ∼1.25 double bonds per lipid molecule and that the ratio of double bonds to CH2 bonds is ∼0.124. For the lipids in triggered cells, there are ∼0.97 double bonds per lipid molecule on average, and the ratio of double bonds to CH2 bonds is ∼0.09. Sharing similar formula CH3(CH2)m(CH═CH)nCOO—, the average chain length of the fatty acids incorporated in the TAGs equals the number of CH2 bonds plus two carbons in each C═C bond and two carbons at the COO— head and —CH3 tail, e.g., 1.25/0.124 + 2 × 1.25 + 2 ∼ 14.6, for the extracted algal lipids. Therefore, based on the ratiometric data summarized above, the average fatty acid chain formula of the lipids extracted from the normal N. oleoabundans cell estimates to be 14.6:1.25, which matches the length of the fatty acid methyl ester reported by using gas chromatography (13).
LTRS measurements show that, for the individual cells cultured in nitrate-depleted media, the average chain length is shortened to n = 14.3, with a decreased degree of unsaturation of ∼0.97 C═C bond per lipid. This difference in the degree of unsaturation associated with the lipid makeup of whole cells in nitrate-depleted media is presumably caused by the lipid synthetic pathways in response to the stress induced by nitrate-depleted growth condition. This finding is consistent with many reports of the lipid profile changes that occur in response to growth conditions, such as nutrient limitation (14, 41–44).
In Situ Measurements of Lipid Tm.
Further insight can be gained from the fact that the degree of unsaturation is directly related with the Tm of fatty acids, another critical factor affecting the quality of biodiesel. Temperature dependence of chain order parameters typically generate sigmoidal curves (45, 46), which can be used to predict the Tm of unknown fatty acids with Raman spectroscopy. Specifically, Tm of the model fatty acid relates to the unsaturation that is quantified by Raman ratio I1,650/I1,440 and can be fitted with a sigmoidal curve (R2 = 0.98) (Fig. 4C) (see SI Appendix, Note S1 for fitting parameters). The Raman ratio I1,650/I1,440 from the lipids extracted from N. oleoabundans and from the lipids in individual nitrate-starved cells are then plotted on the same calibration curve, predicting a transition temperature of approximately −0.3 °C for the extracted lipids and 2.6 °C for the lipids measured in whole cells. When the Tm of the extracted lipids was measured by differential scanning calorimetry (DSC), a difference of <1 °C separated the predicted and measured values (Fig. 4D). Because DSC usually requires milligrams of sample for each measurement, obtaining this level of accuracy from single cells represents a significant advance in characterizing the lipids within microalgae. These results demonstrate unprecedented real-time in situ quantitative oil profiling and Tm prediction within a live cell during growth and oil production (see SI Appendix, Table S1 for predictions for other cell types).
Discussion
The enormous biodiversity of microalgae poses huge challenges in the identification and screening of potential candidates for effective biofuel production (47). The utilization of genetic manipulation and physiological engineering to improve biofuel production only increases the sample diversity. This study of only five algal species in response to changes of light exposure, nitrogen, silicon, CO2, pH, and temperature required culturing a minimum of 30 samples. This number rapidly increases when full-factorial statistically designed experiments are required to understand the important biotic and abiotic factors affecting growth and lipid production. Central to overcoming these challenges is the development of a rapid, facile, and reliable analytical method to characterize the composition and concentration of energy-rich neutral lipids. The work reported here illustrates the promise of single-cell Raman spectroscopy for a rapid, accurate, and quantitative characterization of lipid profiles from single microalgae cells in a nondestructive and label-free manner. The technique is amenable to high-throughput screening and/or a sorting-based assay when coupled with microfluidics, or it can be implemented as a pond- or bioreactor-based in situ screening assay for the routine assessment of algal health and productivity. Microalgae have also been used as a resource for obtaining other compounds, such as dietary fatty acids and proteins, as well as ingredients of cosmetics and pharmaceuticals (48), such as antioxidant carotenoids, which also have distinct Raman signatures. Single-cell Raman spectroscopy would also be useful for in situ and direct monitoring of these components. Thorough quantitative analysis enabled by chemometrics techniques could extract more information from lipids and other important components produced in microalgae (49).
The use of single-cell LTRS affords a simple, quick, and highly effective method to interrogate individual microalgal cells with a high degree of specificity in terms of the information generated, and it promises to enable rapid screening for optimal species in terms of sustainable, as well as economically and environmentally responsible, supplies of liquid fuel and other valuable derivatives. This demonstration of facile, label-free, quantitative in vivo analysis of microalgae is suitable to understanding lipid profiles of any organism or metabolic system where knowledge of fast dynamics of metabolites, pathways, and lipid compositions are desired. Lipid metabolism disorders and their link to numerous diseases have led to a dramatic increase in research and system-level studies of extremely intricate cellular interactions and lipid profiles. Mass spectrometry and chromatography-based techniques on cell extracts have filled significant gaps in our current understanding in terms of lipid metabolism disorders. To date, there are a handful of experimental approaches with living cells using fluorescent lipids that have also shown promise but are limited in their ability because of nonspecificity, providing mostly qualitative signatures (8, 50). This in vivo capability expands the list of available analytical tools to researchers in the emerging field of lipidomics for multiple applications.
Materials and Methods
Algal Strain, Culture Condition, and Lipid Triggering.
From the Culture Collection of Algae at the University of Texas at Austin (UTEX), we obtained B. braunii (nos. 572 and 2441) with its growth medium, Bold 3N, and N. oleoabundans (no. 1185) with its growth medium, soil extract. C. reinhardtii (CC-503 cw92 mt+) and Sueoka's growth medium were obtained from the Chlamydomonas Center at Duke University. Algal cultures (120 mL) were grown in sterile 250-mL glass culture flasks covered loosely with sterile foil. Culture flasks were placed in a diurnal incubator at a temperature of 20.8 °C with a daily cycle of 16 h of fluorescent light followed by 8 h of dark.
Nitrate-depleted growth of N. oleoabundans cultures were obtained by initially growing cultures in soil extract medium. At day 19 of growth, the regular medium was changed to the triggering medium, which was made by following the soil extract recipe from UTEX but eliminating the NaNO3 component entirely. The other components included CaCl2·2H2O, MgSO4 (anhydrous), K2HPO4, KH2PO4, NaCl, and soil–water medium purchased from WARD's Natural Science.
All chemicals were purchased from Sigma-Aldrich, unless otherwise stated. Organic solvents are all HPLC grade.
Lipid Extraction.
Chloroform/methanol extraction of N. oleoabundans was based on the method described by Bligh and Dyer (10) with slight modifications. A mixture of chloroform and methanol (2:1 vol/vol) was used as an extraction solvent. Glassware was used for all of the steps to minimize polymer contamination. We used 1 L of N. oleoabundans culture at confluence. The cells were centrifuged at 2,700 × g for 10 min. The cell pellets were then collected and resuspended in 1 mL of deionized water in a glass vial. The vial was sonicated for 10 min in a 50 °C water bath, and then 3 mL of extraction solvent was added into the vial. The extraction step was processed by placing the vial in a 50 °C water bath for 1 h while vortexing vigorously for 1 min every 10 min and allowing the suspension to settle for 30 s on ice between each vortex. Afterward, the sample was placed at −20 °C for 1 h to accelerate separation. After separation, the bottom oil/chloroform phase was transferred with a needle and stored at −20 °C for future use. To evaluate lipids by Raman measurements, the chloroform was evaporated under a stream of nitrogen, and the sample was placed under vacuum for 2 h to remove residual chloroform. The lipid film was finally resuspended in deionized water at a concentration of 5 mg/mL.
Single-Cell LTRS.
Briefly, a 785-nm laser with 70-mW output power from a continuous-wave diode-pumped solid-state laser (CrystaLaser) was directed through a 4× beam expander and a 785-nm bandpass filter (Omega Filters) to remove any possible plasma emission generated within the laser tube. The beam was then delivered into an inverted Olympus optical microscope (IX71), which was equipped with a dichroic longpass beamsplitter (Chroma Technology) to reflect the laser beam into a 1.2 N.A. 60× water-immersion objective lens, resulting in a diffraction limited spot of 0.5-μm diameter at the laser focus. The beam was focused through a quartz coverslip with a power of ∼10 mW out of the objective. Raman scattering generated at the focus was collected by the same objective, passed through the dichroic beamsplitter and a longpass edge filter (Omega Filters) to block elastic scattering photons, and then focused through a 100-μm pinhole for background signal rejection. The signal is dispersed by a spectrometer (Acton 2300i; Princeton Instruments) equipped with 1-μm blaze wavelength 300 g/cm grating and projected on an air-cooled EM-CCD with 1,340 × 100 pixels (PIXIS; Roper Scientific). Spectral resolution was 3 cm−1 (3,200 cm−1/1,280 pixels).
For measurements of fatty acid standards, a small amount of sample (5 μL for liquid samples or a couple of grains for solid samples) was immersed in a 200-μL water droplet sitting on the cleaned quartz coverslip. The focal point of the laser was moved onto the sample if it was solid. Measurements were done at room temperature. The acquisition time was 1 s for fatty acid standards.
For particles suspended in aqueous buffer, the laser focused by the high-N.A. objective lens forms an optical trap that can immobilize a single micrometer-sized particle. The focused laser can trap the particle and lift it several micrometers above the surface, which reduces surface perturbation. In addition, noise from regions other than the focal point is rejected by the confocal pinhole. For both these reasons, LTRS has an extremely high signal-to-noise ratio.
For measurements of single cells, cell suspension was diluted such that only a few cells would occupy the microscopic field of view. A sample (40 μL) was placed on the cleaned quartz coverslip. Measurements were started 2 min after a cell was trapped, enabling the prerequisite photobleaching of the chlorophyll fluorescence, and acquisitions were made for 10 s. Raman spectra were acquired and processed by WinSpec32 software (Roper Scientific). After background subtraction, the spectra were smoothed by using an averaging filter. The intensity of each peak was calculated by subtracting the average baseline value of that peak from the maximum intensity value. Baselines for each peak were simplified as a line connecting the two lowest points at the left and right shoulders of the peak. Data fitting was done with Origin software (Microcal Software).
DSC Measurements.
Pure fatty acid standards, linoleic acid and palmitoleic acid, were tested as received without further purification. A small volume (30 μL) of each was sealed into separate DSC crucibles. For the algal lipid extracts, ∼1.5 mg of lipids was evaporated from chloroform under a stream of nitrogen directly into the DSC crucible. Residual chloroform was removed under vacuum for at least 24 h before sealing the crucible. Phase transitions were measured with a differential scanning calorimeter (DSC822e; Mettler-Toledo). Samples were scanned from −20 °C to 20 °C, initially at 10 °C/min and on subsequent scans at 1 °C/min. The values reported were taken at 1 °C/min. Lipid phase transitions were taken as the maxima of endothermic peaks on thermal up-scans.
Supplementary Material
Acknowledgments
We thank Drs. Yin Yeh and Thomas Huser for generous support on the Raman instrument setup. We thank Dr. Ryan Davis for helpful discussions. This project was supported by Laboratory Directed Research and Development Program 09-0800 of Sandia National Laboratories (to S.S.). Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the US Department of Energy's National Nuclear Security Administration under Contract DE-AC04-94AL85000. Work at University of California, Davis, was supported by Grant DE-FG02-04ER46173 from the Division of Materials Science and Engineering, Office of Basic Energy Sciences, Department of Energy (to A.E.O. and A.N.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.F.J.V. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009043108/-/DCSupplemental.
References
- 1.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006;103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chisti Y. Biodiesel from microalgae. Biotechnol Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Gordon JM, Polle JE. Ultrahigh bioproductivity from algae. Appl Microbiol Biotechnol. 2007;76:969–975. doi: 10.1007/s00253-007-1102-x. [DOI] [PubMed] [Google Scholar]
- 4.Hu Q, et al. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- 5.Sheehan J, Dunahay T, Benemann J, Roessler PG. A Look Back at the U.S. Department of Energy's Aquatic Species Program: Biodiesel from Algae. Golden, CO: National Renewable Energy Laboratory; 1998. Rep TP-580-24190. [Google Scholar]
- 6.Thompson GA., Jr. Lipids and membrane function in green algae. Biochim Biophys Acta. 1996;1302:17–45. doi: 10.1016/0005-2760(96)00045-8. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Horsman M, Wang B, Wu N, Lan CQ. Effects of nitrogen sources on cell growth and lipid accumulation of green alga Neochloris oleoabundans. Appl Microbiol Biotechnol. 2008;81:629–636. doi: 10.1007/s00253-008-1681-1. [DOI] [PubMed] [Google Scholar]
- 8.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 9.Watson AD. Thematic review series: Systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: A global approach to lipid analysis in biological systems. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Yoon B-D, Oh H-M. Rapid method for the determination of lipid from the green alga Botryococcus braunii. Biotechnol Tech. 1998;12:553–556. [Google Scholar]
- 12.Pruvost J, Van Vooren G, Cogne G, Legrand J. Investigation of biomass and lipids production with Neochloris oleoabundans in photobioreactor. Bioresour Technol. 2009;100:5988–5995. doi: 10.1016/j.biortech.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Gouveia L, Marques AE, da Silva TL, Reis A. Neochloris oleabundans UTEX #1185: A suitable renewable lipid source for biofuel production. J Ind Microbiol Biotechnol. 2009;36:821–826. doi: 10.1007/s10295-009-0559-2. [DOI] [PubMed] [Google Scholar]
- 14.Yu E, et al. Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation. J Appl Phycol. 2009;21:669–681. [Google Scholar]
- 15.Craigie JS, MacKinnon SL, Walter JA. Liquid seaweed extracts identified using H-1 NMR profiles. J Appl Phycol. 2008;20:665–671. [Google Scholar]
- 16.Cooksey KE, Guckert JB, Williams SA, Callis PR. Fluorometric determination of the neutral lipid content of microalgal cells using Nile Red. J Microbiol Methods. 1987;6:333–345. [Google Scholar]
- 17.Chen W, Zhang C, Song L, Sommerfeld M, Hu Q. A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods. 2009;77:41–47. doi: 10.1016/j.mimet.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Liaaen-Jensen S, Andrewes AG. Microbial carotenoids. Annu Rev Microbiol. 1972;26:225–248. doi: 10.1146/annurev.mi.26.100172.001301. [DOI] [PubMed] [Google Scholar]
- 19.Vermaas WFJ, et al. In vivo hyperspectral confocal fluorescence imaging to determine pigment localization and distribution in cyanobacterial cells. Proc Natl Acad Sci USA. 2008;105:4050–4055. doi: 10.1073/pnas.0708090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington KJ. Chemical and physical properties of vegetable oil esters and their effect on diesel fuel performance. Biomass. 1986;9:1–17. [Google Scholar]
- 21.Sadeghi-Jorabchi H, Wilson RH, Belton PS, Edwards-Webb JD, Coxon DT. Quantitative analysis of oils and fats by Fourier transform Raman spectroscopy. Spectrochim Acta [A] 1991;47:1449–1458. [Google Scholar]
- 22.Ashkin A. Acceleration and trapping of particles by radiation pressure. Phys Rev Lett. 1970;24:156–159. [Google Scholar]
- 23.Cherney DP, Conboy JC, Harris JM. Optical-trapping Raman microscopy detection of single unilamellar lipid vesicles. Anal Chem. 2003;75:6621–6628. doi: 10.1021/ac034838r. [DOI] [PubMed] [Google Scholar]
- 24.Chan JW, et al. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys J. 2006;90:648–656. doi: 10.1529/biophysj.105.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoladek A, Pascut F, Patel P, Notingher I. Development of Raman imaging system for time-course imaging of single living cells. Spectroscopy. 2010;24:131–136. [Google Scholar]
- 26.Wu Q, et al. Differentiation of algae clones on the basis of resonance Raman spectra excited by visible light. Anal Chem. 1998;70:1782–1787. doi: 10.1021/ac971098b. [DOI] [PubMed] [Google Scholar]
- 27.Huang YY, Beal CM, Cai WW, Ruoff RS, Terentjev EM. Micro-Raman spectroscopy of algae: Composition analysis and fluorescence background behavior. Biotechnol Bioeng. 2010;105:889–898. doi: 10.1002/bit.22617. [DOI] [PubMed] [Google Scholar]
- 28.Samek O, et al. Raman microspectroscopy of individual algal cells: Sensing unsaturation of storage lipids in vivo. Sensors. 2010;10:8635–8651. doi: 10.3390/s100908635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao H, Arias-Gonzalez JR, Smith SB, Tinoco I, Jr., Bustamante C. Temperature control methods in a laser tweezers system. Biophys J. 2005;89:1308–1316. doi: 10.1529/biophysj.104.054536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lippert JL, Peticolas WL. Laser Raman investigation of the effect of cholesterol on conformational changes in dipalmitoyl lecithin multilayers. Proc Natl Acad Sci USA. 1971;68:1572–1576. doi: 10.1073/pnas.68.7.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschirner N, et al. Raman excitation profiles of β-carotene—novel insights into the nature of the ν1-band. Phys Stat Sol (B) 2008;245:2225–2228. [Google Scholar]
- 32.Wood BR, et al. A portable Raman acoustic levitation spectroscopic system for the identification and environmental monitoring of algal cells. Anal Chem. 2005;77:4955–4961. doi: 10.1021/ac050281z. [DOI] [PubMed] [Google Scholar]
- 33.Chan JW, et al. Nondestructive identification of individual leukemia cells by laser trapping Raman spectroscopy. Anal Chem. 2008;80:2180–2187. doi: 10.1021/ac7022348. [DOI] [PubMed] [Google Scholar]
- 34.Arboleda PH, Loppnow GR. Raman spectroscopy as a discovery tool in carbohydrate chemistry. Anal Chem. 2000;72:2093–2098. doi: 10.1021/ac991389f. [DOI] [PubMed] [Google Scholar]
- 35.Schulte F, Lingott J, Panne U, Kneipp J. Chemical characterization and classification of pollen. Anal Chem. 2008;80:9551–9556. doi: 10.1021/ac801791a. [DOI] [PubMed] [Google Scholar]
- 36.Ramser K, et al. Micro-resonance Raman study of optically trapped Escherichia coli cells overexpressing human neuroglobin. J Biomed Opt. 2007;12:044009. doi: 10.1117/1.2753478. [DOI] [PubMed] [Google Scholar]
- 37.Puppels GJ, et al. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature. 1990;347:301–303. doi: 10.1038/347301a0. [DOI] [PubMed] [Google Scholar]
- 38.Suen Y, Hubbard JS, Holzer G, Tornabene TG. Total lipid production of the green alga Nannochloropsis sp. QII under different nitrogen regimes. J Phycol. 1987;23:289–296. [Google Scholar]
- 39.Giles JH, Gilmore DA, Denton MB. Quantitative analysis using Raman spectroscopy without spectral standardization. Journal of Raman Spectroscopy. 1999;30:767–771. [Google Scholar]
- 40.Freudiger CW, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson GJ, Ingram LO. Effects of temperature and nutritional changes on the fatty acids of Agmenellum quadruplicatum. J Bacteriol. 1975;124:373–379. doi: 10.1128/jb.124.1.373-379.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piorreck M, Pohl P. Formation of biomass, total protein, chlorophylls, lipids and fatty acids in green and blue-green algae during one growth phase. Phytochemistry. 1984;23:217–223. [Google Scholar]
- 43.Roessler PG. Changes in the activities of various lipid and carbohydrate biosynthetic enzymes in the diatom Cyclotella cryptica in response to silicon deficiency. Arch Biochem Biophys. 1988;267:521–528. doi: 10.1016/0003-9861(88)90059-8. [DOI] [PubMed] [Google Scholar]
- 44.Reitan KI, Rainuzzo JR, Olsen Y. Effect of nutrient limitation on fatty-acid and lipid-content of marine microalgae. J Phycol. 1994;30:972–979. [Google Scholar]
- 45.Mantsch HH, McElhaney RN. Phospholipid phase transitions in model and biological membranes as studied by infrared spectroscopy. Chem Phys Lipids. 1991;57:213–226. doi: 10.1016/0009-3084(91)90077-o. [DOI] [PubMed] [Google Scholar]
- 46.Chiantia S, Giannola LI, Cordone L. Lipid phase transition in saccharide-coated cholate-containing liposomes: Coupling to the surrounding matrix. Langmuir. 2005;21:4108–4116. doi: 10.1021/la046974c. [DOI] [PubMed] [Google Scholar]
- 47.Norton TA, Melkonian M, Andersen RA. Algal biodiversity. Phycologia. 1996;35:308–326. [Google Scholar]
- 48.Borowitzka M. Microalgae as sources of pharmaceuticals and other biologically active compounds. J Appl Phycol. 1995;7:3–15. [Google Scholar]
- 49.McCreery RL. Raman Spectroscopy for Chemical Analysis. New York: Wiley; 2000. [Google Scholar]
- 50.North AJ. Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition. J Cell Biol. 2006;172:9–18. doi: 10.1083/jcb.200507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.