Abstract
Myosin X (MyoX), encoded by Myo10, is a representative member of the MyTH4–FERM domain-containing myosins, and this family of unconventional myosins shares common functions in promoting formation of filopodia/stereocilia structures in many cell types with unknown mechanisms. Here, we present the structure of the MyoX MyTH4–FERM tandem in complex with the cytoplasmic tail P3 domain of the netrin receptor DCC. The structure, together with biochemical studies, reveals that the MyoX MyTH4 and FERM domains interact with each other, forming a structural and functional supramodule. Instead of forming an extended β-strand structure in other FERM binding targets, DCC_P3 forms a single α-helix and binds to the αβ-groove formed by β5 and α1 of the MyoX FERM F3 lobe. Structure-based amino acid sequence analysis reveals that the key polar residues forming the inter-MyTH4/FERM interface are absolutely conserved in all MyTH4–FERM tandem-containing proteins, suggesting that the supramodular nature of the MyTH4–FERM tandem is likely a general property for all MyTH4–FERM proteins.
Keywords: complex structure, tail domain
Among the unconventional myosin family members (with respect to the conventional myosin for muscle contractions), there is a subfamily, called MyTH4–FERM myosins, which includes myosin VII, X, and XV (MyoVII, MyoX, and MyoXV). These three myosins are so named because the tail of each motor contains one or a pair of MyTH4–FERM domains. The MyTH4–FERM domains in the tail regions of myosin VII, X, and XV are believed to function as cargo binding domains of these myosins (1–7), although the molecular basis of the motor–cargo interactions are not known.
MyoX is perhaps the best studied MyTH4–FERM myosins (8). Its tail MyTH4–FERM tandem is implicated in binding to cargo proteins including β-integrins, microtubules, and axonal guidance receptor DCC (1–3) (Fig. 1A). MyoX is well known for its capacity in filopodial induction and elongation (9–12). The functional roles of MyoX in diverse cellular processes such as cell adhesion, cell or subcellular structure migrations, and angiogenesis (1, 3, 13–16) are likely to be related to the filopodial formation activity of the motor, because filopodia are focal points that integrate various cell signals to regulate actin cytoskeletal structures (17).
Fig. 1.
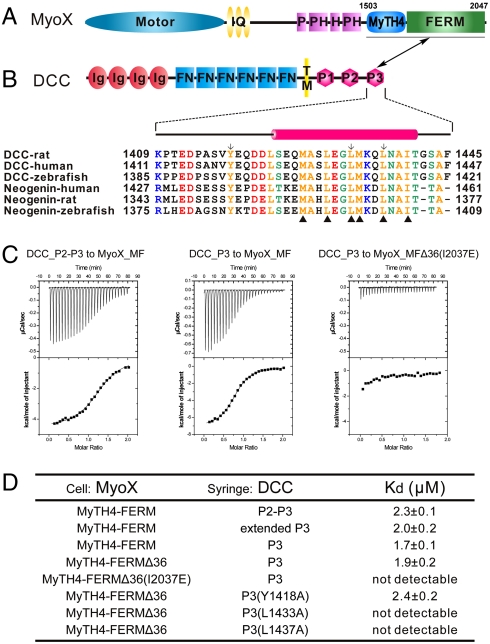
Characterization of the MyoX_MF/DCC_P3 interaction. (A and B) The domain organizations of MyoX and DCC. The boundary of MyoX_MF is indicated. The MyoX_MF/DCC_P3 interaction is indicated by a two-way arrow. The sequence alignment of the P3 motif of DCC and neogenin from different species is included. The residues involved in the MyoX_MF/DCC_P3 interaction are indicated by triangle ups. The residues substituted with Ala to test their role in binding to MyoX_MF are labeled by arrows. (C) The ITC curves show the interactions between MyoX_MF and the different DCC cytoplasmic fragments. The MyoX_MF construct with 36-residue deletion in the FERM domain is indicated as MyoX_MFΔ36. (D) The dissociation constants of the binding reactions of various forms of MyoX_MF and DCC obtained from ITC-based assays. The boundaries for the DCC fragments are residues 1321–1445 for P2–P3, 1370–1445 for the extended P3, and 1409–1445 for P3.
A distinct structural feature of the MyoX tail is that it contains a MyTH4 domain and a FERM domain connected with a short linker. Even though FERM domains in many proteins are known to function autonomously as a versatile protein and/or lipid membrane binding modules (18), the FERM domain in MyoX seems to require its N-terminal MyTH4 domain for its cellular functions. For example, deletion of either the MyTH4 or the FERM domain abolishes the MyoX’s capacity in inducing elongated filopodia formation (9, 11, 12). The MyTH4–FERM tandem, but not either of the isolated domains, of MyoX binds to microtubules (2). Binding of MyoX to DCC or neogenin also requires the covalent connection of the MyTH4 and FERM domains (3). These findings suggest that the integrity of the MyTH4–FERM tandem is critical for MyoX’s diverse cellular activities. Moreover, this integrity seems to be a conserved feature for other MyTH4–FERM domain-containing proteins. Interaction between MyoVIIa and shroom2 requires the second MyTH4–FERM tandem of the myosin motor (19). Formation of the MyoVII/Sans complex requires the covalent connection of the MyTH4 and FERM domains of the first MyTH4–FERM tandem of the motor (6). In the case of MyoXV, the interaction between the motor and the adaptor protein whirlin involves both domains of the motor’s second MyTH4–FERM tandem (4). However, the molecular basis of MyTH4–FERM tandem-mediated cargo recognitions for these myosins is unknown.
Here we report the 2.5-Å crystal structure of MyoX_MF in complex with the DCC P3 motif, providing an atomic picture of the MyTH4–FERM tandem-mediated cargo recognition for the MyTH4–FERM subfamily of myosins. The structure of MyoX_MF shows that the MyTH4 and FERM domains form an integral structural supramodule. The structure of the MyoX_MF/DCC_P3 complex also reveals a noncanonical FERM domain-mediated target recognition mode. Structure-based amino acid sequence analysis suggests that formation of the MyTH4–FERM supramodule is a conserved feature of all MyTH4–FERM-containing proteins.
Results
The MyTH4–FERM tandem of MyoX Binds to the P3 Domain of DCC.
We first characterized biochemical properties of MyoX_MF. Numerous trials with various combinations of domain boundaries and protein expression conditions revealed that neither the isolated MyTH4 domain nor the FERM domain of MyoX could be expressed as soluble proteins in bacteria. In contrast, MyoX_MF was expressed as a soluble monomeric protein.
We next characterized the interaction between MyoX_MF and netrin receptor DCC. Three DCC fragments (the P2-P3 domains, an extended P3 domain, and the P3 domain) displayed essentially identical binding affinities to MyoX_MF (Fig. 1 C and D). DCC_P1 was recently found to interact with ERK1/2 (20), and no interaction between DCC_P1 and MyoX_MF could be detected. The above biochemical data indicate that the P3 domain of DCC (DCC_P3) is the minimal MyoX binding region, and this is consistent with an earlier study (3). Neogenin, which shares high sequence identity with DCC in the corresponding P3 domain (Fig. 1B), binds to MyoX_MF with an affinity comparable to that of the DCC/MyoX_MF complex (Fig. S1). We further showed by analytical ultracentrifugation that MyoX_MF and DCC_P3 form a stable 1∶1 complex in solution (Fig. S2).
The Overall Structure of the MyoX_MF/DCC_P3 Complex.
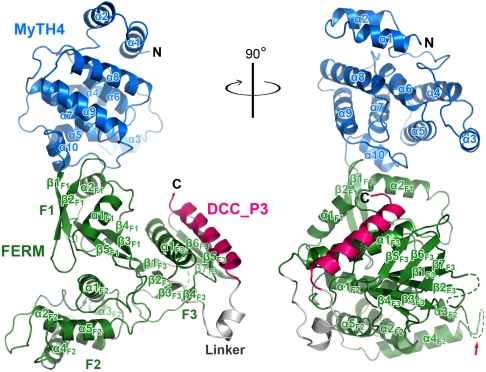
To understand the molecular basis of the MyoX/DCC interaction and the structural properties of the MyTH4–FERM tandem, we tried to solve the MyoX_MF/DCC_P3 complex structure by X-ray crystallography. To prepare stable and stoichiometric complex samples, we fused the DCC_P3 peptide (residues 1409–1445) to either the N- or C-terminal end of MyoX_MF. After extensive trials, we found that deletion of a 36-residue unstructured fragment (residues 1871–1906, Fig. S3) in the FERM domain, which had no impact on the binding between MyoX and DCC (Fig. 1D), was necessary for the crystallization of the MyoX_MF/DCC_P3 complex. We further discovered that fusion of DCC_P3 to the C-, but not to the N-, terminal end of MyoX_MF yielded high-quality crystals of the complex. We determined the 2.5-Å crystal structure of the MyoX_MF/DCC_P3 complex by the single-wavelength anomalous dispersion (SAD) method (Fig. 2 and Table S1).
Fig. 2.
Ribbon representation of the MyoX_MF/DCC_P3 complex structure. The color coding of the domains is used throughout the entire manuscript except as otherwise indicated. The linker region contains the residues of the N-terminal part of DCC_P3 (residues 1409–1421). A disordered loop in the F3 lobe and a truncated region (marked by a red arrow) in the F2 lobe are indicated by dashed lines.
The MyTH4 domain of MyoX adopts an all-helical structure containing 10 α-helices (the blue ribbons in Fig. 2). The FERM domain of MyoX is composed of three lobes: F1, F2, and F3 (the green ribbons in Fig. 2). The three lobes of MyoX FERM form a cloverleaf-like structure seen in other FERM domains (e.g., the FERM domains in moesin, merlin, radixin, ezrin, talin, protein 4.1, and FAK) (18). In the complex, DCC_P3 binds to the F3 lobe of the FERM domain forming an elongated α-helix (Fig. 2). The MyTH4 and FERM domains physically contact each other via a set of highly conserved polar and charged residues, suggesting that the interdomain interaction is very specific (see below for details).
Structure of the MyoX MyTH4 Domain.
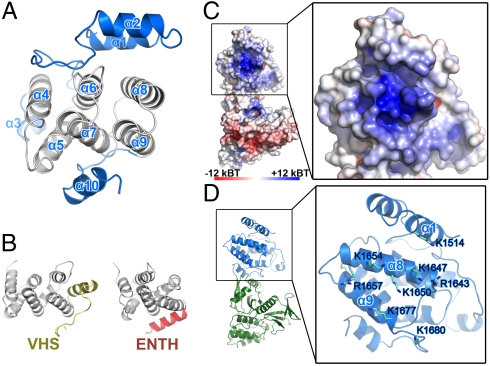
Because MyoX MyTH4 is the first MyTH4 domain structure determined to date, we performed a homologous structure search using DALI (21). The structures with high similarity scores (Z score > 7.0) all belong to the VHS and ENTH domains, although these domains barely show any sequence similarity to MyoX MyTH4 (< 10% sequence identity). MyoX MyTH4, VHS, and ENTH domains share a common six-helix bundle structure, and this six-helix bundle in MyoX MyTH4 is formed by α4-α9 (Fig. 3 A and B). However, MyoX MyTH4 has additional unique features compared with VHS and ENTH domains. First, MyoX MyTH4 contains an N-terminal helix hairpin formed by α1 and α2 and a short α3 helix (Fig. 3A). Second, although MyoX MyTH4, VHS, and ENTH domains all contains an additional helix (α10 in MyoX MyTH4) C-terminal to the six-helix bundle core, the orientation of this last helix in MyoX MyTH4 is distinct from those of VHS and ENTH domains (Fig. 3 A and B). The N-terminal helix hairpin and α10 of MyoX MyTH4 interact with the central six-helix bundle core through extensive hydrophobic interactions as well as several salt bridges (Fig. S4), suggesting that the three parts of MyoX MyTH4 together form an integral structural unit. Additionally, α3 and its N-terminal hairpin loop are also involved in the hydrophobic interactions with the six-helix bundle core (Fig. S4A).
Fig. 3.
The MyoX MyTH4 domain. (A) Ribbon diagram view of MyoX MyTH4. In this drawing, the six-helix bundle core and peripheral regions of MyoX MyTH4 are colored in gray and blue, respectively. (B) The VHS and ENTH domains each contain a similar six-helix bundle (colored in gray) core as MyoX MyTH4 does. (C) Surface analysis of MyoX MyTH4 reveals a highly positively charged surface. The figure is drawn using electrostatistic surface charge potential map. (D) Combined ribbon and stick models showing the residues forming the negatively charged surface shown in C. The molecules in C and D are drawn with the same orientation.
Analysis of the surface properties of MyoX MyTH4 reveals that the domain contains a prominently charged surface, composed of eight conserved positively charged residues in α1, α8, and α9 (Lys1514, Arg1643, Lys1647, Lys1650, Lys1654, Arg1657, Lys1677, and Lys1680; Fig. 3 C and D). This conserved charged surface of MyoX MyTH4 may interact with MyoX’s target proteins that contain complementary negatively charged surfaces (e.g., the acidic tails of tubulins) (2).
Structure of the MyoX FERM Domain.
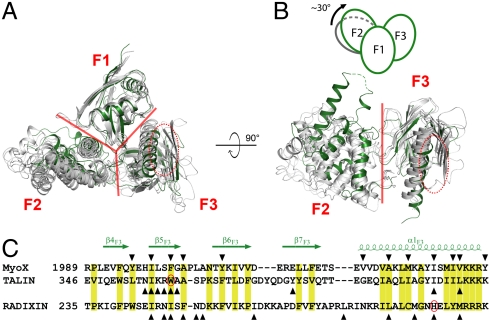
The overall folding of the individual lobes of MyoX FERM is similar to those of other FERM domains (Fig. S5) (18). However, the interlobe orientations of MyoX FERM domain are significantly different to those of other FERM domains (Fig. 4 A and B). The major interlobe orientation difference can be largely attributed to the positions of their F2 lobes. Using the F1 lobes as the reference, the orientations of the F3 lobes of the FERM domains with known structures (including MyoX FERM) are highly similar and can be superpositioned to each other well (Fig. 4 A and B). The orientation variations of the F2 lobes in all of these FERM domains, except for that of MyoX, are within a 15° range. In contrast, the F2 lobe of MyoX FERM is rotated by approximately 30° clockwise from the average position of the F2 lobes in the rest of the FERM domains (Fig. 4 A and B), possibly due to the intimate interactions between the MyTH4 domain and the F1 lobe of MyoX FERM.
Fig. 4.
Comparison of the structure of MyoX FERM with those of other FERM domains. (A and B) MyoX FERM and other FERM domains, including radixin (PDB ID code 1GC6), merlin (1H4R), protein 4.1 (2HE7), and FAK (2AEH) are overlapped by superimposing their F1 lobes. The borders between each pair of the three lobes are indicated with red lines. The MyoX FERM domain is colored in green while other FERM domains are in gray. The αβ-groove in the F3 lobe is indicated by red ovals. In B, the superimposed F1 lobes are omitted for better appreciation of the conformational differences of the F2 lobes in different FERM domains. The schematic diagram shows the approximately 30° rotation of the MyoX F2 lobe. (C) Structural based sequence alignment of the αβ-grooves from MyoX, talin, and radixin. The conserved residues are labeled with yellow boxes. The residues of which side chains are involved in their respective target interactions are marked by triangles. The two residues highlighted in Fig. 5 C and D are also circled.
FERM domains are well known for their capacities in binding to target proteins, and F3 lobes in FERM domains function as the major target binding sites (18). Two target binding grooves are known for F3 lobes. The first groove (αβ-groove) is mainly formed by β5F3 and α1F3, the second groove (ββ-groove) locates on the opposite side of the first one (22) (Fig. 4C and Fig. S5). The αβ-groove is the better characterized binding site in FERM domains including those of moesin (23), radixin (24–28), and talin (29–31). The targets of both radixin and talin, though sharing little sequence similarity, invariably bind to the αβ-groove in a β-strand or β-strand like extended structures, and the bindings are mediated by both hydrophobic interactions and H-bonds (Fig. 4C). We aligned the sequence of the F3 lobes of MyoX FERM with those of radixin and talin and found that the key hydrophobic residues in αβ-grooves of radixin and talin responsible for binding to their respective targets also exist in the αβ-grooves of MyoX (Fig. 4C), suggesting that the F3 lobe of MyoX FERM domain also uses this groove to bind to its targets such as DCC.
The Atypical MyoX/DCC Interaction.
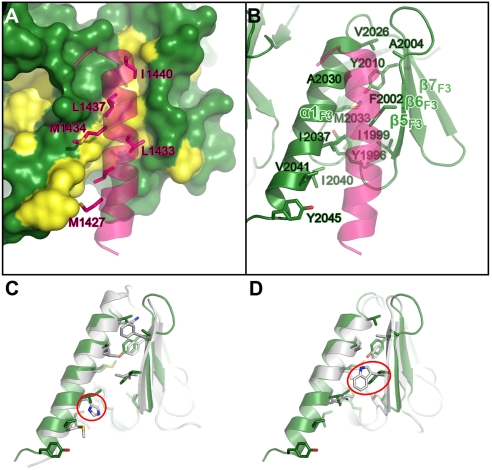
In the MyoX_MF/DCC_P3 complex, DCC_P3 indeed binds to the αβ-groove of the FERM F3 lobe (Fig. 2). To our surprise, MyoX FERM domains interact with DCC_P3 using a recognition mode distinct from those observed in other FERM domains, although both the conformations and the amino acid sequences of the αβ-grooves in these FERM domains are highly similar. In the MyoX_MF/DCC_P3 complex, DCC_P3 binds to the MyoX F3 lobe αβ-groove with an elongated α-helix (Fig. 5). The residues responsible for binding to DCC_P3 in the αβ-groove of MyoX FERM F3 lobe are all highly conserved hydrophobic residues (Fig. 5 A and B and Fig. S3). Correspondingly, the six strictly conserved hydrophobic residues in the DCC_P3 helix (Met1427, Leu1430, Leu1433, Met1434, Leu1437, and Ile1440) are directly responsible for DCC’s binding to the F3 lobe of MyoX (Figs. 1B and 5A and Fig. S6). Consistent with this structural analysis, substitutions of either Leu1433/Leu1437 in DCC with Ala or Ile2037 in MyoX with Glu abolished the binding of DCC_P3 to MyoX_MF (Fig. 1 C and D).
Fig. 5.
The hydrophobic interaction between MyoX_MF and DCC_P3. The interface residues in DCC_P3 (A) and the F3 lobe of MyoX (B) are presented as in the stick mode. The DCC_P3 binding surface of the F3 lobe is illustrated in A with the conserved hydrophobic residues colored in yellow. (C and D) The structural comparison of the αβ-grooves between MyoX and radixin (C) and between MyoX and talin (D). The F3 lobes of radixin and talin are colored in gray. The hydrophobic residues (corresponding to the DCC-binding residues in MyoX) in radixin and talin are drawn in the explicit atomic model for comparison. His288 in radixin and Trp359 in talin are highlighted by red circles.
We compared the structures and sequences of the αβ-grooves of the MyoX, radixin, and talin FERM domains and found that the target binding residues of MyoX αβ-groove are dominated by hydrophobic amino acids, and such hydrophobic residues are particularly concentrated in α1F3. In contrast, the residues in the αβ-grooves of both radixin and talin are less hydrophobic, and the target-interacting residues are concentrated in β5F3 instead of α1F3 (Fig. 4C). Further analysis of the surface residues at the αβ-grooves of MyoX and radixin/talin showed that the αβ-grooves of radixin and talin do not fit for binding to DCC_P3. In radixin, His288 in the middle of α1F3 (the corresponding position in MyoX is Ile2037) reduces the hydrophobicity of the αβ-groove (Fig. 5C). Accordingly, the αβ-groove of radixin binds to a more polar peptide fragment from ICAM-2 (24). In line with this analysis, the Ile2037Glu mutation abolished the binding of MyoX_MF to DCC_P3 (Fig. 1C). Although the αβ-groove of Talin F3 is highly hydrophobic, the bulky Trp359 at the center of α1 physically occludes targets like DCC_P3 from binding to Talin F3 (Fig. 5D). Thus, although the F3 lobes in different FERM domains share high degrees of structural similarity, the unique amino acid residue distributions in their respective αβ-grooves provide binding specificities for their respective targets. Nonetheless, we could not rule out the possibility that some other targets may also bind to the MyoX αβ-groove using β-strand structures. In fact, the F3 lobe of MyoX was reported to interact with the NPXY-motif of integrin-β3 (1), which also binds to the αβ-groove of talin with a β-strand structure (30, 31). Further structural study is required to confirm/refute this issue.
In our 2.5-Å crystal structure, several residues N-terminal to the DCC_P3 α-helix (the linker region, Fig. 2) interact with a crystallographic symmetry related MyoX_MF (Fig. S7A). This interaction is composed of two parts: The sidechain of Tyr1418DCC inserts into a small hydrophobic pocket and Glu1419DCC and Asp1421DCC form two salt bridges with Lys1514 and Lys1650 in the MyTH4 domain of the neighboring MyoX_MF molecule (Fig. S7B). Because the MF/P3 complex forms a stable monomer in solution (Fig. S2), the intermolecular interaction shown in Fig. S7 is likely to be a crystal packing artifact. Consistent with this conclusion, the substitution of Tyr1418DCC with Ala does not appreciably weaken its binding to MyoX (Fig. 1D). In another complex structure solved at the resolution of 2.7 Å, no discernable electron densities for the linker could be traced (Fig. S7 C and D) despite near identical structures solved at the two resolutions (rmsd of 1.5 Å for overall structure and of 0.5–0.7 Å for the individual domains), further confirming that the interaction between the linker region and the MyoX MyTH4 domain is an artifact introduced by the crystal packing. Nonetheless, this conserved, positively charged surface, together with the neighboring hydrophobic pocket, may be involved in binding to negatively charged target proteins such as tubulin tails (1).
The MyTH4/FERM Interaction Is Critical for MyoX’s Function.
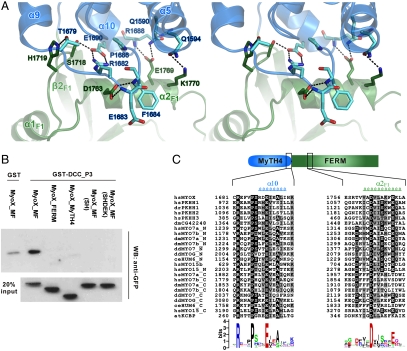
In the MyTH4–FERM tandem of MyoX, the MyTH4 and FERM domains physically contact each other to form a structural supramodule instead of two separate units. A short (four residues) and well-defined loop connects the last helix (α10) of the MyTH4 domain and the first β-strand (β1F1) of the FERM domain (Fig. S3). The MyTH4/FERM interaction is mediated by a group of residues from α5, α10, and the α9/α10-loop of MyTH4, and α2F1 and the β2F1/α1F1-loop of the FERM F1 lobe. These residues bridge the two domains together through the H-bonding interactions involving both side chains as well as main chains (Fig. 6A). The residues forming these inter-MyTH4/FERM interactions are completely conserved in MyoX (Fig. S3), suggesting that the interaction between the MyTH4 and FERM domains is highly specific and conserved in MyoX. To investigate the roles of the residues involved in the interdomain interactions, we created two mutants (termed as “SH” and “SHDEK,” respectively) in which Ser1718 and His1179 in the first mutant and Ser1718, His1719, Asp1763, Glu1769, and Lys1770 in the second mutant (Fig. 6A) were replaced by Ala. The designed mutations are expected to partially (the SH mutant) or entirely (the SHDEK mutant) disrupt the interdomain interaction between the MyTH4 and FERM domains. Interestingly, although the binding region for DCC_P3 is at the F3 lobe of MyoX FERM, both MyoX_MF mutants are not capable of binding to DCC (Fig. 6B), indicating that the MyTH4/FERM interaction is critical for maintaining the proper conformation of the F3 lobe. Likewise, neither the isolated MyTH4 domain nor the FERM domain alone showed any detectable binding to DCC (Fig. 6B). A trivial explanation of the data in Fig. 6B is that the MyoX MF mutants were unfolded and therefore these mutant proteins are not capable of binding to DCC. Because we were not able to express any of the mutant proteins in their soluble forms in bacteria, we took an alternative approach to assess the qualities of these MyoX_MF mutants expressed in mammalian cells. Each of these mutants was expressed in HEK293T cells, and soluble proteins in cell lysates (without addition of any protease inhibitors other than EDTA) were tested for their stabilities by incubating the mixtures for certain periods of time, assuming that unfolded/misfolded proteins would be degraded faster than the well-folded wild-type counterpart (Fig. S8). The MyoX_MF mutants displayed a similar stability profile when compared to the wild-type protein, indicating that the overall folding of these mutants is not grossly changed. Therefore, the loss of DCC binding of the MyoX_MF mutants shown in Fig. 6B is likely due to the compromise of the intactness of the MyTH4–FERM supramodule induced by the mutations.
Fig. 6.
The MyTH4/FERM interface of MyoX. (A) The stereo view of the MyTH4/F1 interface. The residues involved in the MyTH4/F1 interaction are displayed in the explicit stick model. Hydrogen bonds are indicated by dashed lines. (B) The interactions between GFP-tagged MyoX_MF and its mutants with DCC_P3 were assayed by GST pulldown assay. Note that weak background level interaction between GST and MyoX_MF could be detected. (C) Sequence alignment of MyTH4 domains from 24 MyTH4–FERM tandems showing the conserved residues in the MyTH4/FERM interface. As for the names of the MyTH4 domains, the first two lowercase letters represents their species (“hs” for human, “dr” for Danio rerio, “dm” for Drosophila melanogaster, “dd” for Dictyostelium discoideum, “ce” for Caenorhabditis elegans, and “at” for Arabidopsis thaliana), and the following upper cases represent the protein names, including MYO (myosin), PKHH (pleckstrin homology domain containing, family H), HUM (heavy chain of an unconventional myosin), and KCBP (kinesin-like calmodulin binding protein). For proteins containing two MyTH4–FERM tandems, the first and second MyTH4 are discriminated by suffixes (_N and _C, respectively).
Discussion
The MyTH4 domain appears in various proteins, such as several unconventional myosins including MyoVII, MyoX, and MyoXV, plekhh (pleckstrin homology domain-containing family H, also known as MAX-1) (32), as well as a plant kinesin (kinesin-like calmodulin binding protein, KCBP) (33). A common feature of these proteins is that the MyTH4 and FERM domains always appear in tandem. We hypothesize that the MyTH4 and FERM domains in each MyTH4–FERM may fold together to form an integral structural supramodule. We performed a detailed MyoX_MF structure-based sequence analysis of all identified MyTH4–FERM-containing proteins, hoping to establish a correlation for the above hypothesis. We first analyzed the sequences of the FERM domains in all MyTH4–FERM proteins and found that an Asp residue in the α2 of the F1 lobe (Asp1763 in MyoX FERM F1 lobe) is strictly conserved (Fig. 6C). In the MyoX MyTH4–FERM tandem, Asp1763 is located in the MyTH4/FERM interface and forms two strong H bonds with two main chain nitrogen atoms in the α9/α10-loop of the MyTH4 domain (Fig. 6A), pointing to a possibility that corresponding inter-MyTH4/FERM H bonds mediated by this strictly conserved Asp also exist in all other MyTH4–FERM tandems. More interestingly, sequence alignment analysis reveals that every MyTH4 domain contains three strictly conserved residues (Arg1682, Pro1686, and Glu1690 in human MyoX and termed as the RPE-motif) in α10 and the α9/α10-loop (Fig. 6C). A salt bridge formed by the Arg and Glu residues together with the Pro in the RPE-motif defines the conformation of the α9/α10-loop for its interaction with the F1 lobe (e.g., the highly conserved H bonds involving Asp1763 described above) (Fig. 6A). This above structure-based sequence analysis strongly indicates that the strictly conserved RPE-motif in MyTH4 domains and the Asp in α2F1 of FERM domains in the rest of the MyTH4-FERM tandems are likely to form the same inter-MyTH4/FERM interaction network observed in the MyoX MyTH4–FERM tandem. Supporting this notion, mutations of the absolutely conserved Arg and Pro in the RPE-motif of the MyoVII and MyoXV MyTH4 domains are known to cause hereditary deafness in human patients (34–36). Therefore, the supramodular nature of the MyTH4–FERM tandem observed in MyoX is likely a general feature for other MyTH4–FERM-containing proteins.
In conclusion, the MyoX_MF/DCC_P3 complex structure reveals the structure of a MyTH4–FERM tandem, which occurs in a number of unconventional myosins as well as several nonmyosin proteins. The complex structure uncovers a new target binding mechanism for its FERM domain. It is formally possible, although unlikely, that the deletion of the 36 amino acids from the FERM domain or the fusion of the DCC sequence could potentially cause the MyTH4-FERM structure obtained here to differ from that of the native MyTH4-FERM/DCC_P3 complex. Structure-based amino acid sequence analysis indicates that the formation of the MyTH4–FERM supramodule seen in MyoX is likely a general property of all MyTH4–FERM tandems. Thus, the structure of the MyoX MyTH4–FERM tandem described in this study provides guidance for future studies of myosin motors such as myosin VII&XV of which mutations are frequently associated with blindness and/or deafness in humans.
Materials and Methods
Protein expression, Purification, and Crystallization.
The coding sequences of MyoX_MF (residues 1503–2047) and neogenin_P3 (residues 1397–1477) were PCR amplified from a human MyoX and mouse neogenin, respectively. For MyoX_MF/DCC_P3 fusion constructs, DCC_P3 was fused to MyoX_MF at its C terminus. Fusion proteins were expressed as His6-tagged proteins and purified using Ni2+-NTA affinity chromatography. Crystals of the MyoX_MF/DCC_P3 fusion protein (10 mg/ml) were obtained by hanging drop vapor diffusion at 16 °C in approximately 8% PEG8000 and 10% glycerol in 0.1 M HEPES buffer (pH 7.5). An extended method describing protein preparation, crystallization, and structural determination can be found in SI Materials and Methods. The PDB accession code of the MyoX_MF/DCC_P3 structure is 3PZD.
GST Pulldown Assay.
Direct interactions between DCC_P3 and various MyoX MyTH4-FERM mutants were assayed in phosphate-buffered saline (pH 7.4). GST-DCC_P3 fragment (approximately 0.6 nmol each) loaded GSH-Sepharose beads were incubated with GFP-tagged MyoX_MF and its various mutants expressed in HEK293T cells. GST-DCC_P3-bound proteins were separated by SDS/PAGE. The GFP-MyoX_MF proteins were visualized by immunodetection using anti-GFP antibody.
Supplementary Material
Acknowledgments.
We thank Richard Cheney and Wencheng Xiong for providing the myosin X cDNA construct, Wencheng Xiong for the DCC and neogenin constructs, Ling-Nga Chan for helping in cell biology experiments, Yanxiang Zhao for accessing the in-house X-ray diffractor, the BL17U1 beamline of the Shanghai Synchrotron Radiation Facility for the beamline time, and Anthony Zhang for editing the manuscript. This work was supported by grants from the Research Grants Council of Hong Kong to M.Z. (HKUST663808, 664009 CA07/08.SC01, SEG_HKUST06, and AoE/B-15/01-II) and to Z.W. (HKUST662710).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.E.C. is a guest editor invited by the Editorial Board.
Data deposition: The MyoX_MF/DCC_P3 structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3PZD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016567108/-/DCSupplemental.
References
- 1.Zhang H, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 2.Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 3.Zhu XJ, et al. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat Cell Biol. 2007;9:184–192. doi: 10.1038/ncb1535. [DOI] [PubMed] [Google Scholar]
- 4.Delprat B, et al. Myosin XVa and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Hum Mol Genet. 2005;14:401–410. doi: 10.1093/hmg/ddi036. [DOI] [PubMed] [Google Scholar]
- 5.Boeda B, et al. Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adato A, et al. Interactions in the network of Usher syndrome type 1 proteins. Hum Mol Genet. 2005;14:347–356. doi: 10.1093/hmg/ddi031. [DOI] [PubMed] [Google Scholar]
- 7.Bahloul A, et al. Cadherin-23, myosin VIIa and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Hum Mol Genet. 2010;19:3557–3565. doi: 10.1093/hmg/ddq271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sousa AD, Cheney RE. Myosin-X: A molecular motor at the cell’s fingertips. Trends Cell Biol. 2005;15:533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 11.Tokuo H, Mabuchi K, Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J Cell Biol. 2007;179:229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe TM, Tokuo H, Gonda K, Higuchi H, Ikebe M. Myosin-X induces filopodia by multiple elongation mechanism. J Biol Chem. 2010;285:19605–19614. doi: 10.1074/jbc.M109.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang YS, Luo T, Xu Y, Sargent TD. Myosin-X is required for cranial neural crest cell migration in Xenopus laevis. Dev Dyn. 2009;238:2522–2529. doi: 10.1002/dvdy.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pi X, et al. Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J Cell Biol. 2007;179:1569–1582. doi: 10.1083/jcb.200704010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nie S, Kee Y, Bronner-Fraser M. Myosin-X is critical for migratory ability of Xenopus cranial neural crest cells. Dev Biol. 2009;335:132–142. doi: 10.1016/j.ydbio.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattila PK, Lappalainen P. Filopodia: Molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 18.Tepass U. FERM proteins in animal morphogenesis. Curr Opin Genet Dev. 2009;19:357–367. doi: 10.1016/j.gde.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Etournay R, et al. Shroom2, a myosin-VIIa- and actin-binding protein, directly interacts with ZO-1 at tight junctions. J Cell Sci. 2007;120:2838–2850. doi: 10.1242/jcs.002568. [DOI] [PubMed] [Google Scholar]
- 20.Ma W, et al. Phosphorylation of DCC by ERK2 is facilitated by direct docking of the receptor P1 domain to the kinase. Structure. 2010;18:1502–1511. doi: 10.1016/j.str.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 22.Terawaki S, Maesaki R, Hakoshima T. Structural basis for NHERF recognition by ERM proteins. Structure. 2006;14:777–789. doi: 10.1016/j.str.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 24.Hamada K, Shimizu T, Yonemura S, Tsukita S, Hakoshima T. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 2003;22:502–514. doi: 10.1093/emboj/cdg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terawaki S, Kitano K, Hakoshima T. Structural basis for type II membrane protein binding by ERM proteins revealed by the radixin-neutral endopeptidase 24.11 (NEP) complex. J Biol Chem. 2007;282:19854–19862. doi: 10.1074/jbc.M609232200. [DOI] [PubMed] [Google Scholar]
- 26.Takai Y, Kitano K, Terawaki S, Maesaki R, Hakoshima T. Structural basis of PSGL-1 binding to ERM proteins. Genes Cells. 2007;12:1329–1338. doi: 10.1111/j.1365-2443.2007.01137.x. [DOI] [PubMed] [Google Scholar]
- 27.Mori T, et al. Structural basis for CD44 recognition by ERM proteins. J Biol Chem. 2008;283:29602–29612. doi: 10.1074/jbc.M803606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takai Y, Kitano K, Terawaki S, Maesaki R, Hakoshima T. Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins. J Mol Biol. 2008;381:634–644. doi: 10.1016/j.jmb.2008.05.085. [DOI] [PubMed] [Google Scholar]
- 29.Wegener KL, et al. Structural basis for the interaction between the cytoplasmic domain of the hyaluronate receptor layilin and the talin F3 subdomain. J Mol Biol. 2008;382:112–126. doi: 10.1016/j.jmb.2008.06.087. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Alvarez B, et al. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 31.Wegener KL, et al. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 32.Zhong H, et al. Vertebrate MAX-1 is required for vascular patterning in zebrafish. Proc Natl Acad Sci USA. 2006;103:16800–16805. doi: 10.1073/pnas.0603959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdel-Ghany SE, Reddy AS. A novel calcium/calmodulin-regulated kinesin-like protein is highly conserved between monocots and dicots. DNA Cell Biol. 2000;19:567–578. doi: 10.1089/104454900439791. [DOI] [PubMed] [Google Scholar]
- 34.Bharadwaj AK, Kasztejna JP, Huq S, Berson EL, Dryja TP. Evaluation of the myosin VIIA gene and visual function in patients with Usher syndrome type I. Exp Eye Res. 2000;71:173–181. doi: 10.1006/exer.2000.0863. [DOI] [PubMed] [Google Scholar]
- 35.Luijendijk MW, et al. Identification and molecular modelling of a mutation in the motor head domain of myosin VIIA in a family with autosomal dominant hearing impairment (DFNA11) Hum Genet. 2004;115:149–156. doi: 10.1007/s00439-004-1137-3. [DOI] [PubMed] [Google Scholar]
- 36.Jaijo T, et al. Mutation profile of the MYO7A gene in Spanish patients with Usher syndrome type I. Hum Mutat. 2006;27:290–291. doi: 10.1002/humu.9404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.