Abstract
Through as yet undefined proteins and pathways, the SUMO-targeted ubiquitin ligase (STUbL) suppresses genomic instability by ubiquitinating SUMO conjugated proteins and driving their proteasomal destruction. Here, we identify a critical function for fission yeast STUbL in suppressing spontaneous and chemically induced topoisomerase I (Top1)–mediated DNA damage. Strikingly, cells with reduced STUbL activity are dependent on tyrosyl–DNA phosphodiesterase 1 (Tdp1). This is notable, as cells lacking Tdp1 are largely aphenotypic in the vegetative cell cycle due to the existence of alternative pathways for the removal of covalent Top1–DNA adducts (Top1cc). We further identify Rad60, a SUMO mimetic and STUbL-interacting protein, and the SUMO E3 ligase Nse2 as critical Top1cc repair factors in cells lacking Tdp1. Detection of Top1ccs using chromatin immunoprecipitation and quantitative PCR shows that they are elevated in cells lacking Tdp1 and STUbL, Rad60, or Nse2 SUMO ligase activity. These unrepaired Top1ccs are shown to cause DNA damage, hyper-recombination, and checkpoint-mediated cell cycle arrest. We further determine that Tdp1 and the nucleotide excision repair endonuclease Rad16-Swi10 initiate the major Top1cc repair pathways of fission yeast. Tdp1-based repair is the predominant activity outside S phase, likely acting on transcription-coupled Top1cc. Epistasis analyses suggest that STUbL, Rad60, and Nse2 facilitate the Rad16-Swi10 pathway, parallel to Tdp1. Collectively, these results reveal a unified role for STUbL, Rad60, and Nse2 in protecting genome stability against spontaneous Top1-mediated DNA damage.
Author Summary
The failure of cellular DNA repair mechanisms can lead to cancer, neurodegeneration, or premature aging. Although much is known about specific DNA repair mechanisms, an understanding of how these processes are critically orchestrated by post-translational modifiers such as SUMO and ubiquitin is in its infancy. We identified an intriguing family of E3 ubiquitin ligases called STUbLs that act at the interface between the SUMO and ubiquitin pathways, and through undefined proteins and pathways maintain genome stability. Here we show that dysfunction of STUbL, an associated SUMO-like protein called Rad60, or the Nse2 SUMO E3 ligase converts the normally benign topoisomerase I (Top1) activity into a genome destabilizing genotoxin. Normally, Top1 transiently introduces a break in one strand of the DNA duplex allowing DNA to unwind. However, these transient breaks are converted into recombinogenic DNA lesions when STUbL, Rad60, Nse2, and parallel pathways that we identify are compromised. This study reveals important regulatory circuits reliant on STUbL, Rad60, and Nse2 that insulate the genome from the potentially harmful effects of Top1, which may otherwise promote cancer or neurodegeneration. Furthermore, Top1 is a major chemotherapeutic target, and so our findings may aid in the development of more efficacious Top1-based therapies.
Introduction
Efficient DNA repair suppresses spontaneous genetic alterations that otherwise lead to cell death or transformation. Posttranslational modifications (PTMs) can enhance the efficiency of individual repair processes and proteins and/or channel repair through appropriate pathways (e.g. [1], [2]). Among these PTMs, the small proteins ubiquitin and SUMO have gained increasing recognition as key guardians of chromosomal integrity [1]–[3]. Related enzymatic cascades covalently attach either SUMO or ubiquitin to lysine residues within target proteins to modulate their stability, activity and localization [3]. Each cascade employs dedicated E1 activating enzymes, E2 conjugating enzymes and E3 ligases that contribute to substrate selection and transfer of the modifier from the E2 to the target protein [3]. In contrast to the numerous ubiquitin E3 ligases, there are apparently two major SUMO E3 ligases in fission yeast called Pli1 and Nse2 [4], [5].
Novel crosstalk between the SUMO and ubiquitin pathways is provided by the recently discovered SUMO-targeted ubiquitin E3 ligases (STUbLs), which ubiquitinate and thereby target SUMO-modified proteins to the proteasome for degradation [6]–[8]. Through this novel activity, STUbLs play key but largely enigmatic roles in maintaining genome stability [9]–[15]. Fission yeast STUbL was recently shown to physically interact with Nse5/6 and Rad60, providing a potential link between STUbL activity and DNA repair [12]. Nse5/6 are subunits of the Smc5/6 genome stability complex that is architecturally related to the Cohesin and Condensin complexes, but interestingly, contains the SUMO E3 ligase Nse2 [16], [17].
Mimicry of SUMO was recently discovered as a function of members of the Rad60 DNA repair protein family, which contain two SUMO-like domains (SLDs) at their C-termini [18]–[20]. We recently determined that Rad60 SLD2 mimics SUMO by interacting non-covalently with the SUMO E2 conjugating enzyme Ubc9 at the same interface bound by SUMO [21]. Disruption of the Rad60∶Ubc9 interface via a single Rad60 glutamate 380 to arginine mutation (rad60E380R) causes genome instability and phenotypes associated with dysfunction of the SUMO pathway [21]. Interestingly, rad60E380R cells, like STUbL mutant cells, are dependent on both the Holliday junction (HJ) endonuclease Mus81-Eme1 and the RecA recombinase Rhp51 (Rad51) for viability in the absence of exogenous stress [12], [21]. Given the specific role of Mus81-Eme1 in replication fork restart [22], [23], this suggests that for as yet undefined reasons, replication forks are prone to collapse in Rad60 and STUbL mutant cells. A potential source of fork collapse in these mutant cells are stalled covalent topoisomerase I (Top1)-DNA adducts that are encountered during replication [24]. In budding yeast, covalent Top1-DNA adducts called Top1 cleavage complexes (Top1cc) are efficiently removed by several repair factors acting in parallel, including tyrosyl-DNA phosphodiesterase (Tdp1; [24]–[26]). The corresponding fission yeast pathways and their relative contributions to Top1cc repair have not been defined. However, fission yeast Tdp1 was found to process Top1-independent lesions arising from oxidative stress in quiescent fission yeast [27]. In budding yeast, Tdp1 also affects Top1-independent repair processes, such as enhancing the fidelity of non-homologous end-joining by producing a 3′-phosphate at the exposed ends of DNA double strand breaks [28].
Here, we determine that STUbL, together with the physically associated DNA repair protein Rad60 and the Nse2 SUMO E3 ligase, suppresses spontaneous Top1-induced DNA damage. When STUbL, Rad60 or Nse2 functions are compromised, cells require Tdp1 to repair both spontaneous and induced Top1-dependent DNA damage, which otherwise results in genomic instability, cell cycle checkpoint activation and/or cell death. This is a striking result because Tdp1 mutant fission yeast cells are weakly sensitive to the Top1 poison camptothecin (CPT), due to redundancy with as yet unknown factors (our results and [27]). This primary finding provides mechanistic insight on how STUbL, Rad60 and Nse2 dysfunction can negatively impact genome stability. In addition, we show that Tdp1 is redundant with the fission yeast Ercc1-Xpf homologs, Rad16-Swi10, and that the absence of both pathways is lethal due to an inability to repair spontaneous Top1cc. Epistasis analysis suggests that STUbL acts in the Rad16-Swi10-initiated pathway for Top1cc repair. Furthermore, we find that Tdp1 predominates in the repair of replication-independent Top1cc lesions. Collectively, our data support a function for the evolutionarily conserved STUbL, Rad60 and Nse2 proteins in mitigating DNA damage caused by covalent Top1-DNA complexes, which arise as byproducts of normal cellular metabolism.
Results
STUbL and Tdp1 Define Parallel Pathways for the Repair of Top1cc
To probe the cause of replication fork collapse identified in STUbL mutant fission yeast [12], we constructed a double mutant between the hypomorphic STUbL allele, slx8-1, and tdp1Δ, and analyzed their sensitivity to CPT. The slx8-1 allele contains a mutation of a non-conserved cysteine residue (C218) to tyrosine, which is within the RING finger domain but is not expected to affect zinc coordination [29]. Phenotypes of slx8-1 are normally only apparent at the restrictive temperature of 35.5°C [12]. Tdp1 is an enzyme largely dedicated to the removal of stalled Top1cc [25], [26], [30]. Whereas either single mutant exhibited wild-type sensitivity, slx8-1 tdp1Δ cells were synergistically sensitive to CPT, even at the slx8-1 permissive temperature of 25°C (Figure 1A). This result indicates that STUbL and Tdp1 define parallel or non-overlapping pathways for the repair of Top1cc. Importantly, slx8-1 tdp1Δ cells were as sensitive to the replication fork stalling agent hydroxyurea (HU) as the slx8-1 single mutant (Figure 1A), indicating that the genetic interdependency of slx8-1 and tdp1Δ is specific to Top1-dependent lesions.
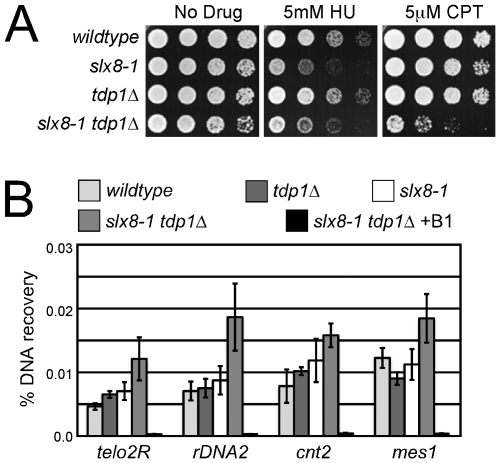
Figure 1. STUbL mutant tdp1 Δ cells are synergistically sensitive to CPT and have increased spontaneous Top1cc levels.
(A) Serial dilutions of the indicated strains were spotted onto media with or without the indicated drugs. (B) ChIP-qPCR assays of an nmt41-inducible Top1-FLAG in the indicated strains, at the subtelomeres of Chr 2 (telo2R), the centromeric inner repeats of Chr 2 (cnt2), the rDNA (rDNA2), and upstream of mes1 on Chr 1. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from four independent experiments. ChIP-qPCR data of nmt41-Top1-FLAG slx8-1 tdp1Δ cells grown in repressing media (+B1) acts as a negative control. Cells were grown at 25°C.
To distinguish between stabilization of Top1ccs or a repair defect downstream of Top1 removal in slx8-1 tdp1Δ cells, we utilized chromatin immunoprecipitation (ChIP) of Top1 in the absence of formaldehyde crosslinking, followed by quantitative PCR (qPCR) to specifically detect Top1cc. To avoid propagating the sick slx8-1 tdp1Δ cells and possible selection of suppressors, we placed Top1 under the repressible nmt41 promoter at its endogenous locus and included the FLAG epitope for ChIP analyses. With Top1 expression induced, a significant elevation of Top1cc was detected in the slx8-1 tdp1Δ double mutant at 3 out of 4 loci tested (Figure 1B). As ChIP-qPCR in the absence of formaldehyde crosslinking is a novel application for high-sensitivity detection of Top1cc, we performed key controls to verify its utility. First, the low background signal under conditions that repress Top1 expression support the specificity of the assay (Figure 1B and Figure S1A). Furthermore, to control for the contribution of non-catalytic Top1 DNA-binding to the ChIP-qPCR signal, we generated and expressed a Top1 catalytic mutant (Top1 Tyrosine 773 mutated to Phenylalanine, Top1Y773F) to compare with the otherwise identical wild-type FLAG-Top1 allele. Despite equivalent levels of protein expression, Top1Y773F generated a weak ChIP-qPCR signal, significantly below that observed for wild-type Top1 in a wild-type background (Figure S1B and S1C). Importantly, the Top1Y773F ChIP-qPCR signal did not change, irrespective of the genetic background e.g. slx8-1 tdp1Δ (Figure S1B). In addition, slx8-1 tdp1Δ top1Y773F triple mutant cells were as insensitive to CPT treatment as slx8-1 tdp1Δ top1Δ triple mutant cells, demonstrating the specificity of the slx8-1 tdp1Δ genetic interaction for Top1cc (Figure S1D and S1E). Finally, because CPT-induced Top1cc would be expected to reverse rapidly under our assay conditions, we measured Top1cc in the presence or absence of CPT. As expected for reversible CPT-induced Top1cc, we did not detect a significant difference in ChIP-qPCR signal between the CPT treated or control samples (Figure S1F). Thus, our controls confirm that in slx8-1 tdp1Δ double mutants there are increased spontaneous covalent Top1-DNA adducts and furthermore, reveal that these Top1cc are not readily reversible, unlike those induced by CPT.
Top1-Dependent DNA Damage Checkpoint Activation in slx8-1 tdp1Δ Cells
Next, we wanted to determine whether STUbL activity was also required to prevent Top1cc-dependent DNA damage during unchallenged growth in Tdp1-deficient cells. Whereas single slx8-1 and tdp1Δ mutants exhibit near wild-type growth, the slx8-1 tdp1Δ double mutant cells are synthetically sick and highly elongated even at the permissive temperature for slx8-1 (Figure 2A). Strikingly, an slx8-1 tdp1Δ top1Δ triple mutant is wild-type in appearance, demonstrating that the sickness of slx8-1 tdp1Δ cells is Top1-mediated (Figure 2A). Consistent with the evolutionary conservation of STUbL function, expression of the human STUbL RNF4 suppresses the slx8-1 tdp1Δ phenotype to a similar extent observed with fission yeast Slx8 (Figure S2; [12]).
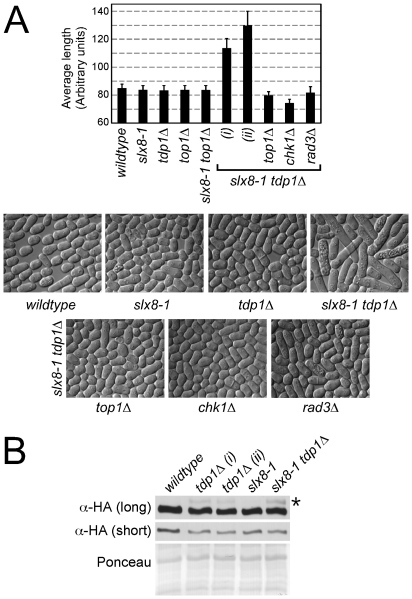
Figure 2. STUbL mutant tdp1 Δ cells activate the DNA damage checkpoint in a Top1-dependent manner.
(A) Upper: graph depicting the average cell length of the indicated strains grown in liquid media at 25°C. The error bars represent 95% confidence intervals between three independent experiments. Lower: representative images of the indicated genotypes are shown. (B) Western blot analysis of whole cell lysates from the indicated strains expressing HA-tagged Chk1 from the endogenous locus (long and short exposures are shown). The uppermost bands (asterisk) in the top panel are phosphorylated Chk1. Ponceau is shown as a loading control.
The elongated phenotype of slx8-1 tdp1Δ cells is reminiscent of fission yeast following exposure to genotoxic agents, activation of the DNA damage checkpoint and consequent delay in cell cycle progression [31]. We therefore assessed checkpoint activation in slx8-1 tdp1Δ cells by monitoring Chk1 phosphorylation and also, abrogated the checkpoint by deleting Chk1. Western analysis of Chk1 revealed no detectable checkpoint activation in slx8-1 single mutant cells, low level activation in tdp1Δ cells and an apparent increase in slx8-1 tdp1Δ double mutant cells (Figure 2B). Deleting Chk1 in slx8-1 tdp1Δ cells dramatically reduced their length, similar to the effect of deleting Top1 (Figure 2A). Deletion of an upstream activator of Chk1, Rad3 (ATR), in slx8-1 tdp1Δ cells also suppressed cell elongation (Figure 2A). The highly elongated cell phenotype of slx8-1 tdp1Δ cells is thus due to activation of the DNA damage checkpoint, indicating the presence of spontaneous DNA lesions in these cells.
As STUbL degrades certain SUMO conjugated proteins and the slx8-1 tdp1Δ phenotype is Top1-dependent, we tested whether STUbL regulates Top1 itself. We first determined that Top1 is sumoylated in a manner dependent on the major E3 SUMO ligase Pli1 (Figure S3). We then assayed Top1 sumoylation in slx8-1, tdp1Δ and slx8-1 tdp1Δ cells versus wild-type at the permissive temperature, or in slx8-1 cells at the restrictive temperature. No detectable change in the sumoylation status of Top1 was observed under any of the conditions tested (Figure S3). However, we cannot exclude deregulation of a minor fraction of sumoylated Top1, such as that on chromatin in the form of Top1cc.
Spontaneous Top1-Dependent DNA Damage in slx8-1 tdp1Δ Cells
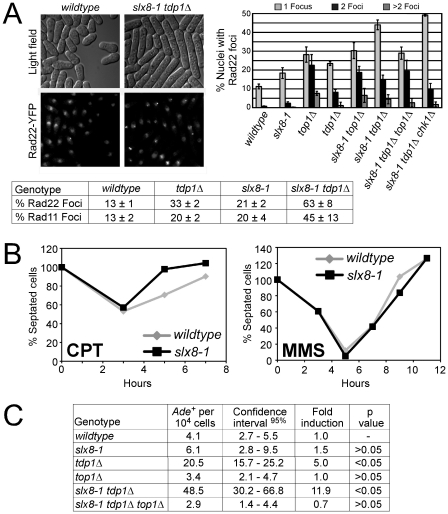
To assay the presence of DNA damage in slx8-1 tdp1Δ cells, we analyzed Rad52 (S. pombe Rad22) and RPA (S. pombe Rad11) DNA repair foci by live cell fluorescence microscopy in strains that either express YFP-tagged Rad22 or Rad11 from their endogenous loci. Compared to wild-type, both slx8-1 and tdp1Δ exhibited elevated levels of repair foci (Figure 3A). Strikingly, greater than 60% of slx8-1 tdp1Δ double mutant cells contained Rad22-YFP foci (Figure 3A). As activation of the G2 DNA damage checkpoint in slx8-1 tdp1Δ double mutants is Top1-dependent (Figure 2A) we analyzed the effect of deleting Top1 on the observed DNA repair foci. Cells deleted for Top1 have a characteristic increase in double Rad22-YFP foci that are associated with nucleoli and likely represent the rDNA loci (Figure 3A). Double mutant slx8-1 tdp1Δ cells that exhibit a profound cell cycle delay have elevated levels of large single Rad22-YFP foci as compared to wild-type and the single mutants, which exhibit no checkpoint-dependent delay in cell cycle progression (Figure 3A). Notably, slx8-1 tdp1Δ top1Δ triple mutant cells have a similar spectrum of Rad22-YFP foci as the top1Δ single mutant and consistently, show no cell cycle delay (Figure 2A and Figure 3A). This observation suggests that the excess of large single DNA repair foci evident in slx8-1 tdp1Δ cells is responsible for checkpoint activation. Furthermore, these results demonstrate that Top1 causes physical DNA damage in slx8-1 tdp1Δ cells. The number and type of Rad22-YFP foci in slx8-1 tdp1Δ was not affected in the chk1Δ background (Figure 3A).
Figure 3. STUbL mutant tdp1 Δ cells exhibit elevated Top1-dependent DNA damage and spontaneous genomic instability.
(A) Upper left: live cell images of the indicated strains expressing Rad22-YFP (Rad52). Upper right: graph depicting the percentage of nuclei containing one, two, or multiple Rad22-YFP foci using live-cell microscopy in the indicated genotypes. Error bars represent the standard deviations from three independent experiments. Base: Table depicting the percentage of nuclei containing one or more Rad22-YFP or Rad11-YFP (RPA) foci in the indicated genotypes. Standard deviations are derived from three independent experiments. (B) Log phase cultures of slx8-1 and wildtype cells were treated with 40 µM CPT or 0.008% MMS. G2 checkpoint arrest and recovery was monitored through determining the percentage of septated cells at the indicated times. (C) Table depicting frequency of spontaneous mitotic recombination between tandem adenine heteroalleles in the indicated strains. Rates represent the mean of the means, between at least three independent assays per strain. All experiments were incubated at 25°C.
The hyper-elongated phenotype of slx8-1 tdp1Δ cells may reflect a role for Slx8 (STUbL) in normal resumption of the cell cycle following checkpoint activation. However, when challenged with the DNA damaging agents CPT or methyl methanesulfonate (MMS), slx8-1 cells showed a wild-type profile of cell cycle arrest (checkpoint activation) and release (checkpoint inactivation; Figure 3B).
Top1cc Stimulates Recombination in slx8-1 tdp1Δ Cells
When STUbL activity is attenuated and Tdp1-based repair is absent, our data indicate that spontaneously occurring Top1cc's generate DNA damage and activate the DNA structure checkpoints. Such DNA damage would be anticipated to be recombinogenic. Therefore, we measured recombination rates in the slx8-1 tdp1Δ double mutant versus the single mutants using an ade6 heteroallele system [32]. We observed a 12-fold increase in recombination in the slx8-1 tdp1Δ double mutant versus 1.5 and 5-fold for slx8-1 and tdp1Δ, respectively (Figure 3C). Notably, the increased recombination rate in the slx8-1 tdp1Δ double mutant is Top1-dependent, consistent with the Top1-dependency of excess DNA repair foci in these cells (Figure 3A, 3C). In addition, we found that slx8-1 tdp1Δ cells depend on the major HR factor Rad51 (Rhp51) for viability (Table 1). Thus, consistent with the accumulation of HR foci and elevated spontaneous recombination in slx8-1 tdp1Δ cells (Figure 3A, 3C), unrepaired Top1cc generates a recombinogenic substrate that requires Rhp51-dependent HR repair.
Table 1. Summary of genetic interactions of tdp1Δ cells.
| Strain | Genetic interaction with tdp1Δ |
| rad60E380R | Lethal* |
| slx8-1 | Synthetic sick* |
| swi10Δ | Lethal* |
| rhp51Δ | None (additive upon CPT treatment) |
| rhp51Δ slx8-1 | Synthetic lethal |
| rad32Δ | None (additive upon CPT treatment) |
| mus81Δ | None (additive upon CPT treatment) |
| rad13Δ | None |
| uve1Δ | None |
| uve1Δ rad13Δ | None |
Interactions were analyzed by tetrad dissection and/or random spore analysis. Where tested, asterisks denote phenotypes suppressed by top1Δ.
Functional Intersection of DNA Repair Protein Rad60 with STUbL
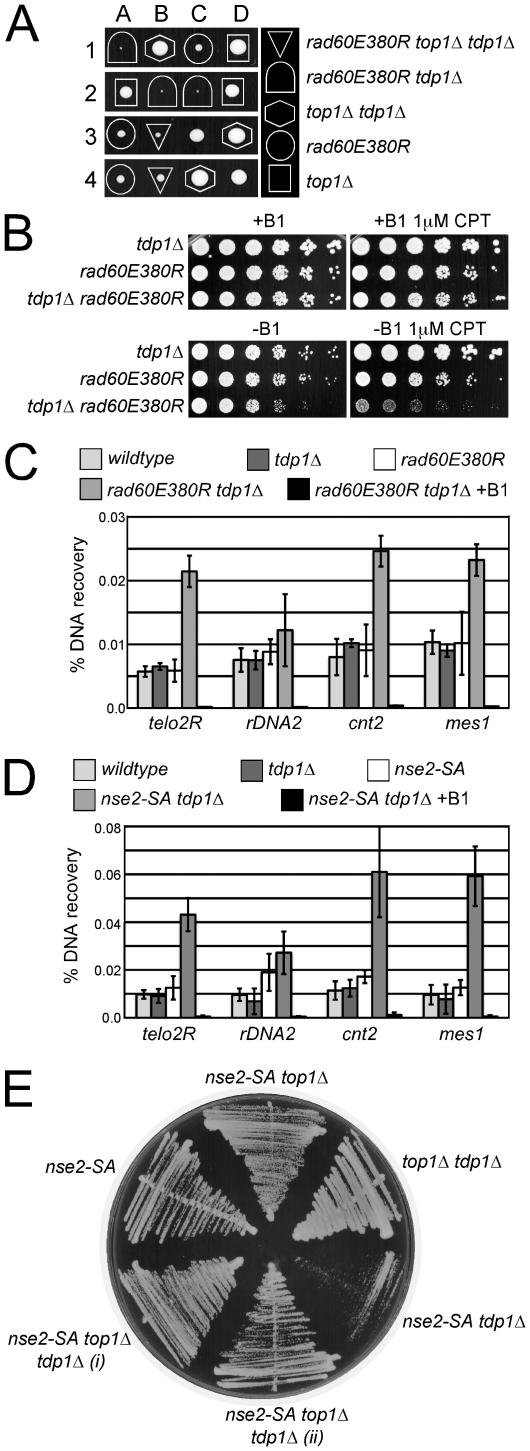
We have previously shown that Rad60 physically interacts with STUbL and shares several mutant phenotypes with the STUbL slx8-1 allele [12], [21]. In particular, a Rad60 mutant unable to interact with the SUMO E2 Ubc9, rad60E380R makes cells prone to replication fork collapse [21]. Thus, we also tested the dependency of rad60E380R cells on Tdp1, and found that the rad60E380R mutation is synthetically lethal with Tdp1 deletion (Figure 4A). Consistent with Top1cc being the major target of Tdp1, the lethality of rad60E380R tdp1Δ double mutants is suppressed by concomitant deletion of Top1 (Figure 4A). We extended this tetrad analysis using a random spore approach that allows many meiotic progeny to be analyzed in one experiment. The genotypes of more than 1000 progeny from a cross between rad60E380R top1Δ and tdp1Δ were analyzed. Importantly, the single mutants, the rad60E380R top1Δ and top1Δ tdp1Δ double mutants, and the rad60E380R tdp1Δ top1Δ triple mutant were all readily recovered. However, no rad60E380R tdp1Δ double mutants were identified indicating that rad60E380R cells require Tdp1 for viability as observed in our tetrad analysis. To further analyze this phenomenon, we used the nmt41-Top1 system to regulate Top1 levels, and constructed an nmt41-top1 rad60E380R tdp1Δ strain under conditions that repress Top1 expression. With Top1 expression repressed, rad60E380R tdp1Δ, rad60E380R and tdp1Δ strains all grew similarly in the absence or presence of a low dose of CPT (Figure 4B). Notably however, upon induction of Top1, rad60E380R tdp1Δ cells grew poorly as compared to either single mutant and exhibited synergistic hypersensitivity to CPT (Figure 4B). These data indicate that in either single mutant Top1cc repair is relatively efficient compared to the rad60E380R tdp1Δ double mutant and that the lethality of rad60E380R tdp1Δ cells is due to Top1 activity.
Figure 4. The Rad60∶Ubc9 complex and the Nse2 SUMO E3 ligase are essential to protect tdp1 Δ cells from Top1-induced DNA damage.
(A) A representative tetrad dissection is shown from a cross between rad60E380R and top1Δ tdp1Δ double mutant cells. The key depicts the genotypes present, which are denoted by various shapes placed around each colony. Wildtype cells do not have a shape placed around them. (B) Serial dilutions of the indicated strains expressing Top1 under a thiamine repressible promoter were spotted onto control or CPT containing media with (+B1) or without (-B1) thiamine to repress or induce Top1 expression, respectively. All strains were incubated at 32°C. (C) ChIP-qPCR assays of an nmt41-inducible Top1-FLAG in the indicated strains at the subtelomeres of Chr 2 (telo2R), the centromeric inner repeats of Chr 2 (cnt2), the rDNA (rDNA2), and upstream of mes1 on Chr 1. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from at least three independent experiments. ChIP-qPCR data of nmt41-Top1-FLAG rad60E380R tdp1Δ grown in repressed media (+B1) is shown as a negative control. Cells were grown at 25°C. (D) ChIP qPCR assays of the indicated strains as in (C). (E) Cells of the indicated genotype were restruck directly from tetrad dissection plates onto YES media. Cells were incubated at 32°C.
Increased Top1cc in rad60E380R tdp1Δ Double Mutant Cells
We next applied the Top1cc ChIP-qPCR assay as for slx8-1 tdp1Δ cells, and detected low levels of Top1cc at the tested loci in wild-type, rad60E380R and tdp1Δ single mutants (Figure 4C). Consistent with a defect in the processive repair of Top1cc in the rad60E380R tdp1Δ double mutant, there was a significant increase in Top1cc at 3 out of the 4 loci tested in these cells (Figure 4C). Specificity of the assay for Top1cc was again confirmed by performing ChIP-qPCR on rad60E380R tdp1Δ cells in which either the expression of Top1 was repressed, or the catalytic mutant Top1Y773F was expressed instead (Figure 4C and Figure S1B-S1D). Western analysis shows equal expression of Top1 in these strain backgrounds and the absence of detectable Top1 in the repressed control strain (Figure S1B-S1D and S1G). Thus, like STUbL, the Rad60∶Ubc9 complex constitutes a key activity in the mitigation of Top1-mediated DNA damage in a pathway distinct from that initiated by Tdp1.
Increased Top1cc in Cells Lacking Nse2 SUMO E3 Ligase Activity and Tdp1
Rad60 and STUbL both physically and functionally interact with the Smc5/6 complex, which contains the Nse2 SUMO E3 ligase [12], [19], [21]. Given the intimate association of STUbL and Rad60 function with the SUMO pathway, we tested the potential role of Nse2-dependent sumoylation in supporting Top1cc repair in tdp1Δ cells. To do this we combined the SUMO ligase defective Nse2 mutant, nse2-SA, with a Tdp1 deletion. Using ChIP-qPCR with nmt41-Top1, we detected significantly elevated Top1cc that were specific to the nse2-SA tdp1Δ double mutant background (Figure 4D and Figure S1B-S1D and S1H). Furthermore, nse2-SA tdp1Δ cells were poorly viable and their growth defects were rescued by deletion of Top1 or expression of the catalytic mutant Top1Y773F (Figure 4E and Figure S1B-S1D). We did not observe any growth defect of cells lacking both Tdp1 and the SUMO E3 ligase Pli1 (not shown). Thus, a functionally related “hub” of proteins, including STUbL, Rad60 and the SUMO E3 ligase Nse2, is required to suppress Top1-dependent DNA damage when Tdp1 activity is compromised.
Top1cc Repair Requires Either the Tdp1 or Rad16-Swi10 Pathway
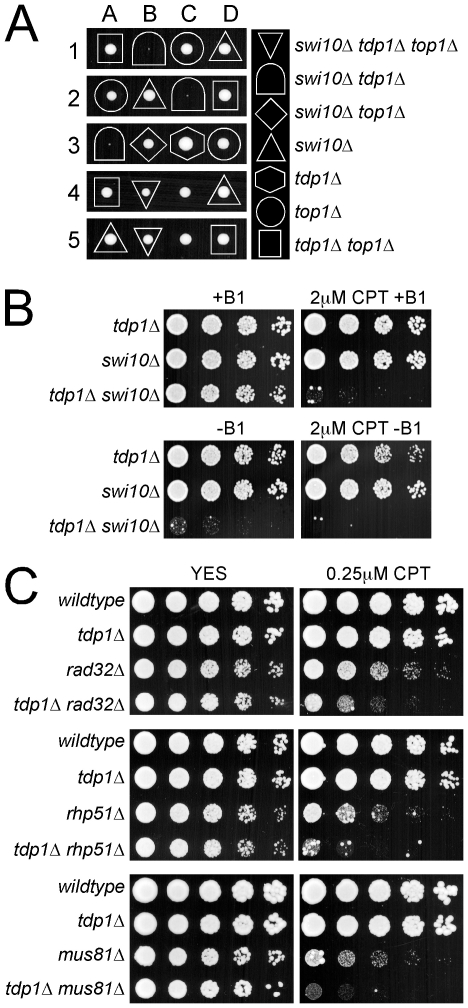
The synthetic sickness and synergistic sensitivity to CPT observed for rad60E380R or slx8-1 with tdp1Δ, indicates that these factors act in non-redundant pathways for the repair of spontaneous and induced Top1cc. In budding yeast, the Xpf-Ercc1 family endonuclease Rad1-Rad10 initiates a major pathway parallel to Tdp1 [33], [34]. We therefore tested the contribution of the fission yeast Xpf-Ercc1 family endonuclease Rad16-Swi10 to the repair of spontaneous Top1cc in the absence of Tdp1. Strikingly, tetrad analyses demonstrated that the tdp1Δ swi10Δ double mutant is inviable due to the presence of irreparable Top1-dependent lesions (Figure 5A). This function of Rad16-Swi10 is independent of its nucleotide excision repair (NER) roles as deletion of another component of NER, Rad13 (XPG) shows no genetic interaction with tdp1Δ (Table 1). Similarly, deletion of the Uve1 DNA repair endonuclease, which incises 5′ to several DNA lesions, is not synthetic sick with tdp1Δ (Table 1). Hence, the role of Rad16-Swi10 in repairing/preventing Top1-induced DNA-damage is likely attributable to its 3′-flap endonuclease activity as concluded in S. cerevisiae [33], [34]. It should be noted, in budding yeast there is apparently additional redundancy in the repair of Top1cc over fission yeast, as cells lacking both Tdp1 and the Rad16-Swi10 homologues Rad1-Rad10 are viable, but exhibit a Top1-dependent growth defect [34].
Figure 5. Tdp1-deficient cells depend on Swi10, but not on homologous recombination repair factors, to prevent Top1-induced cell death.
(A) A representative tetrad dissection is shown of a cross between swi10Δ and the top1Δ tdp1Δ double mutant. The key depicts the genotypes present, which are denoted by various shapes placed around each colony. Wildtype cells do not have a shape placed around them. (B) Serial dilutions of the indicated strains expressing Top1 under a thiamine repressible promoter were spotted onto control or CPT containing media with (+B1) or without (-B1) thiamine to repress or induce Top1 expression, respectively. (C) Five fold serial dilutions of the indicated strains were spotted onto rich media that contained either no drug (YES), or camptothecin (CPT). Plates were incubated at 32°C.
To examine the parallel functions of Tdp1 and Rad16-Swi10 in fission yeast, we employed our nmt41-Top1-Flag system to generate a viable tdp1Δ swi10Δ double mutant. When Top1 expression was repressed, the tdp1Δ swi10Δ double mutant grew slightly slower than either single mutant, likely due to the inability to completely shut off the nmt41 promoter (Figure 5B). Strikingly, even under Top1-repressed conditions the tdp1Δ swi10Δ double mutant was exquisitely sensitive to CPT, whereas the growth of either single mutant was unaffected (Figure 5B, upper panels). Furthermore, induction of Top1 expression in the absence or presence of CPT rapidly killed the tdp1Δ swi10Δ double mutant, but neither single mutant (Figure 5B, lower panels). Consistently, elevated Top1cc were detected by ChIP-qPCR in the swi10Δ tdp1Δ double mutant versus the single mutants (Figure S4A and S4B). As expected, the toxicity of Top1 in tdp1Δ swi10Δ cells depends on Top1 catalytic activity, as the double mutant is refractory to expression of the Top1Y773F mutant (Figure S4C). Collectively, these data indicate that Tdp1 and Rad16-Swi10 define the predominant pathways for the initiation of the repair of spontaneous and induced Top1cc.
The fission yeast Rad32 (Mre11)-Rad50-Nbs1 (MRN) complex, which is a central HR factor, has also been implicated in the direct removal of Top1cc [35]. In light of the finding that Tdp1 and Rad16-Swi10 define the essential parallel pathways for Top1cc removal, we believe it is likely that MRN functions mainly downstream of Top1cc removal in its well-defined HR role. Consistent with this hypothesis and distinct from the synthetic lethality/sickness of rad60E380R, slx8-1, nse2-SA or swi10Δ in combination with tdp1Δ, the rad32Δ tdp1Δ double mutant grows comparably to the rad32Δ single mutant (Table 1 and Figure 5C). Similarly, the rhp51Δ tdp1Δ and mus81Δ tdp1Δ double mutants grow as well as the rhp51Δ and mus81Δ single mutants, respectively (Table 1 and Figure 5C). Thus, HR factors including MRN are not essential for the response to spontaneous Top1cc in tdp1Δ cells. We also tested the CPT sensitivity of rad32Δ tdp1Δ, rhp51Δ tdp1Δ and mus81Δ tdp1Δ, which all exhibited a similar degree of additivity over the respective single HR mutants (Figure 5C). These data are consistent with partially non-overlapping roles of Tdp1 and the HR machinery in CPT-induced Top1cc repair. This could indicate that either Rad51, Mus81 and Rad32 can directly remove Top1cc, or as we suggest, the delayed removal of Top1cc in tdp1Δ cells causes fork collapse, which then engages the HR-dependent replication restart pathway [22].
Tdp1 Pathway Protects Cells from Replication-Independent Top1cc Lesions
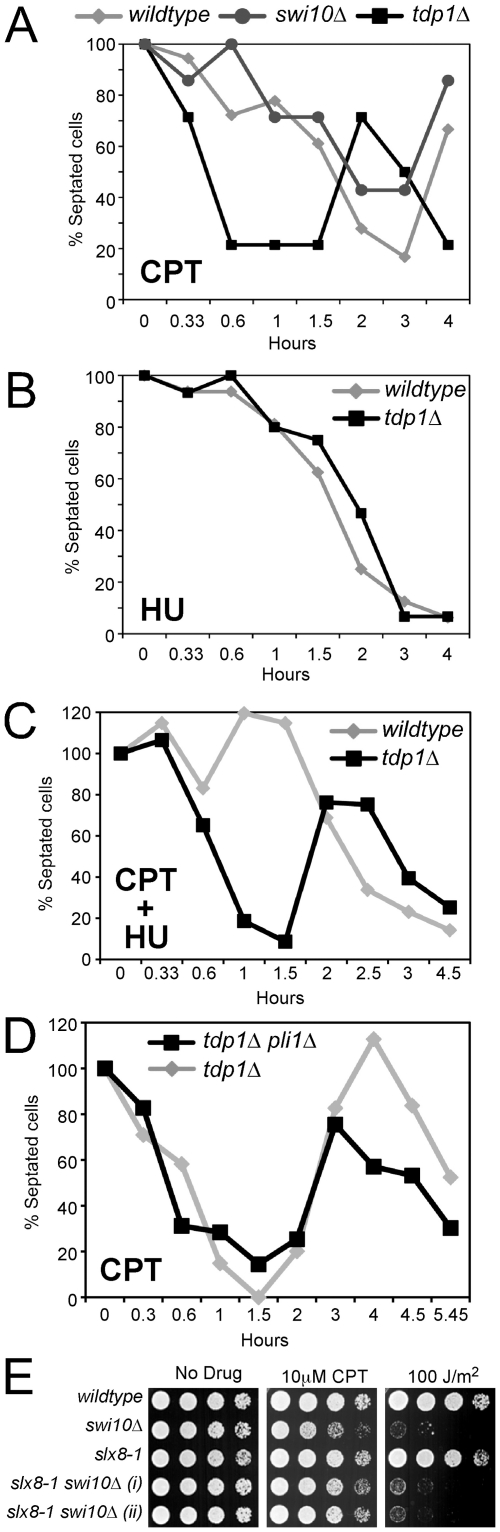
The preferred pathways for the removal of Top1cc during either transcription or replication are poorly defined [30]. In wild-type fission yeast, CPT induces cell cycle arrest at the G2/M boundary in a replication-dependent manner [36]. This data indicates that the generation of DNA damage checkpoint-visible lesions requires replication forks to collide with Top1ccs [30]. Interestingly, while testing the acute response of cells lacking either Tdp1 or the Rad16-Swi10 pathways to CPT treatment, we detected a striking difference in growth inhibition of each mutant. The addition of CPT to asynchronous cultures resulted in a rapid growth arrest of tdp1Δ but not wild-type or swi10Δ cells (Figure 6A). This phenomenon was dependent on both Top1 and the G2/M checkpoint kinase Chk1 (not shown). Both wild-type and swi10Δ cells arrested with kinetics consistent with passage through S phase and arrest in the subsequent G2 phase (Figure 6A). Although the growth of unchallenged tdp1Δ cells is indistinguishable from wild-type (our unpublished observations and [27]), we considered the possibility that tdp1Δ cells may exhibit delayed completion of S phase and thus, addition of CPT would cause first cycle replication-coupled DNA damage and G2/M checkpoint activation. Therefore, we treated asynchronous tdp1Δ and wild-type cells with the replication-blocking agent hydroxyurea (HU) and monitored growth. As anticipated, tdp1Δ and wild-type cells arrested with the same kinetics (Figure 6B), which is consistent with the known requirement for passage into S phase for the action of HU in fission yeast [37]. To confirm that Tdp1 mediates replication-independent Top1cc DNA damage repair, we co-treated wild-type and tdp1Δ cells with both HU and CPT and scored growth. Again, tdp1Δ but not wild-type cells arrested first-cycle, demonstrating that this is a replication-independent phenomenon (Figure 6C). Top1 is a known SUMO substrate in several species; however, the role of SUMO modification is unknown. As described earlier, Top1 is SUMO-modified in a manner solely dependent on Pli1 (Figure S3). It has been suggested that sumoylation of Top1 might be a repair response [38], or required for efficient Top1cc formation [39]. Either of these responses might modulate the rapid arrest kinetics of tdp1Δ cells treated with CPT. However, pli1Δ tdp1Δ double mutant cells that lack Top1 sumoylation arrest with the same rapid kinetics of tdp1Δ cells (Figure 6D), whereas pli1Δ cells arrest with wild-type kinetics (data not shown). Overall, these data reveal a previously undefined dominant role for fission yeast Tdp1 in suppressing replication-independent Top1-induced DNA damage.
Figure 6. STUbL facilitates Top1cc repair in the Rad16-Swi10 pathway.
(A) Log phase cultures of the indicated strains were treated with 40 µM CPT and checkpoint arrest and recovery was monitored through determining the percentage of septated cells at the indicated times. (B) As for (A), except cultures were treated with 15 mM HU. (C) As for (B), except cultures were co-treated with 15 mM HU and 40 µM CPT. (D) The indicated strains were treated with CPT and checkpoint arrest and recovery were monitored as in (A-C). In all of these graphs, the asynchronous septation index was set at 100% for that observed at time zero in each strain studied. (E) The indicated strains were serially diluted and spotted on drug-free or CPT rich media, or were UV-irradiated at the indicated dose, and incubated at 25°C.
A Potential Role for STUbL in Rad16-Swi10 Initiated Top1cc Repair
Given that STUbL is critical in the absence of Tdp1, we tested whether it acts in the Rad16-Swi10 initiated pathway by generating an slx8-1 swi10Δ double mutant and comparing it to either single mutant. In stark contrast to slx8-1 tdp1Δ, the slx8-1 swi10Δ double mutant did not exhibit synthetic sickness and was no more sensitive to CPT than the swi10Δ single mutant (Figure 6E). In keeping with a key role in nucleotide excision repair, Swi10 mutant cells were hypersensitive to UV irradiation, whereas slx8-1 cells were insensitive to this agent as expected (Figure 6E). The absence of synergistic CPT sensitivity in slx8-1 swi10Δ double mutant cells, coupled with the fact that Tdp1 and Rad16-Swi10 initiate the critical Top1cc repair pathways, is consistent with STUbL facilitating the Rad16-Swi10-dependent pathway. Due to the sickness of swi10Δ tdp1Δ, it was not possible to generate a triple mutant with slx8-1 to perform additional confirmatory epistasis analyses. Further supporting their overlapping functions parallel to those of Tdp1, both slx8-1 and swi10Δ cells arrest with similar delayed (wild-type) kinetics in response to CPT treatment (Figure 3B and Figure 6A).
Discussion
Suppression and efficient repair of spontaneous DNA damage is crucial to limit genetic changes that can cause cell death, transformation, or accelerate the aging process. Understanding the molecular basis for these defenses is thus vital to improve our current models of disease and aid novel chemotherapeutic strategies. Our collective results show that STUbL, the STUbL-interacting Rad60∶Ubc9 complex, and the Nse2 SUMO E3 ligase are critical for responding to spontaneous Top1cc-mediated DNA lesions.
Detection of Top1cc in tdp1Δ cells that are also hypomorphic for STUbL, Rad60 or Nse2 by ChIP-qPCR reveals important information about the nature of these lesions. Top1cc induced by CPT are normally readily reversible upon drug removal, due to completion of the Top1 catalytic cycle [24]. This raises the question: why are the Top1cc we detect in the absence of crosslinking or denaturing conditions stable? The answer likely lies in the propensity for Top1 to become irreversibly trapped at lesions in DNA, such as nicks or larger gaps, which are potentially common due to failed or stalled base excision repair (BER) [40], [41]. In addition to our ChIP-qPCR data, our genetic analyses provide strong support for the formation of spontaneous and intrinsically stable Top1ccs. For example, deleting Top1 or mutating the Top1 catalytic site suppresses the synthetic lethality of tdp1Δ and swi10Δ mutants (Figure 5A and Figure S4C). Therefore, even in the absence of exogenous agents, Top1 can form stable Top1ccs that require either Tdp1 or Rad16-Swi10-mediated removal to prevent cell death. Thus, ChIP-qPCR is a valuable novel application for the identification of a subset of Top1ccs.
In contrast to budding yeast, we determined that fission yeast lacking both Tdp1 and Rad16-Swi10 (S. cerevisiae Rad1-Rad10) are inviable due to their inability to repair spontaneous Top1-dependent DNA damage. In budding yeast, Tdp1 or multiple redundant activities, including Rad1-Rad10, initiate Top1cc repair [33], [34]. Thus, the genetic dependency of fission yeast tdp1Δ cells on Rad60, STUbL and Nse2 indicates that this group of SUMO pathway regulators may facilitate Top1cc processing by Rad16-Swi10.
We showed that the human STUbL, RNF4, is able to functionally substitute the Slx8-based fission yeast STUbL in Top1cc repair. Further supporting evolutionary conservation of the STUbL-dependent Top1cc repair pathway, the strongest negative genetic interactors of tdp1Δ in budding yeast are the non-essential STUbL components slx5Δ and slx8Δ [42]. Consistent with redundancy in the processing of spontaneous Top1cc in budding yeast, and in keeping with our findings in fission yeast, Tdp1 mutants show no increased dependency on HR factors during unchallenged growth ([33], [34], [42]; Figure 5C).
The involvement of STUbL, Rad60 and Nse2 in Top1cc repair is intriguing, as each factor is functionally connected with the SUMO pathway [7], [21], [43], and human Top1 is extensively SUMO-conjugated when stalled in the cleavage complex [38], [39], [44], [45]. SUMO conjugation to Top1 may affect many processes including directing Top1 subcellular localization, initiating the repair signal or enhancing Top1cc levels [38], [39], [44], [45]. The human Top1cc is also subject to ubiquitin-dependent proteasomal degradation [46]–[48], which may allow subsequent access of DNA repair factors to the otherwise occluded Top1 active site tyrosyl-DNA linkage [49], [50]. A recent mass spectrometric study found that mammalian Top1cc was modified extensively with SUMO-2/3 and ubiquitin following CPT treatment [45]. Such modifications are consistent with SUMO-modified human Top1cc being targeted by STUbL-dependent ubiquitination prior to proteasomal degradation (see [6], [7]). Unlike human Top1, we find that fission yeast Top1 is not as extensively SUMO modified, and this sumoylation is not increased upon CPT treatment (Figure S3 and data not shown). In addition, we have not detected hyper-sumoylated Top1 in fission yeast STUbL mutants or STUbL tdp1Δ mutants (Figure S3). However, as such a small proportion of total Top1 is sumoylated, functionally important changes could be masked. Because a single unrepaired Top1cc per cell could account for the checkpoint response and death of slx8-1 tdp1Δ cells; STUbL-mediated degradation of Top1cc, stimulated by Rad60∶Ubc9 and Nse2 remains a possibility.
It should be noted that cells lacking the predominant Top1 SUMO E3 ligase Pli1 are not sensitive to CPT, whereas those lacking the Nse2 SUMO E3 ligase are hypersensitive to CPT [4], [51]. Furthermore, Rad60 and STUbL physically and functionally associate with the Smc5–6 complex, of which Nse2 is a core component [12], [18], [19]. Thus, it is conceivable that an Nse2-dependent sumoylation target other than Top1 is at the hub of this Top1cc repair network. In keeping with this, we find that nse2-SA tdp1Δ double mutants accumulate Top1cc that cause the observed severe sickness of these cells (Figure 4D and 4E). Determining whether Top1 or other potential STUbL, Rad60∶Ubc9 or Nse2 targets are responsible for the observed repair defect in tdp1Δ cells will require the development of novel genetic and proteomic approaches, which will be the focus of future endeavors.
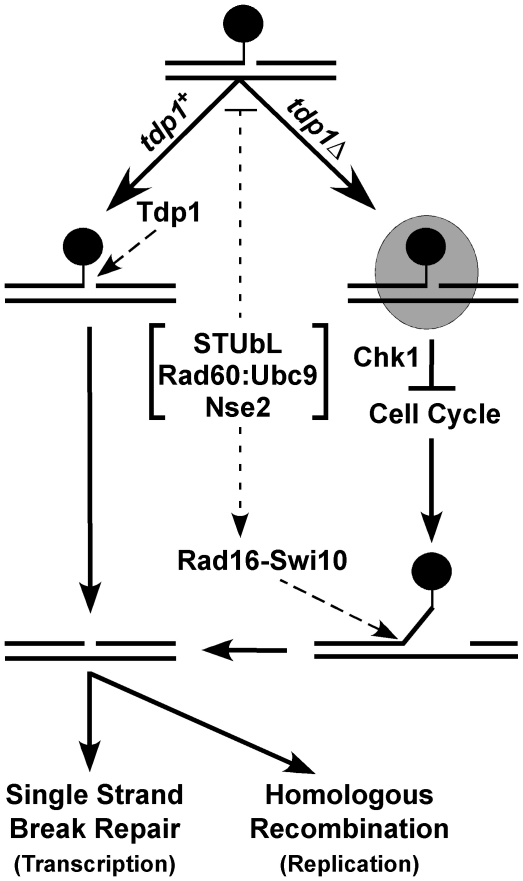
Conceptually, either Tdp1 or Rad16-Swi10 could process Top1cc encountered during transcription, initiating single strand break repair to heal the resulting lesion without the involvement of HR (Figure 7). However, if a replication fork encounters a Top1cc and collapses, HR is required to restart replication. In tdp1Δ cells, the number of lesions encountered during replication may increase due to defective Top1cc removal during transcription, and this could explain the observed additive CPT sensitivity of tdp1Δ rhp51Δ (and other HR factor) double mutants. This is supported by our data indicating that in fission yeast, Tdp1 is the predominant replication-independent Top1cc repair activity, most likely acting in a transcription-coupled manner. However, as tdp1Δ cells are dependent on Rad16-Swi10, but not HR factors for viability, Rad16-Swi10 must be able to initiate HR-independent Top1cc repair. Supporting replication-independent Tdp1 functions, Tdp1 mutation can cause degeneration of post-mitotic cells such as neurons [52]. In the absence of both Tdp1 and Rad16-Swi10, the burden of unrepaired Top1cc leads to lethality. In this scenario, neither single strand break repair nor HR can proceed. In light of the well-characterized role of Rad16-Swi10 in budding yeast (Rad1-Rad10) as a 3′ flap endonuclease, we propose that in tdp1Δ swi10Δ cells HR cannot engage due to the Top1cc blocking the DNA 3′ terminus (Figure 7). Within our model, STUbL, Rad60∶Ubc9 and Nse2 facilitate Top1cc removal by Rad16-Swi10. Our data does not exclude the possibility that STUbL, Rad60∶Ubc9 and Nse2 may also act to suppress genomic lesions that favor stable Top1cc formation. Other factors may act in the removal of Top1cc from 3′ termini such as MRN [35]; however, in light of the synthetic lethality of tdp1Δ and swi10Δ, as opposed to the observed epistasis between tdp1Δ and rad32Δ during normal growth, such contributions appear minor.
Figure 7. Model depicting the parallel actions of Tdp1 and Rad16-Swi10 in Top1cc removal, potentially facilitated by STUbL, Rad60∶Ubc9, and Nse2.
In wild-type cells, Tdp1 efficiently removes Top1cc (black circle), leaving a substrate for single strand break repair (SSBR) or homologous recombination (HR). In the absence of Tdp1, Top1cc is converted into a checkpoint visible lesion (shaded oval) that arrests cell cycle progression in a Chk1-dependent manner. Top1cc can ultimately be removed by Rad16-Swi10, a process that may be facilitated by STUbL, Rad60∶Ubc9 and Nse2. SSBR or HR can then heal the resulting lesion.
By combining genetic, physical and mutational analyses, we here identify a unifying and critical role for STUbL, Rad60∶Ubc9 and Nse2 in DNA repair. While these factors have additional non-overlapping roles (indicated by the Top1-independent lethality of the rad60E380R slx8-1 double mutant [21]), they apparently collaborate in processing potentially lethal or genome destabilizing spontaneous Top1cc lesions. High-throughput observations in budding yeast (see above) and the RNF4 rescue-experiment indicate that the pathways we have defined in fission yeast are likely to contribute to Top1cc repair in other species. Top1 is an important chemotherapeutic target but acquired resistance to chemotherapy, via up regulation of repair factors or reduction in Top1 levels, is a common cause of therapeutic failure ([24], [30], [53] and refs. therein). Thus, our data implicating the evolutionarily conserved STUbL, Nse2 and Rad60∶Ubc9 factors in Top1cc repair may aid therapeutic strategies. Currently, the presented results characterize a critical STUbL-Rad60-Nse2 DNA repair function acting parallel to Tdp1, potentially in a pathway initiated by the Xpf-Ercc1 family DNA endonuclease Rad16-Swi10. More broadly, these STUbL-Rad60-Nse2 results provide key knowledge on how cells deal with the genotoxic effects of spontaneous Top1-induced DNA damage.
Materials and Methods
General Yeast Techniques and Drug Treatments
Standard yeast methods were performed as described [54]. Top1 was N-terminally tagged at its endogenous locus using nmt41-3FLAG, as described previously [55]. The Top1 catalytic mutant was generated by site directed mutagenesis (Quikchange; Stratagene). Drugs were obtained from Sigma–Aldrich. Table S1 lists the strains used in this study.
Microscopy
Cells were either grown on solid media at 25°C for 3 days and resuspended in 1 x PBS before imaging, or cultured in supplemented minimal media to mid-log phase at 25°C, and analyzed with a Nikon Eclipse E800 microscope equipped with a Photometrics Quantix charge-coupled device camera. Images were analyzed with ImageJ (NIH, http://rsbweb.nih.gov/ij/). For cell length comparisons at least 100 cells per strain were measured. Data represents the average cell length in units defined by ImageJ with 95% confidence intervals. For indirect fluorescence at least 300 cells were analyzed for each strain. Data represents the average of three independent experiments with standard deviations.
Determination of Septation Index
Log phase cultures (OD600 of ∼0.3–0.4) were treated with 40 µM CPT, 0.008% MMS, or 15 mM HU. Septation was monitored using a Zeiss Axioscope 20. After 4 h, MMS was washed out and cells were transferred to medium without drug to allow recovery. For comparison between the slx8-1 and wild type strain a minimum of 175 cells were scored for each data point. Data represents the average of four experiments with standard deviations.
S. pombe Protein Lysate Preparation and Immunoblotting
Cultures were grown at the permissive temperature (25°C) for the slx8-1 allele to mid-log phase for all experiments (except where otherwise indicated). Top1-FLAG cells were cultured overnight in minimal media containing thiamine. Cultures were then washed and diluted into minimal media either with or without thiamine, grown for an additional 48 h and harvested. Cells were lysed in 8 M Urea, 50 mM Tris pH 8, 50 mM NaH2PO4, 300mM NaCl, and Complete Protease Inhibitors EDTA-free (Roche, IN). Chk1-HA cells were grown over night in rich media, lysed in 50 mM Tris pH 8, 150 mM NaCl, 2.5 mM EDTA, 10% glycerol, 0.2% Nonidet P-40, 90 mM NaF, Complete Protease Inhibitors EDTA-free, and 5 mM phenylmethylsulfonyl fluoride (PMSF). Protein samples were separated by SDS-PAGE on 4-20% Tris-glycine gels (Invitrogen, CA) for Top1-FLAG and 8% acrylamide gels (99%) for Chk1-HA. Immunoblotting was performed as previously described [21], [23], [56], [57].
Detection of Sumoylated Top1
Top1-TAP cells were grown over night at either 30°C, or at the permissive temperature (25°C), then cultured at the semi-permissive temperature (30°C) on day two, and shifted to the restrictive temperature (36°C) for 6 h on day three, to inactivate the slx8-1 allele. Top1-TAP cells were lysed using the buffer described above for Chk1-HA, supplemented with 60 mM N-ethyl maleimide (NEM) that lacked glycerol. Protein extracts were incubated with IgG-Sepharose beads (GE healthcare) at 4°C for 2 h, washed and separated by SDS-PAGE on 4-8% Tris-glycine gels (Expedeon, CA) followed by western blotting. For denaturing Nickel pull downs, Top1-Myc and 6His-Pmt3 expressing strains were grown overnight in rich media at the permissive temperature (25°C). Cells were lysed in 8 M Urea buffer described above for Top1-FLAG, supplemented with 60 mM NEM. Equal amounts of total protein per strain were incubated with Ni-NTA Superflow beads (Qiagen) for 1.5 hrs rotating at room temperature. Beads were washed and analyzed by SDS-PAGE on 4–8% Tris-glycine gels followed by anti-Myc western blotting.
Chromatin Immunoprecipitation
ChIP experiments were essentially performed as published [57] with minor modifications. In order to capture covalent Top1-DNA complexes, cells were not treated with formaldehyde. 4.5×108 cells were lysed by bead beating in buffer L (50 mM HEPES KOH pH 7.4, 140 mM NaCl, 1 mM EDTA, 0.1% Triton, 0.1% Na-Deoxycholate, Complete Protease Inhibitors EDTA-free, and 5 mM PMSF). After bead beating the Triton-X concentration of the buffer was brought to 1%. DNA was sonicated to 500–800 bp using the Sonicator 3000 (Misonix, NY) equipped with a cup horn for 3×20 seconds at power level 10, in 1 minute intervals. Protein extracts were normalized to the lowest protein concentration (1–2 mgs final) between strains. Lysates were then incubated with Protein-G Dynabeads (Invitrogen, CA) pre-bound with FLAG antibodies (M2, F1804, Sigma). Following a 2 h pull-down at 4°C, immunocomplexes were washed 3 times in buffer L (1% Triton-X), 2 times in Buffer H (buffer L with 500 mM NaCl), 2 times in buffer D (10 mM Tris-HCl pH 8, 250 mM LiCl, 1 mM EDTA, 0.5% NP40, 0.5% Na-deoxycholate), and one time in 10 mM Tris-HCl pH 8, 1 mM EDTA. The DNA was eluted off the Dynabeads by heating in 10 mM Tris-HCl pH 8, 10 mM EDTA, at 70°C for 15 minutes. The protein was digested with 0.5 mg/ml final concentration Proteinase K (Invitrogen, CA) for 2 h at 50°C. The DNA was purified using the PureLink PCR purification Kit (Invitrogen, CA). For each experiment, the percentage DNA recovery of ChIP samples relative to the DNA amount in the input was averaged over triplicate qPCR measurements. Data represents the average of at least three independent experiments with standard deviations. Primer sequences for cnt2, telo2R, mes1 and rDNA2 have been published [58], [59].
Determination of spontaneous recombination rates
Spontaneous mitotic recombination rates between Adenine heteroalleles were determined by fluctuation tests as described [32], using the ade6-L469/pUC8/ura4+/ade6-M375 heteroallele system [60]. For each assay, four independent colonies were analyzed. Each assay was repeated independently at least three times. Assays were performed at 25°C.
Supporting Information
Top1-FLAG expression and functional analysis of strains used for ChIP-qPCR assays. (A) Western blot analysis of whole cell lysates from the indicated strains immunoblotted with anti-FLAG antiserum. The strains were cultured in supplemented minimal media that either induced (-B1), or repressed (+B1) nmt41-Top1-FLAG expression. Cells were induced at 25°C for 48 hours prior to harvesting. Top1-FLAG is similarly expressed in wt, slx8-1, and slx8-1 tdp1Δ strains. (B) ChIP-qPCR analysis of the indicated strains expressing either nmt41-Top1-FLAG (wildtype) or a catalytically inactive nmt41-top1Y773F-FLAG mutant. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from three independent experiments when nmt41-Top1-FLAG is induced. Cells were induced at 25°C for 48 hours prior to harvesting. (C) Western blot analysis of whole cell lysates from the indicated strains immunoblotted with anti-FLAG antiserum. The strains used for the ChIP analysis described in (B) were analysed; each strain shows similar expression of the top1Y773F construct compared to wildtype Top1-FLAG. (D) Five fold serial dilutions of the indicated strains were spotted onto media that induced expression of the nmt constructs, which contained either no drug, or 150 µM CPT. Plates were incubated at 32°C. (E) Five fold serial dilutions of the indicated strains were spotted onto media that induced expression of the nmt constructs, which contained either no drug, or the indicated concentration of CPT. Plates were incubated at 25°C. (F) ChIP-qPCR analysis of Top1-FLAG expressing wildtype cells, either treated or not with 50 µM CPT for 1 hour prior to harvesting. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from three independent experiments when nmt41-Top1-FLAG was induced. Cells were induced at 25°C for 48 hours prior CPT treatment and harvesting. (G) and (H) Top1-FLAG expression analysis as described in (A) repeated for the indicated strains, except in (G) an untagged wildtype control is also shown.
(1.08 MB TIF)
Expression of pSlx8 and Human pRNF4 suppresses the slx8-1 tdp1Δ phenotype. The slx8-1 tdp1Δdouble mutants were transformed with vectors expressing Slx8, human RNF4, or an empty Glutathione-S-transferase (GST) control, and then grown at 25°C, either in inducing conditions (supplemented minimal media without thiamine: left panels) or in rich media to repress expression and allow vector loss (YES: right panels). Cells were cultured for 20 hours until reaching mid-log phase before photographing. The elongated phenotype of slx8-1 tdp1Δ is rescued when pSlx8 and pRNF4 are present, and induced.
(3.59 MB TIF)
Pli1-dependent Top1 SUMO modification appears unchanged in the slx8-1 background. (A) Representative anti-TAP western blots are shown following IgG pulldowns from the indicated strains expressing TAP-tagged Top1 from the endogenous locus. The sumoylated forms of Top1-TAP are indicated by asterisks. Left: expression of a vector containing GFP-tagged Pmt3 retards gel migration of the Top1-SUMO species by the amount expected for a monosumoylated Top1-SUMO-GFP. Center: Top1 monosumoylation is undetectable in pli1Δ cells. Right: Top1 monosumoylation appears unaffected by STUbL inactivation. All strains in this experiment were incubated at the restrictive temperature for slx8-1 (36°C). (B) Western blot analysis of whole cell lysates from the indicated strains expressing Top1-Myc ± 6His-Pmt3 from their endogenous loci, after denaturing Ni-NTA pulldowns (long and short exposures are shown). The monosumoylated species of Top1 is indicated (Top1-Pmt3). All experiments were performed at 25°C.
(3.01 MB TIF)
swi10Δ tdp1Δ have elevated levels of Top1ccs but are not sensitive to oxidative damage caused by H2O2. (A) ChIP-qPCR assay of the indicated strains. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from at least three independent experiments when nmt41-Top1-FLAG is induced. ChIP-qPCR data of nmt41-Top1-FLAG swi10Δ tdp1Δ grown in repressed media (+B1) is shown as a negative control. Cells were induced at 25°C for 48 hours prior to harvesting. (B) Anti-FLAG western blot analysis of the strains used for the ChIP analysis shown in (A). (C) Cells of the indicated genotype were restruck onto plates that either repressed (+B1), or induced expression of the nmt promoter in the presence or absence of the indicated concentration of CPT. Cells were cultured at 32°C. (D) Five fold serial dilutions of the indicated strains were spotted onto media, which either contained no drug, H202, or camptothecin (CPT) at the indicated concentrations. Plates were incubated at 32°C. No profound sensitivity of swi10Δ tdp1Δ top1Δ cells as compared to wild-type or spc1Δ cells was observed. This was anticipated due to the known robust and stress-inducible catalase activity in fission yeast, and the key role of Apn2 phosphodiesterase in processing the 3′ blocked end generated by Nth1-dependent AP site incision e.g. [Hida Y., Ikeda S. (2008) Base excision repair of oxidative DNA damage in a catalase-deficient mutant of schizosaccharomyces pombe. Genes and Environment 30, 86–91. Kanamitsu K., Ikeda S. (2010) Early Steps in the DNA Base Excision Repair Pathway of a Fission Yeast Schizosaccharomyces pombe. J Nucleic Acids 2010].
(5.73 MB TIF)
S. pombe strains used during this study.
(0.08 MB DOC)
Acknowledgments
We thank Eishi Noguchi for plasmids. We also thank The Scripps Cell Cycle Group for support and encouragement and Paul Russell (TSRI) for yeast strains and use of the qPCR cycler.
Footnotes
The authors have declared that no competing interests exist.
This study was funded by the National Institutes of Health (http://www.nih.gov/) grants GM068608 and GM081840 awarded to MNB, who is supported by a Scholar Award from The Leukemia & Lymphoma Society (http://www.leukemia-lymphoma.org/hm_lls). JH is supported by a predoctoral fellowship from Boehringer Ingelheim Fonds (http://www.bifonds.de/cgi-bin/index.pl). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schleker T, Nagai S, Gasser SM. Posttranslational modifications of repair factors and histones in the cellular response to stalled replication forks. DNA Repair (Amst) 2009;8:1089–1100. doi: 10.1016/j.dnarep.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich HD. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 2005;15:525–532. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Watts FZ, Skilton A, Ho JC, Boyd LK, Trickey MA, et al. The role of Schizosaccharomyces pombe SUMO ligases in genome stability. Biochem Soc Trans. 2007;35:1379–1384. doi: 10.1042/BST0351379. [DOI] [PubMed] [Google Scholar]
- 5.Xhemalce B, Riising EM, Baumann P, Dejean A, Arcangioli B, et al. Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc Natl Acad Sci U S A. 2007;104:893–898. doi: 10.1073/pnas.0605442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 7.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Hunter T, Sun H. Crosstalk between the SUMO and ubiquitin pathways. Ernst Schering Found Symp Proc. 2008:1–16. doi: 10.1007/2789_2008_098. [DOI] [PubMed] [Google Scholar]
- 9.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Roberts TM, Yang J, Desai R, Brown GW. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KM, O'Connell MJ. A new SUMO ligase in the DNA damage response. DNA Repair (Amst) 2006;5:138–141. doi: 10.1016/j.dnarep.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, 3rd, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boddy MN, Shanahan P, McDonald WH, Lopez-Girona A, Noguchi E, et al. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol Cell Biol. 2003;23:5939–5946. doi: 10.1128/MCB.23.16.5939-5946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morishita T, Tsutsui Y, Iwasaki H, Shinagawa H. The Schizosaccharomyces pombe rad60 Gene Is Essential for Repairing Double-Strand DNA Breaks Spontaneously Occurring during Replication and Induced by DNA-Damaging Agents. Mol Cell Biol. 2002;22:3537–3548. doi: 10.1128/MCB.22.10.3537-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics. 2005;6:22. doi: 10.1186/1471-2105-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN. Molecular mimicry of SUMO promotes DNA repair. Nat Struct Mol Biol. 2009;16:509–516. doi: 10.1038/nsmb.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, et al. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, et al. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 25.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 26.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 27.Ben Hassine S, Arcangioli B. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J. 2009;28:632–640. doi: 10.1038/emboj.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahmed K, Nitiss KC, Nitiss JL. Yeast Tdp1 regulates the fidelity of nonhomologous end joining. Proc Natl Acad Sci U S A. 2010;107:4057–4062. doi: 10.1073/pnas.0909917107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 30.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhind N, Russell P. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113(Pt 22):3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osman F, Adriance M, McCready S. The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr Genet. 2000;38:113–125. doi: 10.1007/s002940000145. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc Natl Acad Sci U S A. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci U S A. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan S, Capasso H, Walworth NC. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast. 1999;15:821–828. doi: 10.1002/(SICI)1097-0061(199907)15:10A<821::AID-YEA422>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 37.Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 38.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci U S A. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horie K, Tomida A, Sugimoto Y, Yasugi T, Yoshikawa H, et al. SUMO-1 conjugation to intact DNA topoisomerase I amplifies cleavable complex formation induced by camptothecin. Oncogene. 2002;21:7913–7922. doi: 10.1038/sj.onc.1205917. [DOI] [PubMed] [Google Scholar]
- 40.Lebedeva N, Auffret Vander Kemp P, Bjornsti MA, Lavrik O, Boiteux S. Trapping of DNA topoisomerase I on nick-containing DNA in cell free extracts of Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:799–809. doi: 10.1016/j.dnarep.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Lebedeva N, Rechkunova N, Boiteux S, Lavrik O. Trapping of human DNA topoisomerase I by DNA structures mimicking intermediates of DNA repair. IUBMB Life. 2008;60:130–134. doi: 10.1002/iub.5. [DOI] [PubMed] [Google Scholar]
- 42.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 43.Potts PR. The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst) 2009;8:499–506. doi: 10.1016/j.dnarep.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Desai SD, Mao Y, Sun M, Li TK, Wu J, et al. Ubiquitin, SUMO-1, and UCRP in camptothecin sensitivity and resistance. Ann N Y Acad Sci. 2000;922:306–308. doi: 10.1111/j.1749-6632.2000.tb07050.x. [DOI] [PubMed] [Google Scholar]
- 45.Kanagasabai R, Liu S, Salama S, Yamasaki EF, Zhang L, et al. Ubiquitin-family modifications of topoisomerase I in camptothecin-treated human breast cancer cells. Biochemistry. 2009;48:3176–3185. doi: 10.1021/bi802179t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001;61:5926–5932. [PubMed] [Google Scholar]
- 47.Desai SD, Liu LF, Vazquez-Abad D, D'Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 48.Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. J Biol Chem. 2008;283:21074–21083. doi: 10.1074/jbc.M803493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J Biol Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debethune L, Kohlhagen G, Grandas A, Pommier Y. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, et al. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Zhou S, Begum S, Sidransky D, Westra WH, et al. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer. Lung Cancer. 2007;55:303–311. doi: 10.1016/j.lungcan.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 55.Noguchi C, Garabedian MV, Malik M, Noguchi E. A vector system for genomic FLAG epitope-tagging in Schizosaccharomyces pombe. Biotechnol J. 2008;3:1280–1285. doi: 10.1002/biot.200800140. [DOI] [PubMed] [Google Scholar]
- 56.Pebernard S, McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell. 2004;15:4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pebernard S, Schaffer L, Campbell D, Head SR, Boddy MN. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 2008;27:3011–3023. doi: 10.1038/emboj.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, et al. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 2007;26:1327–1339. doi: 10.1038/sj.emboj.7601585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osman F, Fortunato EA, Subramani S. Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics. 1996;142:341–357. doi: 10.1093/genetics/142.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top1-FLAG expression and functional analysis of strains used for ChIP-qPCR assays. (A) Western blot analysis of whole cell lysates from the indicated strains immunoblotted with anti-FLAG antiserum. The strains were cultured in supplemented minimal media that either induced (-B1), or repressed (+B1) nmt41-Top1-FLAG expression. Cells were induced at 25°C for 48 hours prior to harvesting. Top1-FLAG is similarly expressed in wt, slx8-1, and slx8-1 tdp1Δ strains. (B) ChIP-qPCR analysis of the indicated strains expressing either nmt41-Top1-FLAG (wildtype) or a catalytically inactive nmt41-top1Y773F-FLAG mutant. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from three independent experiments when nmt41-Top1-FLAG is induced. Cells were induced at 25°C for 48 hours prior to harvesting. (C) Western blot analysis of whole cell lysates from the indicated strains immunoblotted with anti-FLAG antiserum. The strains used for the ChIP analysis described in (B) were analysed; each strain shows similar expression of the top1Y773F construct compared to wildtype Top1-FLAG. (D) Five fold serial dilutions of the indicated strains were spotted onto media that induced expression of the nmt constructs, which contained either no drug, or 150 µM CPT. Plates were incubated at 32°C. (E) Five fold serial dilutions of the indicated strains were spotted onto media that induced expression of the nmt constructs, which contained either no drug, or the indicated concentration of CPT. Plates were incubated at 25°C. (F) ChIP-qPCR analysis of Top1-FLAG expressing wildtype cells, either treated or not with 50 µM CPT for 1 hour prior to harvesting. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from three independent experiments when nmt41-Top1-FLAG was induced. Cells were induced at 25°C for 48 hours prior CPT treatment and harvesting. (G) and (H) Top1-FLAG expression analysis as described in (A) repeated for the indicated strains, except in (G) an untagged wildtype control is also shown.
(1.08 MB TIF)
Expression of pSlx8 and Human pRNF4 suppresses the slx8-1 tdp1Δ phenotype. The slx8-1 tdp1Δdouble mutants were transformed with vectors expressing Slx8, human RNF4, or an empty Glutathione-S-transferase (GST) control, and then grown at 25°C, either in inducing conditions (supplemented minimal media without thiamine: left panels) or in rich media to repress expression and allow vector loss (YES: right panels). Cells were cultured for 20 hours until reaching mid-log phase before photographing. The elongated phenotype of slx8-1 tdp1Δ is rescued when pSlx8 and pRNF4 are present, and induced.
(3.59 MB TIF)
Pli1-dependent Top1 SUMO modification appears unchanged in the slx8-1 background. (A) Representative anti-TAP western blots are shown following IgG pulldowns from the indicated strains expressing TAP-tagged Top1 from the endogenous locus. The sumoylated forms of Top1-TAP are indicated by asterisks. Left: expression of a vector containing GFP-tagged Pmt3 retards gel migration of the Top1-SUMO species by the amount expected for a monosumoylated Top1-SUMO-GFP. Center: Top1 monosumoylation is undetectable in pli1Δ cells. Right: Top1 monosumoylation appears unaffected by STUbL inactivation. All strains in this experiment were incubated at the restrictive temperature for slx8-1 (36°C). (B) Western blot analysis of whole cell lysates from the indicated strains expressing Top1-Myc ± 6His-Pmt3 from their endogenous loci, after denaturing Ni-NTA pulldowns (long and short exposures are shown). The monosumoylated species of Top1 is indicated (Top1-Pmt3). All experiments were performed at 25°C.
(3.01 MB TIF)
swi10Δ tdp1Δ have elevated levels of Top1ccs but are not sensitive to oxidative damage caused by H2O2. (A) ChIP-qPCR assay of the indicated strains. The data represents the average DNA recovery compared to the input DNA samples with standard deviations from at least three independent experiments when nmt41-Top1-FLAG is induced. ChIP-qPCR data of nmt41-Top1-FLAG swi10Δ tdp1Δ grown in repressed media (+B1) is shown as a negative control. Cells were induced at 25°C for 48 hours prior to harvesting. (B) Anti-FLAG western blot analysis of the strains used for the ChIP analysis shown in (A). (C) Cells of the indicated genotype were restruck onto plates that either repressed (+B1), or induced expression of the nmt promoter in the presence or absence of the indicated concentration of CPT. Cells were cultured at 32°C. (D) Five fold serial dilutions of the indicated strains were spotted onto media, which either contained no drug, H202, or camptothecin (CPT) at the indicated concentrations. Plates were incubated at 32°C. No profound sensitivity of swi10Δ tdp1Δ top1Δ cells as compared to wild-type or spc1Δ cells was observed. This was anticipated due to the known robust and stress-inducible catalase activity in fission yeast, and the key role of Apn2 phosphodiesterase in processing the 3′ blocked end generated by Nth1-dependent AP site incision e.g. [Hida Y., Ikeda S. (2008) Base excision repair of oxidative DNA damage in a catalase-deficient mutant of schizosaccharomyces pombe. Genes and Environment 30, 86–91. Kanamitsu K., Ikeda S. (2010) Early Steps in the DNA Base Excision Repair Pathway of a Fission Yeast Schizosaccharomyces pombe. J Nucleic Acids 2010].
(5.73 MB TIF)
S. pombe strains used during this study.
(0.08 MB DOC)