Abstract
Fe(II)- and α-ketoglutarate (α-KG)-dependent dioxygenases are a large and diverse superfamily of mononuclear, non-heme enzymes that perform a variety of oxidative transformations typically coupling oxidative decarboxylation of α-KG with hydroxylation of a prime substrate. The biosynthetic gene clusters for several nucleoside antibiotics that contain a modified uridine component, including the lipopeptidyl nucleoside A-90289 from Streptomyces sp. SANK 60405, have recently been reported, revealing a shared open reading frame with sequence similarity to proteins annotated as α-KG:taurine dioxygenases (TauD), a well characterized member of this dioxygenase superfamily. We now provide in vitro data to support the functional assignment of LipL, the putative TauD enzyme from the A-90289 gene cluster, as a non-heme, Fe(II)-dependent α-KG:UMP dioxygenase that produces uridine-5′-aldehyde to initiate the biosynthesis of the modified uridine component of A-90289. The activity of LipL is shown to be dependent on Fe(II), α-KG, and O2, stimulated by ascorbic acid, and inhibited by several divalent metals. In the absence of the prime substrate UMP, LipL is able to catalyze oxidative decarboxylation of α-KG, although at a significantly reduced rate. The steady-state kinetic parameters using optimized conditions were determined to be Kmα-KG = 7.5 μm, KmUMP = 14 μm, and kcat ≈ 80 min−1. The discovery of this new activity not only sets the stage to explore the mechanism of LipL and related dioxygenases further but also has critical implications for delineating the biosynthetic pathway of several related nucleoside antibiotics.
Keywords: Alzheimer Disease, Cell Death, Neurodegeneration, Synapses, Tau
Introduction
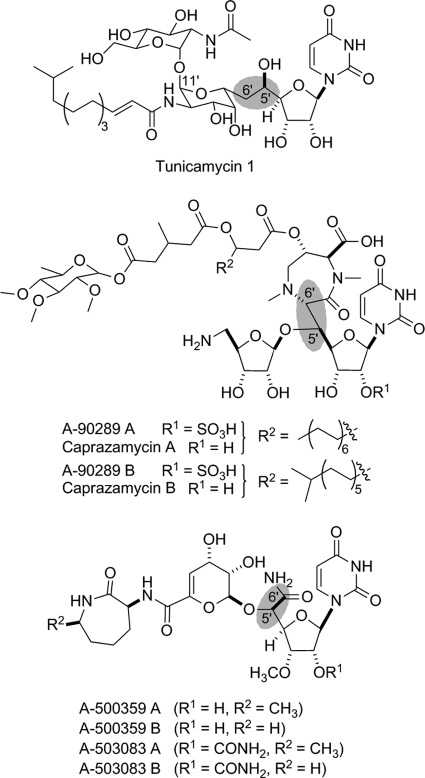
Several nucleoside antibiotics from Streptomyces sp. and related actinomycetes have been discovered that inhibit bacterial translocase I (MraY) involved in the biosynthesis of peptidoglycan cell wall, an essential component for the survival of all bacteria and a proven target for antibiotics (1). These nucleoside antibiotics have been classified into minimally four groups based on structural differences: peptidyl nucleosides represented by pacidamycin from Streptomyces coeruleorubidus (2) and mureidomycin from Streptomyces flavidovirens (3), lipodisaccharyl nucleosides represented by tunicamycins from Streptomyces lysosuperificus (4), lipopeptidyl nucleosides represented by A-90289 from Streptomyces sp. SANK 60405 (5) and caprazamycin from Streptomyces sp. MK730–62F2 (6), and glycosyl-peptidyl nucleosides represented by the capuramycin-type antibiotics A-500359s from Streptomyces griseus SANK 60196 (7), A-503083s from Streptomyces sp. SANK 62799 (8), and A-102395 from Amycolatopsis sp. SANK 60206 (9). However, despite several notable structural variations, the last three of these four groups share one signature feature that is critical for biological activity: a uracil-containing nucleoside component that consists of a high-carbon furanoside, wherein the typical ribosyl moiety is replaced by a hexafuranoside (C6), heptafuranoside (C7), or, in the case of tunicamycin, an undecafuranoside (C11) (Fig. 1) (10).
FIGURE 1.
Structures of representative nucleoside antibiotics that inhibit bacterial translocase I. Highlighted in blue is the unusual C-C bond resulting in the classification as high carbon nucleosides.
The biosynthetic gene clusters for the C7 nucleoside antibiotics A-90289 (5), liposidomycins (11), and caprazamycins (12) have recently been reported. Interestingly, an open reading frame (orf) encoding a protein with similarity to serine hydroxymethyltransferases was uncovered in all of the gene clusters. Serine hydroxymethyltransferases catalyze the interconversion of Gly to Ser utilizing N5,N10-methylene tetrahydrofolate as a hydroxymethyl donor (13). Because the C7 nucleosides contain a C5′-glycyl unit (Fig. 1), this enzyme was an obvious candidate to catalyze an aldol-type condensation using uridine-5′-aldehyde as the “hydroxymethyl” donor in place of N5,N10-methylene tetrahydrofolate. Rather unexpected was the uncovering of a homologous serine hydroxymethyltransferases within the biosynthetic gene clusters for the C6 nucleosides A-500359 (14) and A-503083 (15), which are characterized by a 5′-C-carbamoyl-uridine (Fig. 1). This discovery suggested that a common mechanism is utilized to form the high carbon scaffold, potentially through a common 5′-C-glycyluridine intermediate, but subsequent tailoring step(s) are required to eliminate a carbon unit during the biosynthesis of the C6 nucleosides.
Upon further comparative analysis of the identified gene clusters, we noticed that only one additional orf was shared among all of the putative biosynthetic genes. This orf encodes a protein with sequence similarity to proteins annotated as α-ketoglutarate:taurine dioxygenases (TauD),2 which catalyze the conversion of taurine to aminoacetaldehyde and sulfite (supplemental Fig. S1) (16). TauD is the model member of a large and diverse superfamily of mononuclear, non-heme Fe(II)- and α-ketoglutarate (α-KG)-dependent enzymes that are generally agreed to follow identical reaction coordinates involving the oxidative decarboxylation of α-KG to give an enzyme-bound Fe(IV)-oxo intermediate that is utilized to abstract a hydrogen atom from a prime substrate, taurine for TauD, to yield a carbon-centered radical (17, 18). Typically, hydrogen abstraction is followed by oxygen rebound of the Fe(III)-hydroxo species, resulting in hydroxylation at this carbon, which for TauD results in the formation of 1-hydroxy-2-aminoethanesulfonic acid that decomposes to the products (16). The formation of an aldehyde product is also observed with AtsK, a member of this dioxygenase superfamily that catalyzes hydroxylation of an alkyl sulfate ester that decomposes to sulfate and an aliphatic aldehyde (supplemental Fig. S1B) (19).
The realization that the product of both TauD and AtsK is an aldehyde due to hydroxylation of a carbon bonded to a relatively good leaving group led us to hypothesize that the shared dioxygenase catalyzes a mechanistically similar reaction. Thus, it was envisioned that the putative dioxygenase would function prior to the serine hydroxymethyltransferases-like enzyme by catalyzing the hydroxylation at C-5′ of a uridine-related metabolite leading to a geminal diol or an unstable geminal-hydroxy phosphoester that decomposes to phosphate and uridine-5′-aldehyde. Alternatively, it could not be excluded a priori that these newly discovered dioxygenases function as authentic taurine dioxygenases to give aminoacetaldehyde that would serve as an electrophile for an aldol-type condensation with an unknown, modified uridine component. Therefore, it was of general interest to elucidate the function of the TauD-like dioxygenases as an initial step to delineating the biosynthetic mechanism for high carbon nucleosides.
Using the hypothetical TauD (LipL) from the A-90289 biosynthetic gene cluster as a model enzyme, we now demonstrate that uridine-5′-aldehyde is likely a shared, pathway intermediate by functionally assigning recombinant LipL as a non-heme, Fe(II)-dependent α-KG:uridine-5′-monophosphate (UMP) dioxygenase that catalyzes the formation of succinate, phosphate, and uridine-5′-aldehyde. The biochemical characterization of LipL presented herein reveals a novel functional role for a member of the Fe(II)- and α-KG-dependent dioxygenase superfamily, setting the stage for in-depth mechanistic studies on these new enzyme catalysts.
EXPERIMENTAL PROCEDURES
Chemicals and General Methods
5,5′-Dithiobis(2-nitrobenzoic acid) (Ellman's reagent), UMP disodium, α-KG sodium salt, sodium succinate, sodium l-ascorbate, taurine, nucleosides, and nucleotides were purchased from Sigma. Buffers and salts, including Fe(II) chloride tetrahydrate, were purchased from Fisher Scientific. Uridine-5′-aldehyde was synthesized from uridine by protection of the 2′,3′-hydroxyl group with p-methoxybenzaldehyde, and Pfitzner-Moffatt oxidation of the resulting product provided 2′,3′-O-p-methoxybenzylideneuridine-5′-aldehyde; the oxidized product was deprotected to afford uridine-5′-aldehyde (see supplemental Methods and supplemental Fig. S2) (20, 21). The SensoLyte® MG Phosphate Assay kit was purchased from AnaSpec (San Jose, CA). The uncoupled reaction was analyzed using a succinate detection kit from Megazyme International following the provided instructions. UV/visible spectroscopy was performed with a Bio-Tek μQuant microplate reader using MicrotestTM 96-well plates (BD Biosciences) or a Shimadzu UV/visible-1800 spectrophotometer. Synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). DNA sequencing was performed using the BigDyeTMTerminator version 3.1 Cycle Sequencing kit from Applied Biosystems and analyzed at the University of Kentucky Advanced Genetic Technologies Center. HPLC was performed with a Waters Alliance 2695 separation module (Milford, MA) equipped with a Waters 2998 diode array detector and an analytical Apollo C-18 column (250 mm × 4.6 mm, 5 μm) purchased from Grace (Deerfield, IL). Electrospray ionization-MS was performed using an Agilent 6120 Quadrupole MSD mass spectrometer (Agilent Technologies, Santa Clara, CA) equipped with an Agilent 1200 Series Quaternary LC system and an Eclipse XDB-C18 column (250 mm × 4.6 mm, 5 μm, 80 Å). NMR data were collected using a Varian Unity Inova 500 MHz Spectrometer (Varian, Inc., Palo Alto, CA).
Cloning and Expression of TauD and LipL
The genes were amplified by PCR using the Expand Long Template PCR system from Roche Applied Science with supplied buffer 2, 200 mm dNTPs, 5% dimethyl sulfoxide, 10 ng of DNA template, 5 units of DNA polymerase, and a 10 mm concentration of each of the following primer pairs: EctauD (forward), 5′-GGTATTGAGGGTCGCatgagtgaacgtctgagcattac-3′/(reverse), 5′-AGAGGAGAGTTAGAGCCttaccccgcccgataaaacg-3′ and lipL (forward), 5′-GGTATTGAGGGTCGCatgtccgtgctggggcgg-3′/(reverse),5′-AGAGGAGAGTTAGAGCCtcatgagggcttcttcggtg-3′. DNA templates for PCR cloning were Escherichia coli DH5α genomic DNA (tauD) and cosmid pN1 (lipL) (7). The PCR program included an initial hold at 94 °C for 2 min, followed by 30 cycles of 94 °C for 10 s, 56 °C for 15 s, and 68 °C for 50 s. The gel-purified PCR product was inserted into pET-30 Xa/LIC using ligation-independent cloning as described in the provided protocol to yield pET30-EctauD and pET30-lipL, and the genes were sequenced to confirm PCR fidelity.
Plasmids were introduced into E. coli BL21(DE3) cells, and the transformed strains were grown in LB supplemented with 30 μg/ml kanamycin. Following inoculation of 500 ml of LB with 30 μg/ml kanamycin, the cultures were grown at 18 °C until the cell density reached an A600 = 0.5 when expression was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested after an overnight incubation at 18 °C and lysed using a French press with one pass at 15,000 p.s.i. Following centrifugation, the protein was purified using affinity chromatography with nickel-nitrilotriacetic acid-agarose from Qiagen (Valencia, CA), and the recombinant proteins were desalted into 50 mm Tris-HCl (pH 8), 100 mm NaCl, and 5% glycerol using a PD-10 desalting column (GE Healthcare). The purified protein was concentrated using an Amicon Ultra 10000 MWCO centrifugal filter (Millipore) and stored as glycerol stocks (40%) at −20 °C. Protein purity was assessed by 12% acrylamide SDS-PAGE; His6-tagged proteins were utilized without further modifications.
α-KG:Taurine Dioxygenase Activity
Standard reactions consisted of 50 mm Tris-HCl (pH 7.5), 0.5 mm taurine, 1 mm α-KG, 0.2 mm ascorbate, 0.1 mm FeCl2, and 20 μg/ml LipL or 2.5 μg/ml EcTauD at 30 °C (16). Control reactions were performed without the addition of either α-KG or enzyme. At desired time points, 80 μl of the reaction mixture was added to 20 μl of 0.25 m EDTA and 10 μl of Ellman's reagent (1 mg/ml in 0.1 m phosphate (pH 7.0)). The mixture was incubated at room temperature for 5 min prior to obtaining the absorbance at 415 nm. Sodium sulfite was used for calibration curves yielding a linear response from 25 to 400 μm sulfite. The specific activity of EcTauD, calculated from end point assays under initial velocity conditions, was determined using the reaction mixture above except with 0.3 μg/ml EcTauD.
Metal Content
Iron and zinc content in the proteins was determined using size exclusion chromatography (Agilent, 1200 series LC) with refractive index (Optilab rEX; Wyatt, Goleta, CA), multiangle laser light scattering (DAWN Heleos III; Wyatt), and inductively coupled plasma mass spectrometric (ICP-MS; Agilent 7500CX) detection. The refractive index detector was used to determine protein concentration in the eluting peaks assuming a dN/dC (differential refractive index) value of 0.185, and the multiangle laser light scattering was used in conjunction with the refractive index detector to verify the molar masses of the eluting peaks using a partial Zimm model (22). The ICP-MS was used as an element-specific detector to identify V, Cr, Mn, Fe, Co, Ni, Cu, and Zn and to quantify Fe and Zn content (23). Fe and Zn content were calibrated using bovine hemoglobin and carbonic anhydrase as standards, respectively. The mobile phase was 100 mm NH4NO3 at a flow rate of 0.5 ml min−1 through a Superdex 200HR SEC column (GE Healthcare).
Activity Assays Using HPLC
Reactions consisted of 50 mm Tris-HCl (pH 7.5), 1 mm UMP, 1.25 mm α-KG, 200 μm ascorbate, 100 μm FeCl2, and 100 nm LipL at 30 °C. Reactions were terminated by the addition of cold trichloroacetic acid to 5% (w/v) or by ultrafiltration using a Microcon YM-3. Following centrifugation to remove protein, the reaction was analyzed by HPLC using a C-18 reversed-phase column and ion-pairing conditions. A series of linear gradients was developed from 40 mm phosphoric acid-triethylamine pH 6.5 (A) to 90% acetonitrile (B) in the following manner (beginning time and ending time with linear increase to % B): 0–8 min, 0% B; 8–18 min, 60% B; 18–25 min, 95% B; 25–32 min, 95% B; and 32–35 min, 0% B. The flow rate was 1.0 ml/min, and elution was monitored at 260 nm. LC-MS was performed using a linear gradient from 0.1% formic acid in water to 0.1% formic acid in acetonitrile over 20 min. The flow rate was 0.4 ml/min, and elution was monitored at 254 nm.
Activity Assays by Phosphate Detection and Activity Optimization
The activity of LipL was detected by UV/visible spectroscopy by monitoring the formation of inorganic phosphate with the malachite green binding assay (24). Routine reactions consisted of 50 mm Tris-HCl (pH 7.5), 1 mm UMP, 1.25 mm α-KG, 100 μm ascorbate, 50 μm FeCl2, and 100 nm LipL. Optimal iron concentration for activity was determined with assays containing 100 μm ascorbate, 40 nm LipL, and 0.1–750 μm FeCl2. After identifying the optimal iron concentration, the optimal ascorbate concentration was determined with reactions containing 20 μm FeCl2, 30 nm LipL, and 0.5–50 mm ascorbate. All reactions were carried out at 30 °C, and product formation was analyzed under initial velocity conditions.
Kinetic Analysis
Assays consisted of 50 mm Tris-HCl (pH 7.5), 1 mm ascorbate, 20 μm FeCl2, LipL, and near saturating UMP (2 mm) with variable α-KG (5 μm–3 mm) or near saturating α-KG (1 mm) and variable UMP (2 μm–1 mm). The reaction was performed at 30 °C for 3 min and analyzed under initial velocity conditions. Product formation was determined using either the malachite green binding assay or HPLC. Each data point represents a minimum of two (HPLC assay) or three (malachite green binding assay) replicate end point assays; kinetic constants were obtained by nonlinear regression analysis using GraphPad Prism (GraphPad Software, La Jolla, CA).
Dependence of the Reaction on O2 and Fate of Oxygen Atoms
A 1-ml reaction containing 10 mm Tris-HCl (pH 8.0), 1 mm UMP, 1.5 mm α-KG, 1 mm fresh ascorbate, and 100 μm fresh FeCl2 was placed under a N2 atmosphere and purged twice by bubbling in N2 for 10 min. The atmosphere was maintained in N2 or exchanged with 16O2 (control) or 18O2, and the reaction was initiated by the addition of 2.5 μl of LipL (final concentration of 1 μm). The reaction was incubated at room temperature in a hermetically sealed flask for 90 min. The reaction was terminated by the addition of cold trichloroacetic acid (5%) and the components analyzed by ion pairing chromatograph and LC-MS as described above.
RESULTS
Bioinformatics Analysis
BLAST analysis of LipL revealed closest sequence similarity to proteins primarily annotated as α-KG:taurine dioxygenases (22–28% identity). Sequence similarity of LipL to E. coli TauD, a well characterized α-KG:taurine dioxygenase for which the structure is known (25), is significantly less (15% identity/24% similarity). Similarly, low levels of sequence identity were also found with other structurally elucidated dioxygenases of this superfamily, such as AtsK (19) and clavaminic acid synthase from Streptomyces clavuligerus (26).
All members of this dioxygenase superfamily contain an essential Fe(II)-binding motif HX(D/E)XnH, where X is any amino acid (18). The iron-binding motif of LipL was putatively assigned by alignments with EcTauD, which revealed the potential triad within LipL (His123, Asp125, and His266) (supplemental Fig. S3). In addition to the three conserved metal-binding residues, an Arg residue involved in α-KG binding is also conserved (Arg279 for LipL). Thus, based on sequence analysis, LipL apparently contains the essential residues to function as an Fe(II)- and α-KG-dependent dioxygenase (17–19), and these residues are conserved in the other TauD-like enzymes from the A-500359, A-503083, liposidomycin, and caprazamycin gene clusters (supplemental Fig. S3).
Functional Assignment and Assay Development
LipL was cloned and expressed in E. coli BL21(DE3), and the recombinant protein was purified to near homogeneity as judged by SDS-PAGE (supplemental Fig. S4A). Given the closest sequence similarity to proteins annotated as TauD, the tauD gene from E. coli was also cloned, and the recombinant protein was purified to serve as a control (supplemental Fig. S4B). Using Ellman's reagent for spectroscopic detection, recombinant EcTauD catalyzed the expected α-KG- and taurine-dependent formation of sulfite with a specific activity of 1.4 mmol/min per mg, which is nearly identical to the reported value (supplemental Fig. S5) (16). In contrast, no reaction was observed when LipL was incubated under the identical conditions, consistent with LipL having a unique activity.
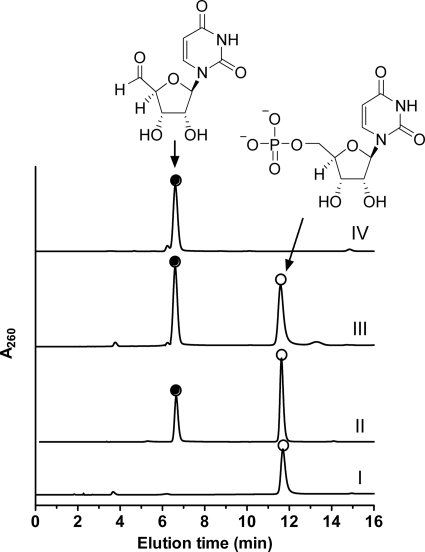
Activity of LipL was next tested with alternative prime substrates in place of taurine. When the enzyme was incubated with UMP, a new peak was observed by HPLC with concomitant loss in the peak corresponding to UMP (Fig. 2). The new compound was unstable upon column purification under a variety of conditions; however, LC-MS analysis of the crude reaction mixture consisting of ∼95% conversion of UMP to product (as determined by ion-pairing HPLC) revealed the new peak had an (M−H)− ion at m/z = 240.8, consistent with the molecular formula for uridine-5′-aldehyde (expected (M−H)− ion at m/z = 241.1) (supplemental Fig. S6). In contrast, only substrates UMP and α-KG were observed by LC-MS when the enzyme was omitted (supplemental Fig. S7). To confirm the identity of the new peak, uridine-5′-aldehyde was synthesized and confirmed by 1H NMR and 13C NMR spectroscopy (supplemental Fig. S2). HPLC and LC-MS analysis with co-injections of authentic material, which was likewise previously demonstrated to be unstable upon several column purification methods (21), revealed uridine-5′-aldehyde was indeed the product of LipL (Fig. 2).
FIGURE 2.
Identification of the LipL product. HPLC analysis of the LipL reaction using authentic UMP without α-KG (I), a 15-min reaction containing all of the necessary components (II), the 15-min reaction mixture spiked with synthetic uridine-5′-aldehyde (III), and synthetic uridine-5′-aldehdye (IV) are shown. A260, absorbance at 260 nm.
TauD and related Fe(II)- and α-KG-dependent dioxygenases produce CO2 and succinate upon oxidative decarboxylation of α-KG (16–18). Succinate was confirmed as a product for LipL by utilizing LC-MS with commercial succinate as a control (supplemental Fig. S6). Alternatively, an enzymatic coupled assay was used to confirm that succinate was produced during the LipL-catalyzed reaction (supplemental Fig. S8) (27). Analogous to sulfite production by TauD, phosphate was also colorimetrically detected by employing a malachite green binding assay (24). Thus, LipL was directly shown to catalyze the formation of succinate, phosphate, and uridine-5′-aldehyde.
Activity Optimization, Specificity, and General Characterization
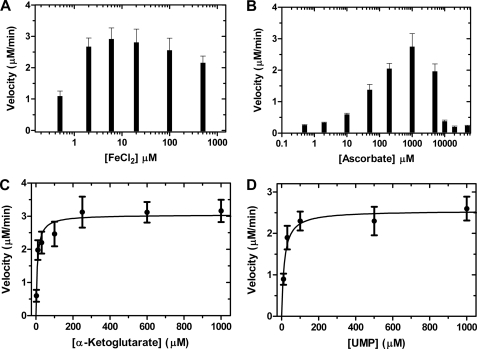
The requirements for LipL activity was assessed by HPLC analysis following uridine-5′-aldehyde formation and colorimetric analysis by detecting phosphate formation, with the latter assay utilized for analysis under initial velocity conditions. The activity was absolutely dependent upon α-KG and O2 (Fig. 2 and supplemental Fig. S9). The addition of Fe(II) or ascorbate was not essential for activity; however, inclusion of EDTA abolished activity, suggesting that Fe(II) is required (supplemental Fig. S10). Subsequently, ICP-MS analysis revealed LipL co-purified with 14 ± 2% iron (mol/mol), thus explaining the residual activity in the absence of additional Fe(II). LipL was found to have optimal activity in the range of 2–100 μm FeCl2 (Fig. 3A). Using 20 μm FeCl2, optimal activity with respect to ascorbate was determined to be 1 mm for LipL (Fig. 3B).
FIGURE 3.
Activity optimization and kinetic analysis of LipL using the malachite green binding assay. A, optimal activity of LipL with respect to varied FeCl2 in reactions containing 100 μm ascorbate and 40 nm LipL. B, optimal activity of LipL with respect to varied ascorbate in reactions containing of 20 μm FeCl2 and 30 nm LipL. C, single-substrate kinetic analysis using variable α-KG (2 μm–3 mm), near saturating UMP (2 mm) and 34 nm LipL. D, single-substrate kinetic analysis using variable UMP (2 μm–1 mm), near saturating α-KG (1 mm) and 34 nm LipL. Data represent the average of minimally three independent replicates obtained using end point assays under initial velocity conditions; S.E. was <15% in all cases.
The specificity of LipL toward different nucleosides and nucleotides was next analyzed. Uridine and UDP were not recognized as substrates, demonstrating that LipL is specific for the monophosphorylated nucleoside (supplemental Fig. S11). LipL was unable to catalyze dephosphorylation and aldehyde formation using the remaining canonical, monophosphorylated ribo- or deoxyribonucleotides (data not shown). Similar to the high specificity toward the prime substrate UMP, pyruvate, α-ketoadipate, α-ketobutyrate, α-ketovalerate, and oxaloacetate could not substitute for α-KG in the reaction.
Finally, the activity with different metals was tested. Potassium ions, Fe(III), or divalent metals including Mg2+, Ca2+, and third-row transition metals were not able to replace Fe(II) or increase the specific activity when included in the reaction with FeCl2 (supplemental Fig. S12). Instead, several metals inhibited the reaction including Zn2+, which was the only metal in addition to Fe that was found to co-purify with LipL (19 ± 2% mol/mol) based on ICP-MS analysis. Further analysis revealed that Zn2+ strongly inhibited LipL activity (supplemental Fig. S12), and therefore co-purification with Zn2+ is likely due to adventitious binding as observed with other non-heme, iron-dependent oxygenases (28). Inhibition by divalent transition metals has been reported for TauD (16) and clavaminic acid synthase (29).
Kinetic Analysis
Steady-state kinetic analysis was performed using both the malachite green binding assay to detect phosphate and HPLC to detect uridine-5′-aldehyde. Single-substrate kinetic experiments with phosphate detection revealed typical Michaelis-Menten kinetics, yielding kinetic constants of Km = 7.5 ± 2.4 μm and kcat = 92 ± 9 min−1 with respect to α-KG and Km = 14 ± 3 μm, kcat = 76 ± 8 min−1 with respect to UMP (Fig. 3, C and D). Although detection limits precluded a more accurate kinetic analysis, similar plots and extracted kinetic constants were obtained using HPLC detection (supplemental Fig. S13).
Uncoupled Oxidative Decarboxylation of α-KG
Many enzymes of the α-KG-dependent dioxygenase superfamily including TauD catalyze oxidative decarboxylation of α-KG without the inclusion of the prime substrate (18, 27, 30). Utilizing LC-MS for detection, succinate was similarly observed for LipL without including UMP in the reaction (supplemental Fig. S14). The enzyme-coupled assay to detect succinate production was employed to confirm the LC-MS result (supplemental Fig. S8), and kinetic analysis with UMP yielded nearly an identical kcat (78 min−1) relative to kinetic analysis by detection of phosphate or uridine-5′-aldehyde. In contrast, the rate of succinate formation by LipL was significantly reduced in the absence of UMP (supplemental Fig. S8), yielding a kcat = 0.4 min−1 for LipL, which corresponds to a relative turnover of 0.5% compared with the UMP-containing reaction.
Although succinate was readily detected, no other hypothetical reaction intermediates (for example, uridine) or shunt products were detected by HPLC with photodiode array or MS detection. Furthermore, hydrogen peroxide was not detected using an enzyme-coupled reaction with horseradish peroxidase (31). In addition, UMP or uridine was not detected when the reverse reaction was assayed.
Isotope Enrichment Studies
The fate of oxygen atoms derived from O2 was examined. After saturating a reaction mixture with 18O2 (97 atom %), excess LipL was added, and the reaction was allowed to proceed for 90 min under an 18O2 atmosphere at room temperature. Detection by LC-MS revealed 97% of the total succinate had an (M−H)− ion at m/z = 119.0, consistent with the incorporation of one atom of oxygen 18O2 (expected (M−H)− ion at m/z = 119.0) (supplemental Fig. S15). Thus, incorporation into succinate from O2 was essentially 100%. When the reaction was performed in H218O (final of 80 atom %), detection by LC-MS revealed mass peaks for succinate that correspond to ∼79% incorporation of an oxygen atom from H218O and ∼83% incorporation for the uncoupled reaction. When LC was bypassed for direct analysis of the uncoupled reaction, the mass corresponding to residual α-KG revealed 75% 18O incorporation, an expected result due to nonenzymatic exchange of the α-keto acid (32). Unfortunately, the rapid and reversible formation of a geminal diol in water did not allow the analysis of putative oxygen incorporation into uridine-5′-aldehyde.
DISCUSSION
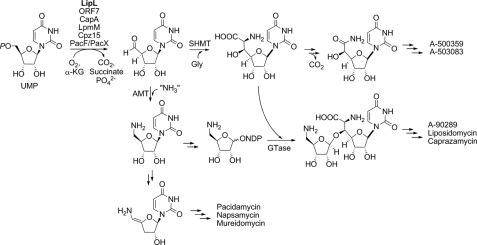
Enzymes of the Fe(II)- and α-KG-dependent dioxygenase superfamily catalyze a diverse array of biotransformations in natural product biosynthesis, including hydroxylation of Arg in viomycin biosynthesis (33, 34); O-demethylation in morphine biosynthesis (35); and hydroxylation, cyclization, and desaturation by clavaminic acid synthase in the biosynthesis of the β-lactam clavulonic acid (29, 36). Bioinformatics analysis of several recently uncovered gene clusters for minimally two different families of nucleoside antibiotics revealed a shared open reading frame encoding a protein with low sequence similarity to TauD, the best studied enzyme of Fe(II)- and α-KG-dependent dioxygenase superfamily. We have characterized a representative enzyme from the A-90289 gene cluster to reveal yet another unusual reaction catalyzed by enzymes of the dioxygenase superfamily: the net dephosphorylation and two-electron oxidation of UMP. Thus, during the biosynthesis of C7 high carbon nucleosides, the modified nucleoside can now be described to begin with the conversion of UMP to uridine-5′-aldehyde (Fig. 4), results that are consistent with prior feeding experiments with other model high carbon nucleoside antibiotics (37–39). The uridine-5′-aldehyde intermediate is subsequently condensed with glycine to form C-5′-glycyluridine, which remains intact during the formation of the lipopeptidyl nucleosides. The identification of a homologous enzyme within the A-500359 and A-503083 gene clusters suggest that an identical reaction occurs during the biosynthesis of C6 high carbon nucleosides, although in this case the resulting C-5′-glycyluridine undergoes oxidative decarboxylation catalyzed by a distinct, unidentified enzyme to yield the final nucleoside (Fig. 4). The lipopeptidyl nucleosides contain an additional aminoribosyl moiety, and the pathway to this component is proposed to also proceed through uridine-5′-aldehyde, which is diverted into this alternative pathway upon transamination, and subsequent processing yields the activated NDP-amino sugar for attachment to the aglycon. In addition, the recent identification of the biosynthetic gene cluster for pacidamycin has revealed a homologous dioxygenase and aminotransferase, suggesting that the unusual enamide-containing nucleoside originates by the tandem action of these two enzymes (40, 41). Thus, this newly discovered enzyme activity is likely essential for the biosynthesis of a number of structurally distinct nucleoside antibiotics.
FIGURE 4.
Proposed biochemical pathway leading to several high carbon nucleosides inhibiting bacterial translocase I. Highlighted in bold is the enzyme characterized in this study; enzymes with likely identical function as LipL include ORF7 from the A-500359 gene cluster, CapA from the A-503083 gene cluster, LpmM from the liposidomycin gene cluster, Cprz15 from the caprazamycin gene cluster, and PacF/PacX from the pacidamycin gene cluster. AMT, aminotransferase; GTase, glycosyltransferase.
Most of the biochemical characteristics for LipL have been reported for other enzymes of the Fe(II)- and α-KG-dependent dioxygenase superfamily. For instance, activity is stimulated by ascorbate, certain divalent metals inhibit activity, activity is specific for both the prime substrate and α-KG, and oxidative decarboxylation of α-KG still occurs in the absence of the prime substrate, all characteristics realized for TauD and AtsK, among others (18). Clearly, the unique feature of LipL is the specific utilization of UMP and the mechanism by which the net dephosphorylation and oxidation occurs. Previously, it was logically speculated that this transformation occurs via tandem reactions catalyzed by a phosphatase and oxidoreductase. Common catalytic strategies for phosphatases include (i) formation of a phosphoenzyme intermediate utilizing a conserved Cys as the nucleophile or (ii) the use of a binuclear center consisting of Mg2+, Mn2+, Fe2+, and/or Zn2+(42). However, our cumulative biochemical data suggest that LipL catalyzes dephosphorylation via a cryptic hydroxylation germinal to a good leaving group and thus likely employs the prototypical Fe(II)- and α-KG-dependent dioxygenase mechanism reminiscent of TauD and AtsK, wherein decarboxylative oxidation of α-KG is coupled to the formation of an Fe(IV)-oxo intermediate that serves as the strong oxidizing agent for hydrogen abstraction of the prime substrate (supplemental Fig. S16). Alternatively, a few additional mechanisms could be envisioned for LipL, including sequential hydrogen abstractions at C-5′ and C-4′ of UMP analogous to the proposed desaturation mechanism of certain Fe(II)- and α-KG-dependent dioxygenases (18), which in this case would be followed by phosphate hydrolysis and enol tautomerization to yield the final product (supplemental Fig. S17). A clear distinction between hydroxylation versus dephosphorylation/oxidation mechanisms is the fate of the bridging phosphoester oxygen, and experiments are under way to examine this feature of the reaction.
The cumulative data suggest that LipL and by extension the homologous enzymes involved in the biosynthesis of C6 and C7 high carbon nucleosides are new members of the superfamily of Fe(II)- and α-KG-dependent dioxygenases, once again highlighting the versatility and significance of these biocatalysts in nature. Interestingly, enzymatic phosphate release as a mechanism of phosphate salvage analogous to sulfite and sulfate formation for TauD and AtsK, respectively, has been prognosticated as a potential function for this dioxygenase superfamily (18), and we have demonstrated that this is indeed possible by providing the enzyme precedence for this chemistry. It is also foreseeable that the biosynthesis of other high carbon nucleoside antibiotics such as the uracil-containing tunicamycin (10) and A-94964 (43), the cytosine-containing ezomycin A1 (44), and the adenine-containing griseolic acid (45) and amipurimycin (46) employ a similar enzymatic strategy for assembling a high carbon sugar skeleton (supplemental Fig. S18). Regardless of these possibilities, we have clearly demonstrated a novel enzyme function that has significance for the biosynthesis of several nucleoside antibiotics, which now sets the stage to interrogate the mechanism of the newfound dioxygenase family and delineate downstream catalytic events in the biosynthesis of several compounds of potential therapeutic significance.
Supplementary Material
This work was supported in part by the Kentucky Science and Technology Corporation (S. G. V. L.) and a National Science Foundation predoctoral fellowship (J. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S18, Methods, and additional references.
- TauD
- α-ketoglutarate:taurine dioxygenases
- ICP-MS
- inductively coupled plasma mass spectrometric
- α-KG
- α-ketoglutarate.
REFERENCES
- 1. Winn M., Goss R. J., Kimura K., Bugg T. D. (2010) Nat. Prod. Rep. 27, 279–304 [DOI] [PubMed] [Google Scholar]
- 2. Chen R. H., Buko A. M., Whittern D. N., McAlpine J. B. (1989) J. Antibiot. 42, 512–520 [DOI] [PubMed] [Google Scholar]
- 3. Isono F., Inukai M. (1991) Antimicrob. Agents Chemother. 35, 234–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tamura G., Sasaki T., Matsuhashi M., Takatsuki A., Yamasaki M. (1976) Agric. Biol. Chem. 40, 447–449 [Google Scholar]
- 5. Funabashi M., Baba S., Nonaka K., Hosobuchi M., Fujita Y., Shibata T., Van Lanen S. G. (2010) ChemBioChem 11, 184–190 [DOI] [PubMed] [Google Scholar]
- 6. Igarashi M., Takahashi Y., Shitara T., Nakamura H., Naganawa H., Miyake T., Akamatsu Y. (2005) J. Antibiot. 58, 327–337 [DOI] [PubMed] [Google Scholar]
- 7. Muramatsu Y., Muramatsu A., Ohnuki T., Ishii M. M., Kizuka M., Enokita R., Tsutsumi S., Arai M., Ogawa Y., Suzuki T., Takatsu T., Inukai M. (2003) J. Antibiot. 56, 243–252 [DOI] [PubMed] [Google Scholar]
- 8. Muramatsu Y., Ohnuki T., Ishii M. M., Kizuka M., Enokita R., Miyakoshi S., Takatsu T., Inukai M. (2004) J. Antibiot. 57, 639–646 [DOI] [PubMed] [Google Scholar]
- 9. Murakami R., Fujita Y., Kizuka M., Kagawa T., Muramatsu Y., Miyakoshi S., Takatsu T., Inukai M. (2007) J. Antibiot. 60, 690–695 [DOI] [PubMed] [Google Scholar]
- 10. Price N. P., Tsvetanova B. (2007) J. Antibiot. 60, 485–491 [DOI] [PubMed] [Google Scholar]
- 11. Kaysser L., Siebenberg S., Kammerer B., Gust B. (2010) ChemBioChem 11, 191–196 [DOI] [PubMed] [Google Scholar]
- 12. Kaysser L., Lutsch L., Siebenberg S., Wemakor E., Kammerer B., Gust B. (2009) J. Biol. Chem. 284, 14987–14996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schirch V., Szebenyi D. M. (2005) Curr. Opin. Chem. Biol. 9, 482–487 [DOI] [PubMed] [Google Scholar]
- 14. Funabashi M., Nonaka K., Yada C., Hosobuchi M., Masuda N., Shibata T., Van Lanen S. G. (2009) J. Antibiot. 62, 325–332 [DOI] [PubMed] [Google Scholar]
- 15. Funabashi M., Yang Z., Nonaka K., Hosobuchi M., Fujita Y., Shibata T., Chi X., Van Lanen S. G. (2010) Nat. Chem. Biol. 6, 581–586 [DOI] [PubMed] [Google Scholar]
- 16. Eichhorn E., van der Ploeg J. R., Kertesz M. A., Leisinger T. (1997) J. Biol. Chem. 272, 23031–23036 [DOI] [PubMed] [Google Scholar]
- 17. Bollinger J. M., Jr., Price J. C., Hoffart L. M., Barr E. W., Krebs C. (2005) Eur. J. Inorg. Chem. 21, 4245–4254 [Google Scholar]
- 18. Hausinger R. P. (2004) Crit. Rev. Biochem. Mol. 39, 21–68 [DOI] [PubMed] [Google Scholar]
- 19. Müller I., Kahnert A., Pape T., Sheldrick G. M., Meyer-Klaucke W., Dierks T., Kertesz M., Usón I. (2004) Biochemistry 43, 3075–3088 [DOI] [PubMed] [Google Scholar]
- 20. Pfitzner K. E., Moffatt J. G. (1963) J. Am. Chem. Soc. 85, 3027 [Google Scholar]
- 21. Jones G. H., Taniguchi M., Tegg D., Moffatt J. G. (1979) J. Org. Chem. 44, 1309–1317 [Google Scholar]
- 22. Wyatt P. J. (1993) Anal. Chim. Acta 272, 1–40 [Google Scholar]
- 23. Unrine J. M., Jackson B. P., Hopkins W. A., Romanek C. (2006) Environ. Toxicol. Chem. 25, 1864–1867 [DOI] [PubMed] [Google Scholar]
- 24. Fisher D. K., Higgins T. J. (1994) Pharm. Res. 11, 759–763 [DOI] [PubMed] [Google Scholar]
- 25. Elkins J. M., Ryle M. J., Clifton I. J., Dunning Hotopp J. C., Lloyd J. S., Burzlaff N. I., Baldwin J. E., Hausinger R. P., Roach P. L. (2002) Biochemistry 41, 5185–5192 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Z., Ren J., Stammers D. K., Baldwin J. E., Harlos K., Schofield C. J. (2000) Nat. Struct. Biol. 7, 127–133 [DOI] [PubMed] [Google Scholar]
- 27. Luo L., Pappalardi M. B., Tummino P. J., Copeland R. A., Fraser M. E., Grzyska P. K., Hausinger R. P. (2006) Anal. Biochem. 353, 69–74 [DOI] [PubMed] [Google Scholar]
- 28. Yan F., Munos J. W., Liu P., Liu H. W. (2006) Biochemistry 45, 11473–11481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Busby R. W., Chang M. D., Busby R. C., Wimp J., Townsend C. A. (1995) J. Biol. Chem. 270, 4262–4269 [DOI] [PubMed] [Google Scholar]
- 30. McCusker K. P., Klinman J. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19791–19795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holt A., Sharman D. F., Baker G. B., Palcic M. M. (1997) Anal. Biochem. 244, 384–392 [DOI] [PubMed] [Google Scholar]
- 32. Lindblad B., Lindstedt G., Lindstedt S. (1970) J. Am. Chem. Soc. 92, 7446–7449 [DOI] [PubMed] [Google Scholar]
- 33. Ju J., Ozanick S. G., Shen B., Thomas M. G. (2004) ChemBioChem 5, 1281–1285 [DOI] [PubMed] [Google Scholar]
- 34. Yin X., Zabriskie T. M. (2004) ChemBioChem 5, 1274–1277 [DOI] [PubMed] [Google Scholar]
- 35. Hagel J. M., Facchini P. J. (2010) Nat. Chem. Biol. 6, 273–275 [DOI] [PubMed] [Google Scholar]
- 36. Marsh E. N., Chang M. D., Townsend C. A. (1992) Biochemistry 31, 12648–12657 [DOI] [PubMed] [Google Scholar]
- 37. Ohnuki T., Muramatsu Y., Miyakoshi S., Takatsu T., Inukai M. (2003) J. Antibiot. 56, 268–279 [DOI] [PubMed] [Google Scholar]
- 38. Isono K., Sato T., Hirasawa K., Funayama S., Suzuki S. (1978) J. Am. Chem. Soc. 100, 3937–3939 [Google Scholar]
- 39. Tsvetanova B. C., Kiemle D. J., Price N. P. J. (2002) J. Biol. Chem. 277, 35289–35296 [DOI] [PubMed] [Google Scholar]
- 40. Rackham E. J., Grüschow S., Ragab A. E., Dickens S., Goss R. J. (2010) ChemBioChem 11, 1700–1709 [DOI] [PubMed] [Google Scholar]
- 41. Zhang W., Ostash B., Walsh C. T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16828–16833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cleland W. W., Hengge A. C. (2006) Chem. Rev. 106, 3252–3278 [DOI] [PubMed] [Google Scholar]
- 43. Murakami R., Fujita Y., Kizuka M., Kagawa T., Muramatsu Y., Miyakoshi S., Takatsu T., Inukai M. (2008) J. Antibiot. 61, 537–544 [DOI] [PubMed] [Google Scholar]
- 44. Knapp S., Shieh W.-C., Jaramillo C., Trilles R. V., Nandan S. R. (1994) J. Org. Chem. 59, 946–948 [Google Scholar]
- 45. Takahashi S., Nakagawa F., Kawazoe K., Furukawa Y., Sato S., Tamura C., Naito A. (1985) J. Antibiot. 38, 830–834 [DOI] [PubMed] [Google Scholar]
- 46. Iwasa T., Kishi T., Matsuura K., Wakae O. (1977) J. Antibiot. 30, 1–10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.