Abstract
The protein inhibitor of activated STAT (PIAS) family proteins regulates innate immune responses by controlling transcription induced by Toll-like receptor, RIG-I-like receptor signaling, and JAK/STAT pathways. Here, we show that PIASy negatively regulates type I interferon (IFN) transcription. Virus infection led to enhanced type I IFN induction in PIASy null cells, and conversely PIASy overexpression reduced IFN transcription. A mutation in the LXXLL motif of the SAP domain abolished inhibition of IFN-stimulated gene expression but did not affect virus or Toll-like receptor/RIG-I-like receptor-stimulated IFN transcription, indicating that PIASy employs distinct mechanisms to inhibit virus-induced and IFN-stimulated transcription. SUMO E3 activity was not required for PIASy inhibition of IFN transcription; however, PIASy relied on the SUMO modification mechanism to inhibit IFN transcription, because the activity of the SUMO-interacting motif was required for inhibition, and knockdown of SUMO E2 enzyme UBC9 decreased inhibitory activity of PIASy. Our results demonstrate that PIASy negatively regulates both IFN transcription and IFN-stimulated gene expression through multiple mechanisms utilizing the function of different domains.
Keywords: Innate Immunity, Interferon, Negative-strand RNA Viruses, Post-translational Modification, Sumoylation
Introduction
Infection of RNA viruses is recognized by two classes of pathogen recognition receptors, Toll-like receptors (TLR)3 and RIG-I-like receptors (RLR), both of which bind viral RNAs (1–7). Once viral RNAs are recognized by these receptors, downstream signaling cascades are activated, triggering transcription of proinflammatory cytokines important for the establishment of innate and adaptive immunity (5). Among these cytokines, type I interferons (IFNs) play a major role in conferring antiviral and antimicrobial activities (8–11). TLR- and RLR-mediated production of type I IFNs and proinflammatory cytokines are regulated both positively and negatively at multiple steps of signaling cascades to minimize harmful excess inflammatory responses and to achieve fine-tuning of the effects (12–14). There is a growing list of proteins that function as a negative regulator of TLR and RLR signaling (15, 16).
PIAS (protein inhibitor of activated STAT) family proteins are encoded by four genes, PIAS1, PIAS3, PIASx (PIAS2), and PIASy (PIAS4) (17–21). The PIAS family was first discovered as an interacting partner of signal transducer and activator of transcription (STAT) (21–24). By associating with transcriptionally activated STATs, PIAS proteins negatively regulate some STAT-dependent genes (20, 21, 23–25). In addition to STATs, PIAS proteins also regulate large numbers of transcription factors involved in the broad range of gene expression that affects cell cycle regulation, immune responses, and development (18–21, 26). PIAS proteins function as a SUMO E3 ligase for a growing list of substrates, most of which are transcription factors (27–31). The conserved RING-like domain at the central portion of PIAS proteins is essential for E3 ligase activity (19–21, 32, 33). Conjugation of SUMO peptides to transcription factors alters transcriptional activity by changing conformation of substrates and creating a new surface for protein-protein interactions (26–29, 34, 35). In addition to regulating SUMO modification by its SUMO ligase activity, PIAS proteins regulate transcription through SUMO-independent mechanisms, including blocking DNA binding activity of transcription factors, recruiting transcriptional co-repressors, and translocation of transcription factors to nuclear subdomains (17–21).
Among PIAS proteins, PIAS1 and PIASy regulate the specificity and magnitude of cytokine-induced gene expression mediated by STAT1 (36, 37). In addition, PIAS1 and PIASy regulate LPS-induced cytokine production by inhibiting NFκB activity (37, 38). Thus, PIAS1 and PIASy play an important role not only in cytokine-mediated JAK/STAT pathways but also in pathogen-activated TLR/RLR pathways of cytokine production (20, 39). Prompted by these reports, we asked whether type I IFN production mediated by TLR and RLR signaling is also regulated by PIASy. We report here that PIASy inhibits virus-induced type I IFN transcription more potently than the other three PIAS members. PIASy targeted IRF3 and IRF7, transcription factors required for activation of the type I IFN promoter. Detailed domain analysis revealed that PIASy inhibits IRF3/IRF7-activated type I IFN transcription by a mechanism distinct from that of PIASy inhibition of IFN-stimulated gene (ISG) induction by STAT1. Additionally, we show that PIASy relies on the SUMO conjugation mechanism through the SUMO-interacting motif (SIM) to inhibit type I IFN transcription, but without relying on its own E3 ligase activity. Together, our findings highlight the diversity and complexity of PIASy-mediated negative regulation of IFN and IFN-stimulated transcription, thus influencing innate immunity.
EXPERIMENTAL PROCEDURES
Cells Culture
Mouse embryonic fibroblasts (MEFs) from PIASy+/+ and PIASy−/− mice were grown in Dulbecco's modified Eagle's medium (DMEM)/high glucose supplemented with 10% FCS and antibiotics (Invitrogen). Human embryonic kidney (HEK) 293T cells were grown in DMEM containing 10% fetal bovine serum.
Plasmids and Reagents
cDNA fragments of murine PIAS1, PIAS3, PIASxa, PIASxb, and PIASy were generated from total RNA of NIH3T3 cells by the standard RT-PCR technique and were inserted into pcDNA3.1/HA and pMSCV/HA retroviral vector. To construct mutants for PIASy, appropriate substitutions were introduced into the pcDNA3.1-PIASy-HA and pMSCV-PIASy-HA using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). T7-SUMO1/GA was generated from the T7-SUMO1 expression plasmid (40). Murine IRF3 and IRF7 constructs were described previously (40). Human VISA cDNA was amplified from total RNA of HEK293T cells and inserted into pcDNA-2×Myc and pcDNA-V5 vector. Myc-TRIF, FLAG-IKKϵ, FLAG-TBK1, and IFNα1 promoter-luciferase reporter plasmids were gifts from Dr. Rongtuan Lin (McGill University, Montreal, Canada). 2×FLAG-RIG-IN (constitutively active RIG-I) and IFNβ promoter-luciferase reporter plasmids were gifts from Dr. Takashi Fujita (University of Kyoto, Kyoto, Japan). For constructing shRNA retroviral vector for human UBC9, an oligonucleotide fragment (target sequence: 5′-aacagatcctattaggaatac-3′) was inserted into pSUPER.retro (Oligoengine, Seattle). Retroviral preparations were made according to the manufacturer's instructions. As a control, a retroviral vector with a scrambled oligonucleotide fragment was prepared and tested in parallel. Mouse monoclonal antibodies against FLAG M2 and α-tubulin were purchased from Sigma. Rabbit and mouse antibodies for the T7 tag were purchased from Abcam (Cambridge, MA) and Novagen (Gibbstown, NJ), respectively. Rabbit antibody against murine IRF3 was from Zymed Laboratories Inc.. Rabbit antibodies against phospho-IRF3 (Ser(P)-396) and phospho-IRF3 (Ser(P)-386) were from Cell Signaling Technology (Danvers, MA) and EPITOMICS (Burlingame, CA), respectively. Rat antibody against HA was from Roche Diagnostics. Mouse monoclonal antibody against UBC9 was from Transduction Laboratories (Lexington, KY), and goat antibodies against rabbit IgG Alexa Fluor 488 and mouse IgG Alexa Fluor 546 were from Invitrogen. Recombinant human IFNβ was from Toray Industries, Inc. (Tokyo, Japan).
GFP-Sendai Virus Infection
Recombinant Sendai virus expressing GFP was generated and titrated as described elsewhere (41, 42). GFP signals in Sendai virus-infected cells were monitored by Axiovert 200 fluorescent microscope (Carl Zeiss Japan, Tokyo, Japan).
Quantitative (q) RT-PCR
PIASy+/+ and PIASy−/− MEFs with or without murine PIASy (1 × 106) were infected with vesicular stomatitis virus (VSV) or EMCV for the indicated time period at an m.o.i. of 5. Total RNA prepared using TRIzol reagent (Invitrogen) was reverse-transcribed with the Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics). The amounts of IFNβ, IFNα4, and hypoxanthine-guanine phosphoribosyltransferase cDNA were measured by using Universal ProbeLibrary and LightCycler 480 (Roche Diagnostics) according to the manufacture's instructions. Primers for qRT-PCR were designed by the Probe Finder software (Roche Diagnostics).
Luciferase Reporter Assay
HEK293T cells were plated in 24-well plates at 3 × 104/0.5 ml and transiently transfected with the indicated combinations of plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. Eighteen h post-transfection, cells were lysed, and luciferase activity was measured by using the Dual-Luciferase reporter assay kit (Promega) according to the manufacturer's procedure. Alternatively, cells were treated with 103 units/ml IFNβ for 6 h starting at 18 h post-transfection. Renilla luciferase activity was used for normalization.
Immunoblot Analysis
Whole cell extracts were prepared using lysis buffer containing 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 4 mm EDTA, 0.1% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, complete protease inhibitor mixture (Roche Diagnostics). For Phos-tag PAGE, cells were lysed using lysis buffer without EDTA. Extracts were separated by SDS-PAGE with or without Phos-tag AAL-107 (NARD Institute, Hyogo, Japan) and immunoblotted as described previously (40).
Immunoprecipitation
293T cells (3 × 106) were transfected with a total of 3.3 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen). Twelve h later, cells were lysed using lysis buffer containing 20 mm N-ethylmaleimide (Sigma). Lysates were centrifuged, and supernatants were incubated with anti-T7-agarose overnight with gentle rotation at 4 °C. Immune complexes were washed four times with N-ethylmaleimide containing lysis buffer, separated on SDS-PAGE, and subjected to immunoblot analysis.
Detection of SUMO-conjugated Proteins
To detect SUMO-conjugated proteins, 293T cells were transfected with the indicated plasmids, and extracts were prepared as above. Extracts were separated by SDS-PAGE and analyzed by immunoblot using the indicated antibodies.
Immunofluorescence Analysis
MEFs retrovirally transduced with or without murine PIASy-HA were plated on coverslips and incubated for 24 h. Cells were then infected with Sendai virus at an m.o.i. of 5 for 8 h and fixed with 3.7% paraformaldehyde. After two washes, cells were permeabilized with 0.2% Triton X-100 for 5 min and incubated at 4 °C with a blocking solution (PBS containing 3% BSA) for 30 min. The primary antibodies were added into the blocking solution at a 1:500 dilution, and cells were incubated for 4 h at 4 °C. After three washes, cells were then incubated with secondary antibodies for 4 h at 4 °C and counterstained with Hoechst for 1 min. Stained cells were viewed on a BZ8000 fluorescence microscope (Keyence, Osaka, Japan).
RESULTS
PIASy Negatively Regulates Virus-induced Type I IFN Expression
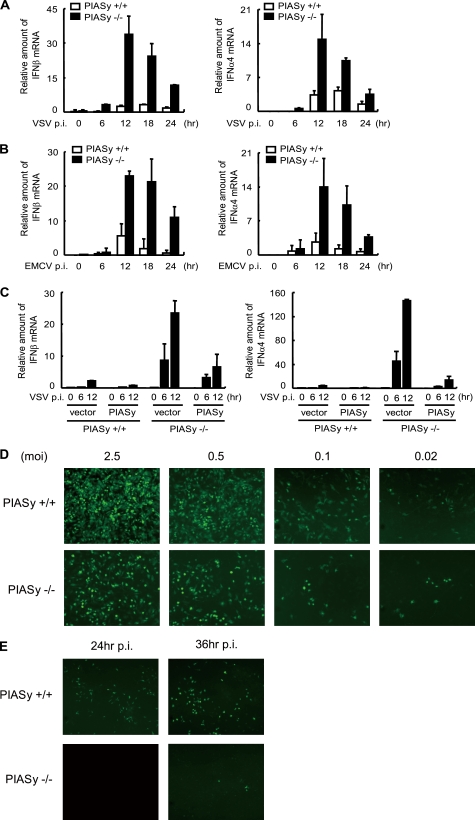
To investigate whether PIASy is involved in the regulation of virus-induced type I IFN production, PIASy−/− and PIASy+/+ MEFs were infected by VSV or EMCV. Infection by these viruses is recognized by RIG-I and MDA-5, respectively (2, 5). The amount of IFNβ and IFNα4 mRNA was measured at various time points during 24 h of infection by quantitative real time-reverse transcription PCR (qRT-PCR). As shown in Fig. 1A, levels of both IFNβ and IFNα4 mRNA were markedly higher in PIASy−/− MEFs than in PIASy+/+ MEFs at all time points tested. Similar increased production of type I IFN mRNA was observed after EMCV infection (Fig. 1B), indicating that PIASy inhibits both RIG-I- and MDA-5-driven type I IFN induction. We next tested whether increased induction of type I IFNs by PIASy deficiency can be reversed by ectopic expression of PIASy. HA-tagged PIASy was retrovirally transduced into PIASy+/+ and PIASy−/− MEFs, and VSV-induced type I IFN production was measured (Fig. 1C). Overexpression of PIASy decreased VSV-induced type I IFN production in both PIASy+/+ and PIASy−/− cells. Thus, PIASy acted as a negative regulator of type I IFN induction for both “loss of function” and “gain of function” experiments. We next tested whether loss of PIASy affected virus growth (Fig. 1D). PIASy+/+ and PIASy−/− MEFs were infected with GFP-Sendai virus at various m.o.i. values, and viral growth was monitored by GFP fluorescent signals. GFP signals in PIASy+/+ cells were clearly much greater than in PIASy−/− cells at 12 h post-infection with m.o.i. of 2.5, 0.5, and 0.1, although a difference was not clear with an m.o.i. of 0.02. Similarly, GFP signals were much higher in PIASy+/+ cells than PIASy−/− cells at 24 and 36 h post-infection (Fig. 1E). These results indicate that PIASy−/− cells are more resistant to Sendai virus infection, presumably due to greater production of type I IFNs.
FIGURE 1.
PIASy inhibits virus-induced activation of type I IFN promoters. A and B, PIASy+/+ and PIASy−/− MEFs were infected with VSV (A) or EMCV (B). IFNβ (left panel) and IFNα4 (right panel) mRNA at indicated time post-infection (p.i.) were measured by qRT-PCR, and data were normalized by hypoxanthine-guanine phosphoribosyltransferase mRNA. The values represent the average of three samples ± S.D. C, PIASy+/+ and PIASy−/− MEFs expressing PIASy or empty vector were infected with VSV, and IFNβ (left panel) and IFNα4 (right panel) mRNA at the indicated time post-infection (p.i.) were quantified as in A. D and E, PIASy+/+ and PIASy−/− MEFs were infected with GFP-Sendai virus at the indicated m.o.i. for 12 h (D) or at an m.o.i. of 0.02 for the indicated time post-infection (p.i.) (E). GFP signals were detected by microscopic inspection.
PIASy Inhibits Type I IFN Promoter Activity Stimulated by the Activated Form of IRF3 and IRF7
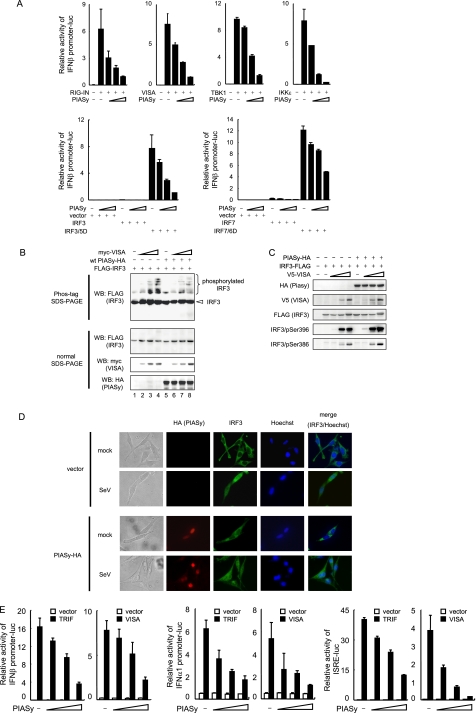
Recognition of viral RNA by RIG-I leads to the activation of downstream signaling molecules such as VISA, TBK1, IKKϵ, IRF3, and IRF7 (5, 6, 43, 44). We next sought to determine a step within the signaling cascade that PIASy targets. As shown in Fig. 2A, IFNβ promoter activity was activated by transfection of an active form of RIG-I (RIG-IN), VISA, TBK1, IKKϵ, and activated forms of IRF3 and IRF7 (IRF3/5D and IRF7/6D, respectively). In all cases, co-expression of PIASy decreased IFNβ promoter activity in a dose-dependent manner. These results suggested that PIASy inhibited a step downstream from IRF3 and IRF7 phosphorylation. If so, phosphorylation of IRF3 and IRF7 and their subsequent nuclear translocation would not have been affected by PIASy overexpression. To test this possibility, 293T cells were transfected with Myc-tagged VISA, and FLAG-tagged IRF3 along with PIASy and phosphorylation of IRF3 were detected by Phos-tag PAGE. As shown in Fig. 2B, slow migrating phosphor-IRF3 bands were detected in Myc-VISA-expressed cells in the presence and absence of PIASy expression (lanes 6–8 versus 2–4). We also tested VISA-induced phosphorylation of IRF3 by using anti-phospho-IRF3 antibodies. Co-expression of VISA increased phosphorylation of IRF3 at Ser-396 and Ser-386 and was not affected by PIASy (Fig. 2C). Similar results were observed when IRF3 phosphorylation was induced by TRIF (data not shown). We next tested whether PIASy affects virus-induced nuclear translocation of IRF3. Cells transfected with HA-tagged PIASy or empty vector were infected with Sendai virus and stained with anti-HA and anti-IRF3 antibodies at 8 h post-infection (Fig. 2D). As expected, PIASy localized to the nucleus before and after virus infection (45). After infection, IRF3 translocated to the nucleus irrespective of PIASy transfection. These results indicate that PIASy inhibits type I IFN promoter activation without interfering with phosphorylation and nuclear translocation of IRF3.
FIGURE 2.
PIASy inhibits transcriptional activity of phosphorylated IRF3. A, 293T cells were transfected with indicated activators of IFNβ promoter, IFNβ promoter-luciferase reporter, and increasing doses of PIASy for 18 h. The luciferase activities were quantified by normalizing with Renilla luciferase activities. B and C, 293T cells were transfected with IRF3-FLAG, with or without PIASy-HA, and increasing doses of Myc-VISA (B) or V5-VISA (C) for 24 h. Whole cell extracts were tested in the Phos-tag SDS-PAGE (B, top panels) or standard normal SDS-PAGE (B, lower panels and C) by Western blot (WB) with the indicated antibodies. D, MEFs retrovirally transduced by empty vector or PIASy-HA were infected with Sendai virus (SeV) for 8 h. Fixed cells were stained with anti-HA, and anti-IRF3 antibodies and were viewed by the fluorescent microscope. E, 293T cells were transfected with indicated promoter-luciferase reporter with or without TRIF or VISA and increasing amounts of PIASy for 18 h. Promoter activities were quantified as in A.
We next tested whether PIASy inhibits IFN promoter activity that was also activated by TRIF. As shown in Fig. 2E, TRIF and VISA overexpression increased IFNβ and IFNα1 promoter activity, which was inhibited by co-expression of PIASy. In addition, PIASy inhibited TRIF- and VISA-induced activation of ISRE promoter activity, again in a dose-dependent manner. These results indicate that PIASy negatively regulates TLR- and RLR-induced IFN transcription as well as ISRE promoter activity. We tested whether other PIAS proteins also regulate TLR/RLR-mediated type I IFN promoter activity and found that other PIAS proteins, except PIASy, do not significantly inhibit IFNβ promoter activity, although modest inhibition was detected by the highest dose of PIAS1 in TRIF-induced promoter activity (supplemental Fig. S1).
LXXLL Motif in the SAP Domain Is Not Involved in the Inhibition of Type I IFN Transcription
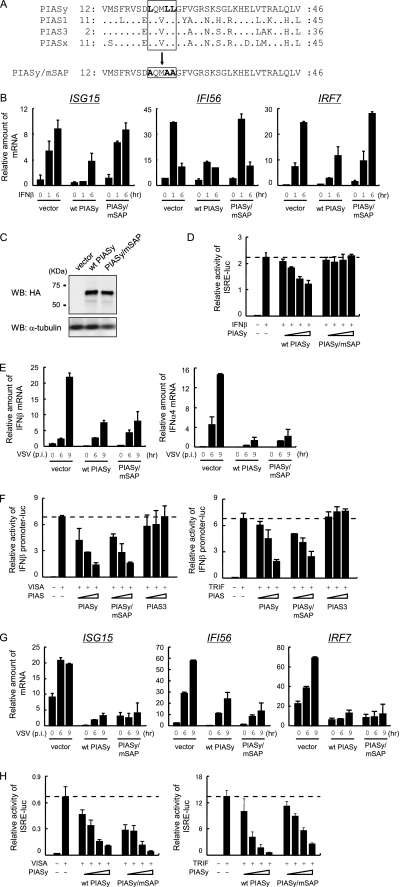
PIASy has been reported to inhibit IFNγ-mediated activation of IFN stimulated genes (ISGs) by preventing STAT1-dependent activation of ISRE promoter activity (37, 45). Although type I IFN transcription is initially activated by IRF3/7, it is further enhanced by the subsequent IFN-positive feedback loop activated by STAT1 and the ISGF3 complex (46–48). Thus, it was possible that inhibition of type I IFN promoter activity was due to PIASy inhibition of STAT1 activity. It was reported that the conserved LXXLL motif in the N-terminal SAP domain of PIASy critically contributes to the inhibition of STAT1-dependent transcription (45). We generated a mutant PIASy in which all three leucine residues in the LXXLL motif were substituted to alanine (referred to PIASy/mSAP, Fig. 3A), which was retrovirally introduced to PIASy−/− cells. The effect of these mutations was first tested on IFNβ-mediated ISG transcription. As shown in Fig. 3B, mRNA levels of three ISGs, ISG15, IFI56, and IRF7, were reduced upon wild type (WT) PIASy expression. In contrast, ISG mRNA levels were comparable in PIASy/mSAP-expressing cells and control cells. The protein levels of WT and PIASy/mSAP were also comparable in these cells (Fig. 3C). We also tested the effect of WT and PIASy/mSAP on IFNβ-stimulated ISRE promoter activity and found that PIASy/mSAP did not inhibit ISRE promoter activity, although WT PIASy did (Fig. 3D). These results indicate that the LXXLL motif is required for the inhibition of IFN-dependent ISG expression. We next investigated whether the PIASy/mSAP also fails to inhibit virus-induced type I IFN induction. As shown in Fig. 3E, mRNA levels of IFNβ and IFNα4 were reduced in PIASy/mSAP-expressing cells to a degree similar to that in WT PIASy-expressing cells. Consistent with these data, activation of IFNβ promoter activity by VISA and TRIF was similarly inhibited by PIASy/mSAP and WT PIASy but not by PIAS3 (Fig. 3F). Thus, the LXXLL motif is dispensable for the inhibition of type I IFN induction by PIASy. These results indicate that PIASy inhibits type I IFN transcription by a mechanism distinct from that by which PIASy inhibits STAT1-mediated ISG induction. Supporting this idea, virus-induced ISG15, ifi56, and IRF7 mRNA levels were reduced by PIASy/mSAP as well as by WT PIASy, in contrast to those induced by IFNβ (Fig. 3G). In addition, both WT PIASy and PIASy/mSAP inhibited VISA- or TRIF-induced ISRE activity in a dose-dependent manner (Fig. 3H). These results lead us to conclude that the LXXLL motif is required for inhibition of STAT1-mediated ISG transcription but not for inhibition of IRF3- and IRF7-mediated IFN transcription.
FIGURE 3.
LXXLL motif is not involved in the regulation of IRF3- and IRF7-mediated promoter activity. A, alignment of SAP domain of murine PIAS proteins. Conserved LXXLL motif and PIASy/mSAP mutation are boxed. B, PIASy−/− MEF were transfected with WT PIASy or PIASy/mSAP. IFNβ was treated for indicated times, and mRNA for ISG15, IFI56, and IRF7 were quantified by qRT-PCR and by normalizing with hypoxanthine-guanine phosphoribosyltransferase mRNA. C, whole cell extracts in B were Western blotted (WB) with the indicated antibodies. D, 293T cells were transfected with ISRE reporter and increasing doses of WT PIASy or PIASy/mSAP. Cells were treated with IFNβ for 6 h, and luciferase activities were quantified by normalizing with Renilla luciferase activities. E, cells in B were infected with VSV for the indicated times, and IFNβ (left panel) and IFNα4 (right panel) mRNAs were quantified. F, 293T cells were transfected with VISA (left panel) or TRIF (right panel), IFNβ promoter reporter, and increasing amount of WT PIASy, PIASy/mSAP, or PIAS3. Luciferase activities were quantified as in D. G, cells in B were infected with VSV for the indicated times, and mRNAs were quantified as in B. H, 293T cells were transfected with VISA (left panel) or TRIF (right panel), ISRE reporter, and increasing doses of WT PIASy or PIAS/mSAP. Luciferase activities were quantified as in D.
SUMO E3 Activity of PIASy Is Not Required for the Inhibition of Type I IFN Transcription
To study whether SUMO E3 ligase activity of PIASy is required for the inhibition of type I IFN transcription, we constructed a mutant PIASy in which the third cysteine residue of the RING-like domain was substituted by phenylalanine (PIASy/C335F). WT PIASy and PIASy/C335F were expressed in 293T cells together with T7-tagged SUMO1, SUMO1/GA, a conjugation-defective mutant (26), or SUMO2. Immunoblot data in Fig. 4A showed that WT PIASy, but not PIASy/C335F, produced slow migrating SUMO1- or SUMO2-conjugated proteins. These bands were not generated when T7-SUMO1 was replaced by T7-SUMO1/GA, as expected. Thus, PIASy/C335F no longer has a SUMO ligase activity. We next tested whether PIASy/C335F inhibits VISA- and TRIF-mediated type I IFN transcription. As shown in Fig. 4B, VISA- and TRIF-induced IFNβ promoter activity was inhibited by PIASy/C335F to a similar degree as WT PIASy in a dose-dependent manner. Cyan fluorescent protein expression tested as a negative control did not inhibit IFNβ promoter activity. These results indicate that PIASy inhibits VISA- and TRIF-induced type I IFN induction independent of its SUMO E3 ligase activity.
FIGURE 4.
SUMO E3 activity is not required for inhibition of TLR and RLR signaling by PIASy. A, HA-tagged WT PIASy or PIASy/C335F was transfected to 293T cells together with T7-tagged WT SUMO1, SUMO1/GA, or SUMO2. Whole cell extracts were subjected to Western blot (WB) analysis with the indicated antibodies. B, 293T cells were transfected with VISA (left) or TRIF (right), IFNβ promoter reporter, and increasing amounts of WT PIASy, PIASy/C335F, or cyan fluorescent protein (CFP). Luciferase activities were quantified by normalizing with Renilla luciferase activities.
C-terminal SUMO-interacting Motif of PIASy Is Involved in the Inhibition of Type I IFN Transcription
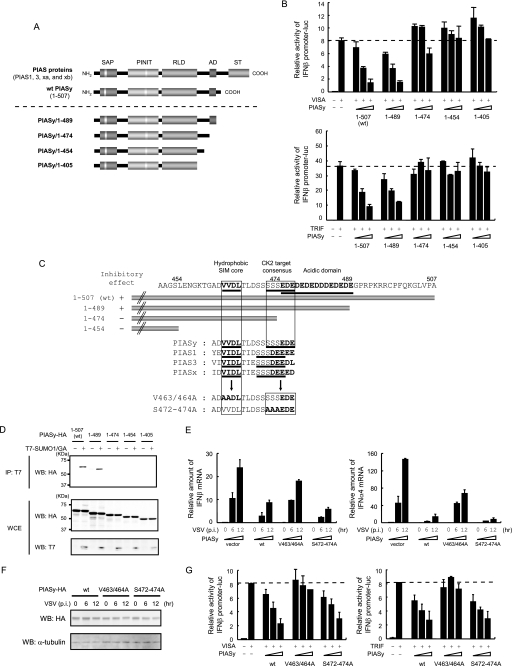
We tested all four PIAS members for their ability to inhibit type I IFN transcription. As shown in supplemental Fig. S1, only PIASy had significant inhibitory activity. Among the PIAS proteins, the C-terminal region after the central RING-like domain is variable, and PIASy differs most from other members in this region (Fig. 5A). Thus we considered it possible that the C-terminal portion of PIASy is responsible for its inhibitory activity. To test this possibility, a series of C-terminal deletion mutants were generated and tested for VISA- or TRIF-induced IFNβ promoter activity. As shown in Fig. 5, A and B, transfection of the smallest deletion, PIASy/1–489 led to inhibition of IFNβ promoter activity in a manner similar to full-length PIASy. However, no inhibition was observed when the deletion was extended further, indicating that the region around amino acid 474 is critical for the inhibition of IFNβ promoter activity. This region contains a cluster of acidic amino acids (Fig. 5C). Although the functional significance of the acidic cluster has yet to be fully studied, a report suggested that serine residues juxtaposed to the acidic cluster are a potential phosphorylation site of CK2 protein kinase required for activity of the SUMO-interacting motif (SIM) (49). We therefore investigated the ability of these C-terminal deletion mutants to interact with SUMO peptides in a noncovalent manner. In Fig. 5D, HA-tagged WT PIASy and deletion mutants were transfected along with T7-tagged SUMO1/GA, and lysates were immunoprecipitated with anti-T7 antibody. Whereas full-length PIASy and PIASy/1–489 co-precipitated SUMO1/GA, all other mutants did not, verifying that the acidic cluster is required for noncovalent association of PIASy with SUMO. Given that SUMO binding activity and the IFNβ promoter inhibitory activity correlated well, we next tested whether noncovalent association of PIASy with SUMO peptides is important for inhibition of type I IFN production. To this end, two mutants were generated; first, hydrophobic amino acids in the SIM core were substituted to alanine, and second, potential target serine residues of CK2 were substituted to alanine (Fig. 5C, V463/464A and S472–474A, respectively). These mutants were introduced into PIASy−/− MEF cells and tested for possible inhibition of VSV-induced type I IFN mRNA expression. As shown in Fig. 5E, WT PIASy and S472/474A strongly inhibited type I IFN mRNA expression, although V463/464A caused noticeably less inhibition both in IFNβ and IFNα4 mRNA expression. Expression levels of transfected PIASy were similar and did not change during virus infection (Fig. 5F). The activity of these mutants was further examined in an IFNβ reporter assay (Fig. 5G). WT PIASy and S472/474A repressed VISA- and TRIF-induced IFN promoter activity in a dose-dependent manner. However, V463/464A did not appreciably inhibit IFNβ promoter activity. These results indicate that the core hydrophobic residues in the putative SIM, but not the serine residues juxtaposed to the acidic cluster, are required for PIASy inhibition of type I IFN transcription.
FIGURE 5.
C-terminal region of PIASy contributes to the inhibition of IRF3- and IRF7-mediated transcription. A, schematic structure of PIAS proteins. Conserved SAP domain, PINIT domain, RING-like domain (RLD), acidic domain (AD), and serine/threonine-rich domain (S/T) are shown on top. A series of PIASy C-terminal deletion mutants are shown. B, 293T cells were transfected with VISA (top) or TRIF (bottom), IFNβ promoter reporter, and increasing doses of WT or indicated PIASy mutant. Luciferase activities were quantified by normalizing with Renilla luciferase activities. C, structure of the C-terminal region of PIASy. Deletion mutants in B are shown on top. The SIM, CK2 target motif, and acidic domain are underlined. Alignment of SIM and CK2 target consensus motif is indicated in the middle. Mutants with disrupted SIM and CK2 consensus motifs are on the bottom. D, HA-tagged WT or mutant PIASy were transfected to 293T cells with or without T7-SUMO1/GA, and extracts were immunoprecipitated (IP) with anti-T7 antibody. Immunoprecipitated (top panel) and whole cell extracts (WCE, lower panels) were analyzed by Western blot (WB) with the indicated antibodies. E, PIASy−/− MEF expressing WT or indicated PIASy mutants were infected with VSV.IFNβ (left panel) and IFNα4 (right panel) mRNA at indicated time post-infection (p.i.) were quantified by qRT-PCR and by normalizing with hypoxanthine-guanine phosphoribosyltransferase mRNA. The values represent the average of three samples ± S.D. F, whole cell extracts in E were tested for Western blot (WB) with the indicated antibodies. G, 293T cells were transfected with VISA (left panel) or TRIF (right panel) and IFNβ promoter reporter and increasing amounts of WT or indicated PIASy mutants. Luciferase activities were quantified as in B.
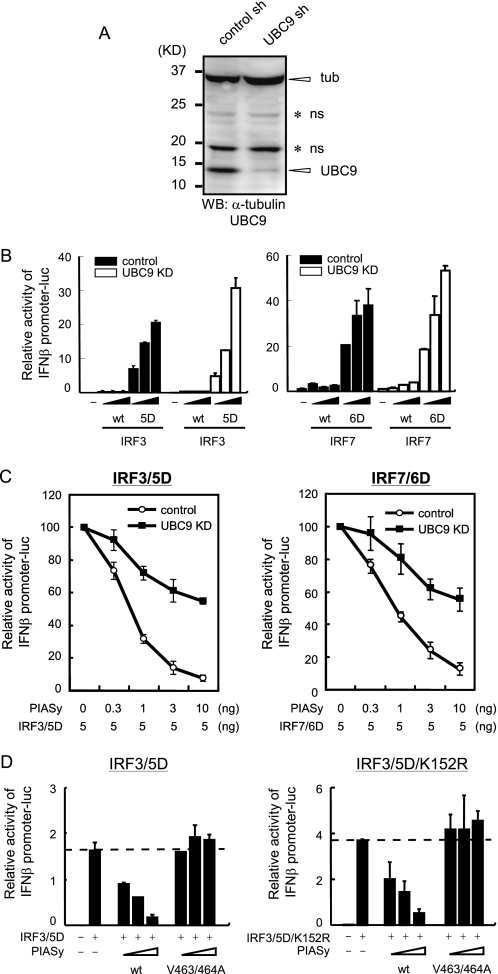
SUMO Conjugation Mechanism Is Required for PIASy Inhibition of Type I IFN Transcription
Given that hydrophobic residues in the PIASy SIM are critical for inhibition of type I IFN induction, we next investigated whether the SUMO modification mechanism is involved in the PIASy inhibition of IFN transcription. To this end, we constructed an shRNA vector for UBC9, the sole E2 enzyme of the SUMO conjugation cascades. Data in Fig. 6A show that this vector stably knocked down UBC9 protein expression of IFNβ reporter activity by the activated forms of IRF3 and IRF7 (Figs. 5D and 6D, respectively) and was slightly higher in UBC9 knockdown (KD) cells than that in control shRNA-expressing cells (Fig. 6B). We then tested IFNβ reporter activity in UBC9 KD cells expressing PIASy. As shown in Fig. 6C, in UBC9 KD cells, IFNβ reporter activities stimulated by both IRF3/5D and IRF7/6D were higher than in control cells at all doses of PIASy tested. These results indicate that SUMO conjugating activity is required for the inhibition of type I IFN production by PIASy. Thus, it is likely that PIASy inhibits type I IFN promoter activity by interacting with a SUMOylated factor through the SIM. We previously showed that SUMO is covalently conjugated to IRF3 mainly through Lys-152 (40). We asked whether SUMOylated IRF3 is a main factor that binds to PIASy SIM, leading to PIASy-mediated inhibition of type I IFN induction. In Fig. 6D, IRF3/5D and the SUMOylation-defective mutant IRF3/5D/K152R were co-transfected with PIASy, and IFNβ promoter activity was measured. PIASy inhibited both IRF3/5D- and IRF3/5D/K152R-induced promoter activity, whereas the SIM mutant, V463/464A did not. These results indicate that IRF3 is not a major factor required for binding to SIM to confer inhibitory activity upon PIASy.
FIGURE 6.
SUMO modification is required for inhibition of type I IFN induction by PIASy. A, whole cell extracts from control or UBC9 knockdown (KD) 293T cells were analyzed by Western blot (WB) with the indicated antibodies. UBC9, α-tubulin (tub) and nonspecific bands (ns) are marked on the right. B, UBC9 KD or control 293T cells were transfected with WT or the activated form of IRF3 (left panel) or IRF7 (right panel) and IFNβ promoter reporter. Luciferase activities were quantified by normalizing with Renilla luciferase activities. C, UBC9 KD or control 293T cells were transfected with the activated form of IRF3 (left panel) or IRF7 (right panel) and IFNβ promoter reporter and increasing doses of PIASy. Luciferase activities were quantified as in B. The promoter activities are expressed as the ratio of PIASy-transfected versus untransfected cells. D, 293T cells were transfected with IRF3/5D (left panel) or IRF3/5D/K152R (right panel), IFNβ promoter reporter, and increasing doses of PIASy. Luciferase activities were quantified as in B.
DISCUSSION
In this study, we investigated the contribution of PIASy to the regulation of type I IFN transcription. We show that VSV and EMCV infection results in greater type I IFN induction in PIASy−/− cells than PIASy+/+ cells, although ectopic expression of PIASy led to reduced IFN expression. Accordingly, PIASy−/− cells exhibited greater antiviral activity against Sendai virus. PIASy inhibited both TRIF- and VISA-induced activation of type I IFN promoter activities, indicating that it affects both TLR- and RLR-mediated type I IFN gene activation. It is likely that PIASy does not act on an early step of TLR/RLR signaling cascades but rather acts on a step after IRF3 and IRF7 are phosphorylated and translocated into the nucleus, considering that PIASy predominantly localizes to the nucleus and PIASy efficiently inhibited type I IFN promoter activity by the activated forms of IRF3 and IRF7. Supporting this view, ectopic PIASy expression did not change VISA- and TRIF-stimulated IRF3 phosphorylation. Given that PIASy is capable of interacting with IRF3 and IRF7, it likely binds to activated IRF3 and IRF7 in the nucleus, leading to the inhibition of IFN transcription (50).
The SAP domain binds to AT-rich DNA sequences present in the scaffold attachment regions/matrix attachment regions (51). This domain is conserved in all PIAS members and shown to be required for the interaction with nuclear receptors and their co-regulators (52). We show that the mutant PIASy with an altered LXXLL motif in the SAP domain, although unable to inhibit IFN-stimulated ISG transcription, nevertheless retained the ability to inhibit type I IFN transcription. Our results illustrate that PIASy employs different mechanisms to inhibit virus-mediated IFN induction and IFN-stimulated ISG expression. This dichotomy may be accounted for by the difference in the transcriptional pathways by which virus-induced IFN transcription and IFN-stimulated transcription is controlled, namely the former is triggered by the TLR/RLR pathway leading to the activation of IRF3 and IRF7, and the latter is activated by the JAK/STAT pathway activating STAT1/STAT2 and IRF9. In this scenario, PIASy may select SAP-dependent and -independent processes according to the types of transcription factors activated and recruited to the promoter. The significance of the differential domain usage is at present unclear. However, it seems clear that PIASy by adopting diverse mechanisms governs specificity, magnitude, and timing of antiviral effects, thereby fine-tuning innate immunity.
PIAS1 and PIASy do not inhibit expression of all ISGs (21, 36, 37). It is likely that ISGs are divided into distinct groups, partly according to different types of negative regulation under which they are controlled. Because IRF3 and IRF7 are involved not only in type I IFN transcription, but TLR/RLR-mediated ISG induction, the mechanism observed in this study likely contributes to negative regulation of ISG transcription. SAP domain independent inhibition of ISRE activity by PIASy is likely to operate in a relatively early stage of virus infection, prior to the initiation of IFN feedback-based ISG expression (46–48). In a later stage of virus infection, ISG expression is boosted by type I IFN induced through the JAK/STAT pathway, controlled by cooperation of PIAS1 and PIASy (37).
Inhibitory activity of PIAS1 is regulated by a phosphorylation-dependent switch by activated IKKα (53). This mechanism is not likely to apply to PIASy given that PIASy does not have a corresponding phosphorylation target. Interestingly, we observed that virus infection induced a slowly migrating band in PIASy Phos-tag PAGE analysis, indicating that PIASy is also phosphorylated during virus infection (data not shown). Although residues targeted for phosphorylation and the kinase that phosphorylates PIASy have not been identified, it is possible that PIASy is activated upon virus and/or IFN induction through phosphorylation.
In addition to SAP domain-independent inhibition, PIASy inhibition of type I IFN transcription did not require its SUMO E3 ligase activity, because the mutation in the RING-like domain did not abolish IRF3- and IRF7-mediated IFN promoter activity. Some transcription factors are known to be regulated by PIASy via SUMO-independent mechanisms (19, 20). For example, PIASy suppresses LEF1-mediated transcription by recruiting LEF1 to the promyelocytic leukemia protein nuclear body (54). Furthermore, PIASy is reported to recruit HDAC1 and HDAC2 without involving its SUMO E3 activity in Smad4 and androgen receptor-mediated transcription (55, 56). These reports, combined with our observation, further support the view that PIASy utilizes varying mechanisms to repress transcription, depending on the signaling pathways activated and the availability of molecules with which it cooperates.
We show that deletion of the acidic cluster juxtaposed with the hydrophobic SIM core resulted in the loss of PIASy function, leading to the loss of inhibition of IFN transcription. The loss of inhibitory activity coincided with the loss of SUMO peptide binding activity. Furthermore, the SIM sequence was required for the inhibition of IFNβ promoter activity. Thus the acidic cluster may confer SUMO binding activity upon PIASy, playing an important role in inhibiting type I IFN induction. On the other hand, serine residues presumed to be phosphorylated by CK2 were dispensable for inhibition of IFN promoter activity (49). Consistent with these data, treatment of CK2 inhibitors, TBB or emodin, did not change PIASy inhibition of IFNβ promoter activity (data not shown). These results suggest that phosphorylation of PIASy, possibly targeted by CK2, is not required for SUMO binding activity of the SIM, although it may be dependent on the acidic cluster (34). There are five consecutive serine residues between the SIM and the acidic cluster in PIASy, and three serine residues are present in the corresponding region of other PIAS members (Fig. 5C). The discrepancy on the requirement of CK2-dependent phosphorylation for the SIM activation among PIAS1, PIAS3, and PIASy may be due to a structural difference caused by these residues. PIASy may have a unique mode of SIM activation, different from that utilized by other PIAS proteins. Given that intact SUMO binding activity appears to be required, the SUMO modification mechanism is expected to play a critical role in negative regulation of type I IFN transcription, despite that the PIASy SUMO E3 ligase domain is dispensable. The role for the SUMOylation machinery in PIASy inhibition of IFN transcription is supported by our data that general down-regulation of SUMO conjugation pathways by UBC9 knockdown led to a profound reduction in the inhibitory effect of PIASy. Our results indicate that PIASy inhibits IRF3- and IRF7-mediated IFN transcription by associating with a factor yet to be unraveled that is conjugated to SUMO. Because SUMOylation of IRF3 was not required for inhibition by PIASy, IRF3 (and presumably IRF7) is not the factor required for SIM binding. The interaction of this unidentified factor may facilitate the formation of a repressor complex. Identification of a PIASy-interacting partner(s) that is modified by SUMO would thus advance our understanding of the mechanism by which PIASy inhibits type I IFN transcription.
In summary, this work shows that among PIAS family members PIASy is the major negative regulator of type I IFN transcription. It inhibits IFN transcription by mobilizing a SUMO modification mechanism through the SIM domain, without relying on its SUMO E3 ligase activity.
Acknowledgments
We thank Dr. Ke Shuai (UCLA) for kindly providing PIASy−/− MEF, Dr. Takashi Fujita for the FLAG-RIG-IN and IFNβ promoter-luciferase plasmids, and Dr. Rongtuan Lin for the FLAG-TRIF, FLAG-IKKϵ, FLAG-TKB1, and IFNα1 promoter-luciferase plasmids.
This work was supported, in whole or in part, by National Institutes of Health grant from the Intramural Research Program of NICHD and Trans NIH Biodefense Program. This work was also supported by Grant-in-aid 22590424 for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- TLR
- Toll-like receptor
- RLR
- RIG-I-like receptor
- RIG-I
- retinoic acid-inducible gene I
- IRF
- interferon regulatory factor
- SUMO
- small ubiquitin-related modifiers
- SIM
- SUMO-interacting motif
- IKK
- IκB kinase
- ISG
- interferon stimulated gene
- VSV
- vesicular stomatitis virus
- EMCV
- encephalomyocarditis virus
- m.o.i.
- multiplicity of infection
- shRNA
- short hairpin RNA
- qRT-PCR
- quantitative RT
- MEF
- mouse embryonic fibroblast
- STAT
- signal transducer and activator of transcription
- ISRE
- interferon-stimulated response element
- TRIF
- TIR domain-containing adaptor-inducing interferon β.
REFERENCES
- 1. Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 2. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 3. Takeda K., Akira S. (2005) Int. Immunol. 17, 1–14 [DOI] [PubMed] [Google Scholar]
- 4. Andrejeva J., Childs K. S., Young D. F., Carlos T. S., Stock N., Goodbourn S., Randall R. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawai T., Akira S. (2006) Nat. Immunol. 7, 131–137 [DOI] [PubMed] [Google Scholar]
- 6. Yoneyama M., Fujita T. (2009) Immunol. Rev. 227, 54–65 [DOI] [PubMed] [Google Scholar]
- 7. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 8. Hiscott J., Lin R., Nakhaei P., Paz S. (2006) Trends Mol. Med. 12, 53–56 [DOI] [PubMed] [Google Scholar]
- 9. Durbin J. E., Fernandez-Sesma A., Lee C. K., Rao T. D., Frey A. B., Moran T. M., Vukmanovic S., García-Sastre A., Levy D. E. (2000) J. Immunol. 164, 4220–4228 [DOI] [PubMed] [Google Scholar]
- 10. Hengel H., Koszinowski U. H., Conzelmann K. K. (2005) Trends Immunol. 26, 396–401 [DOI] [PubMed] [Google Scholar]
- 11. Levy D. E., García-Sastre A. (2001) Cytokine Growth Factor Rev. 12, 143–156 [DOI] [PubMed] [Google Scholar]
- 12. McGettrick A. F., O'Neill L. A. (2010) Curr. Opin. Immunol. 22, 20–27 [DOI] [PubMed] [Google Scholar]
- 13. Moore C. B., Ting J. P. (2008) Immunity 28, 735–739 [DOI] [PubMed] [Google Scholar]
- 14. O'Neill L. A. (2008) Immunity 29, 12–20 [DOI] [PubMed] [Google Scholar]
- 15. Komuro A., Bamming D., Horvath C. M. (2008) Cytokine 43, 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J., Hu Y., Deng W. W., Sun B. (2009) Microbes Infect. 11, 321–327 [DOI] [PubMed] [Google Scholar]
- 17. Rytinki M. M., Kaikkonen S., Pehkonen P., Jääskeläinen T., Palvimo J. J. (2009) Cell. Mol. Life Sci. 66, 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt D., Müller S. (2003) Cell. Mol. Life Sci. 60, 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharrocks A. D. (2006) Genes Dev. 20, 754–758 [DOI] [PubMed] [Google Scholar]
- 20. Shuai K. (2006) Cell Res. 16, 196–202 [DOI] [PubMed] [Google Scholar]
- 21. Shuai K., Liu B. (2005) Nat. Rev. Immunol. 5, 593–605 [DOI] [PubMed] [Google Scholar]
- 22. Arora T., Liu B., He H., Kim J., Murphy T. L., Murphy K. M., Modlin R. L., Shuai K. (2003) J. Biol. Chem. 278, 21327–21330 [DOI] [PubMed] [Google Scholar]
- 23. Chung C. D., Liao J., Liu B., Rao X., Jay P., Berta P., Shuai K. (1997) Science 278, 1803–1805 [DOI] [PubMed] [Google Scholar]
- 24. Liu B., Liao J., Rao X., Kushner S. A., Chung C. D., Chang D. D., Shuai K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao J., Fu Y., Shuai K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5267–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verger A., Perdomo J., Crossley M. (2003) EMBO Rep. 4, 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gill G. (2003) Curr. Opin. Genet. Dev. 13, 108–113 [DOI] [PubMed] [Google Scholar]
- 28. Gill G. (2005) Curr. Opin. Genet. Dev. 15, 536–541 [DOI] [PubMed] [Google Scholar]
- 29. Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 30. Kotaja N., Karvonen U., Jänne O. A., Palvimo J. J. (2002) Mol. Cell. Biol. 22, 5222–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ouyang J., Valin A., Gill G. (2009) Methods Mol. Biol. 497, 141–152 [DOI] [PubMed] [Google Scholar]
- 32. Jackson P. K. (2001) Genes Dev. 15, 3053–3058 [DOI] [PubMed] [Google Scholar]
- 33. Johnson E. S., Gupta A. A. (2001) Cell 106, 735–744 [DOI] [PubMed] [Google Scholar]
- 34. Kerscher O. (2007) EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palvimo J. J. (2007) Biochem. Soc. Trans. 35, 1405–1408 [DOI] [PubMed] [Google Scholar]
- 36. Liu B., Mink S., Wong K. A., Stein N., Getman C., Dempsey P. W., Wu H., Shuai K. (2004) Nat. Immunol. 5, 891–898 [DOI] [PubMed] [Google Scholar]
- 37. Tahk S., Liu B., Chernishof V., Wong K. A., Wu H., Shuai K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11643–11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu B., Yang R., Wong K. A., Getman C., Stein N., Teitell M. A., Cheng G., Wu H., Shuai K. (2005) Mol. Cell. Biol. 25, 1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu B., Shuai K. (2008) Trends Pharmacol. Sci. 29, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kubota T., Matsuoka M., Chang T. H., Tailor P., Sasaki T., Tashiro M., Kato A., Ozato K. (2008) J. Biol. Chem. 283, 25660–25670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akaike T., Fujii S., Kato A., Yoshitake J., Miyamoto Y., Sawa T., Okamoto S., Suga M., Asakawa M., Nagai Y., Maeda H. (2000) FASEB J. 14, 1447–1454 [DOI] [PubMed] [Google Scholar]
- 42. Sakai Y., Kiyotani K., Fukumura M., Asakawa M., Kato A., Shioda T., Yoshida T., Tanaka A., Hasegawa M., Nagai Y. (1999) FEBS Lett. 456, 221–226 [DOI] [PubMed] [Google Scholar]
- 43. Zhao T., Yang L., Sun Q., Arguello M., Ballard D. W., Hiscott J., Lin R. (2007) Nat. Immunol. 8, 592–600 [DOI] [PubMed] [Google Scholar]
- 44. Honda K., Taniguchi T. (2006) Nat. Rev. Immunol. 6, 644–658 [DOI] [PubMed] [Google Scholar]
- 45. Liu B., Gross M., ten Hoeve J., Shuai K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3203–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Honda K., Yanai H., Takaoka A., Taniguchi T. (2005) Int. Immunol. 17, 1367–1378 [DOI] [PubMed] [Google Scholar]
- 47. Marié I., Durbin J. E., Levy D. E. (1998) EMBO J. 17, 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Immunity 13, 539–548 [DOI] [PubMed] [Google Scholar]
- 49. Stehmeier P., Muller S. (2009) Mol. Cell 33, 400–409 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J., Xu L. G., Han K. J., Wei X., Shu H. B. (2004) FEBS Lett. 570, 97–101 [DOI] [PubMed] [Google Scholar]
- 51. Kipp M., Göhring F., Ostendorp T., van Drunen C. M., van Driel R., Przybylski M., Fackelmayer F. O. (2000) Mol. Cell. Biol. 20, 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 53. Liu B., Yang Y., Chernishof V., Loo R. R., Jang H., Tahk S., Yang R., Mink S., Shultz D., Bellone C. J., Loo J. A., Shuai K. (2007) Cell 129, 903–914 [DOI] [PubMed] [Google Scholar]
- 54. Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R. (2001) Genes Dev. 15, 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gross M., Yang R., Top I., Gasper C., Shuai K. (2004) Oncogene 23, 3059–3066 [DOI] [PubMed] [Google Scholar]
- 56. Long J., Matsuura I., He D., Wang G., Shuai K., Liu F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9791–9796 [DOI] [PMC free article] [PubMed] [Google Scholar]